Axon injury triggers EFA-6 mediated destabilization of axonal microtubules via TACC and doublecortin like kinase
Figures
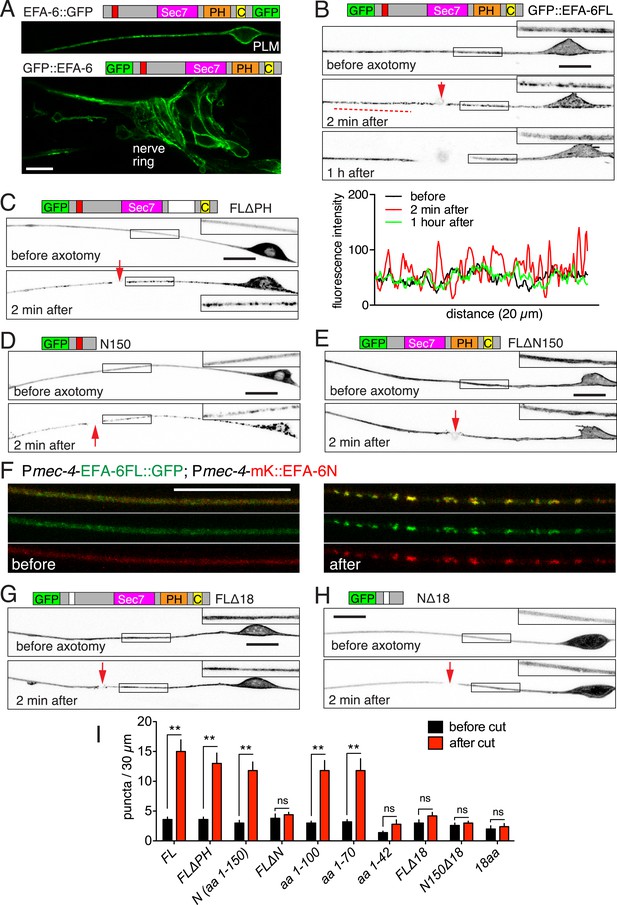
Axon injury triggers rapid relocalization of Exchange Factor for ARF-6 (EFA-6), mediated by its N-terminal domain.
(A) Single focal plane images of PLM (top) and nerve ring (bottom) showing membrane localization of EFA-6. Transgenes: Pmec-4-EFA-6::GFP (juEx6467) (top) and Prgef-1-GFP::EFA-6 (juEx6374) (bottom). (B) Localization of full length EFA-6 (Pmec-4-GFP::EFA-6, juEx6160) before, 2 min after, and 1 hr post axotomy. Projections of confocal z stacks, inverted grayscale; enlargements in inserts. Bottom, fluorescence intensity along line scan. (C–E) Localization of GFP::EFA-6 fusion protein lacking the PH domain (FLΔPH) (Pmec-4-GFP::EFA-6 FLΔPH, juEx6453), EFA-6 N-terminal 150 aa (N150) (Pmec-4-GFP::EFA-6N150, juEx3531), and EFA-6 lacking the N-terminus (FLΔN150) (Pmec-4-GFP::EFA-6FLΔN150, juEx6154). (F) Colocalization of EFA-6FL and EFA-6N150 puncta after injury. Localization of EFA-6FL::GFP and EFA-6N150::mKate2 (juEx6522) in PLM before and after axotomy. EFA-6 full length protein and N terminus relocalize to overlapping puncta. (G, H) Requirement for the 18-aa motif for relocalization of EFA-6FL and EFA-6N150. Localization of GFP::EFA-6FLΔ18aa (juEx6156), and GFP::EFA-6N150Δ18aa (juEx3535) in touch neurons before and 2 min after axotomy. (I) Quantitation of puncta before and 2 min after injury in axons expressing different EFA-6 fragments. Statistics, one-way ANOVA with Bonferroni post test; n = 5 for each bar; **p < 0.01, ns, not significant. Scale, 10 μm.
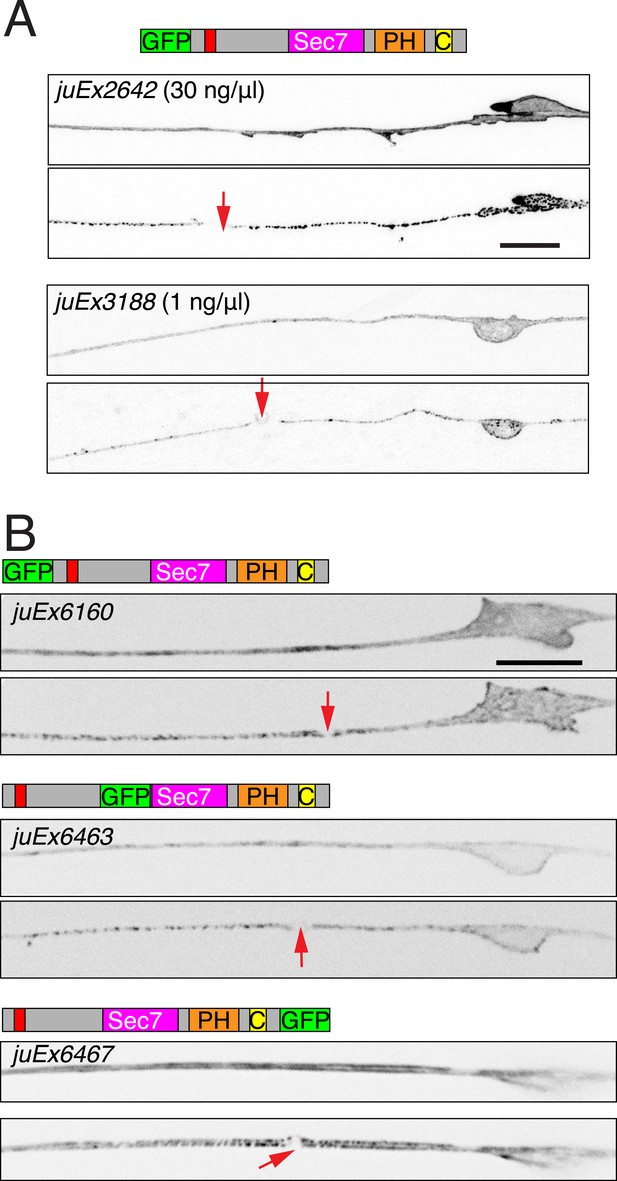
Injury induced relocalization of EFA-6 is independent of expression level or location of tag.
(A) Pmec-4-GFP::EFA-6 transgenic lines generated at different concentrations of injected plasmid (juEx2642 was generated at 30 ng/μl, juEx3188 was generated at 1 ng/μl) displayed similar localization, before and 2 min post axotomy. (B) The relocalization of EFA-6 is independent of the site of GFP tagging. Transgenes: Pmec-4-GFP::EFA-6(juEx6160). Pmec-4-EFA-6N150::GFP::EFA-6C(juEx6463) (GFP inserted between aa 363–364) and Pmec-4-EFA-6::GFP(juEx6467). All three transgenes cause premature PLM termination to similar extents (∼30% undershooting). Red arrows, site of axotomy. Scale, 10 μm.
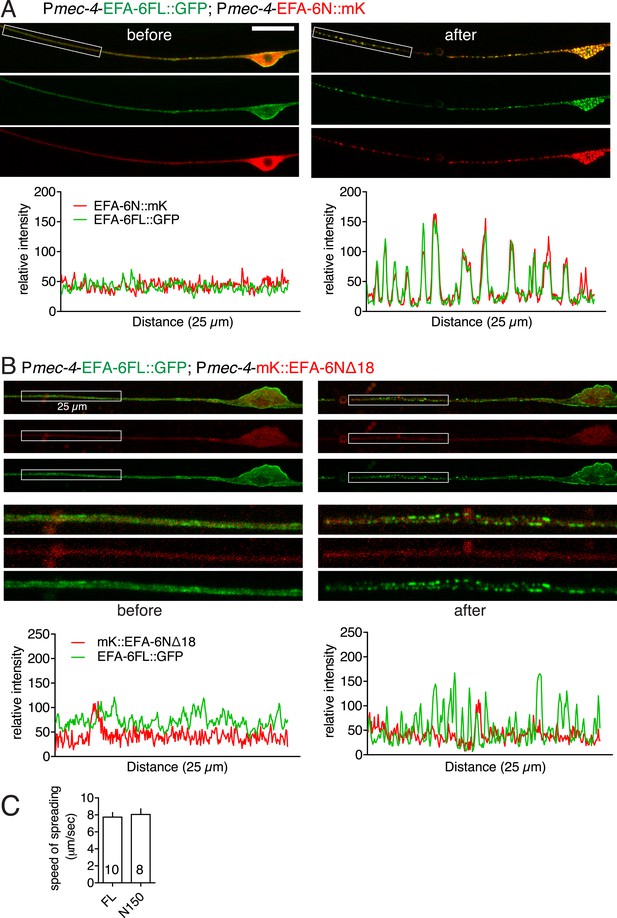
Full-length EFA-6 and N terminus relocalize to the same puncta after injury.
(A) Colocalization of EFA-6FL::GFP and EFA-6N150::mKate2 in PLM before and after axotomy. Enlarged images of the regions in boxes (25 μm length) are shown in Figure 1F. Graphs of line scans are shown below. EFA-6FL::GFP was primarily localized to the plasma membrane and EFA-6N150::mKate2 was diffuse in soma and axon before injury; both became punctate after injury, and these puncta co-localized. (B) Localization of EFA-6FL::GFP and EFA-6N150Δ18::mKate2 in PLM before and after axotomy. Enlarged images of the regions in boxes and graphs of line scans are shown below. EFA-6N150Δ18::mKate2 was diffuse before and after axotomy. (C) Velocity of relocalization spread for GFP::EFA-6FL and GFP::EFA-6N150, calculated by measuring the distance between injury site and the boundary between punctate and even GFP distribution in the distal or proximal axon at 2.3 s (10 frames) post axotomy.
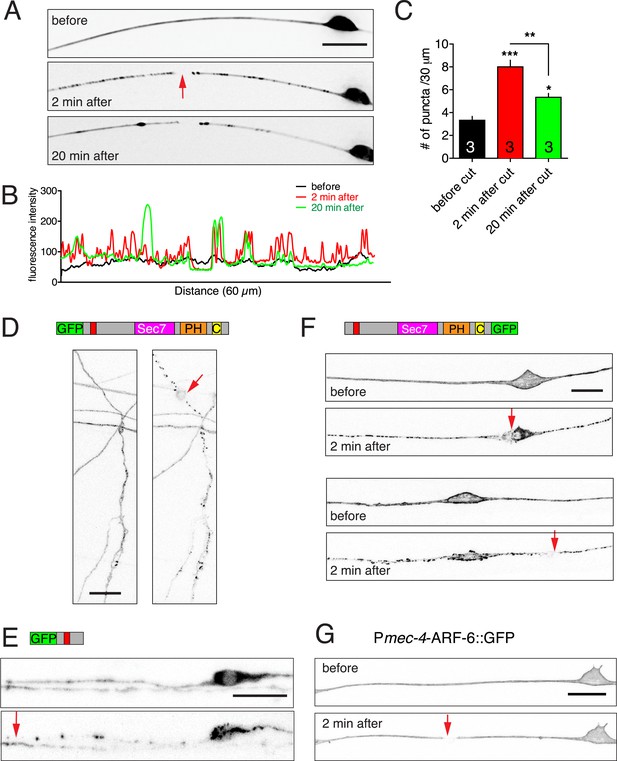
Injury induced relocalization of EFA-6.
(A) Representative images of GFP::EFA-6N150 before and post axotomy. Before axotomy, GFP::EFA-6N150 was diffuse in the axon; by 2 min post axotomy, it became punctate; 20 min post axotomy, GFP recovered to a diffuse pattern. (B) Line scan along axon at different time points. (C) Quantitation of GFP::EFA-6N150 puncta at different times (before, 2 min and 20 min after axotomy). Transgene: Pmec-4-GFP::EFA-6N150(juEx3531). Statistics: One-way ANOVA with Bonferroni post test. ***p < 0.001, **p < 0.01, *p < 0.05. (D) GFP::EFA-6 expressed in motor neurons commissures showed similar relocalization response to injury. Transgene: Prgef-1-GFP::EFA-6(juEx6374). (E) GFP::EFA-6N150 expressed in motor neurons displayed similar injury-induced relocalization. Transgene: Punc-25-GFP::EFA-6N150(juEx6229). (F) Injury to the soma or dendrite of PLM led to similar EFA-6 relocalization. (G) ARF-6::GFP localization before and after axotomy. Transgene: Pmec-4-ARF-6::GFP(juEx5906). Images of PLM before injury and 2 min post injury are shown. Red arrows indicate axotomy sites. Scale, 10 μm.
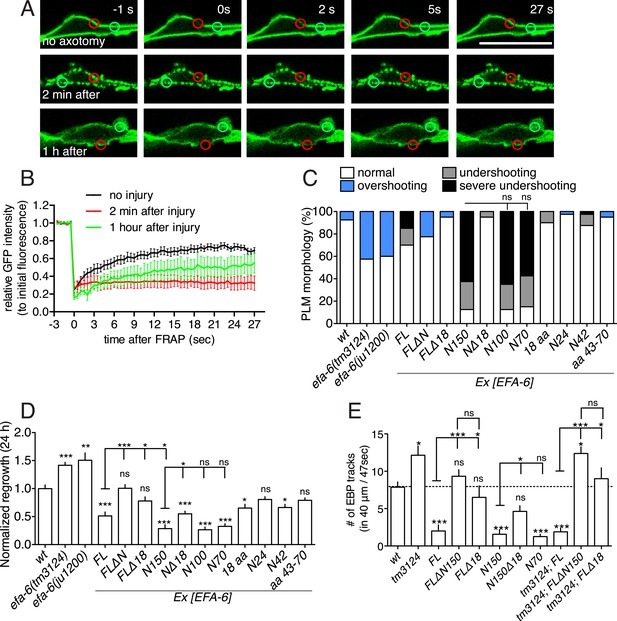
Injury-induced relocalization of EFA-6 correlates with its ability to regulate regrowth and microtubule (MT) dynamics.
(A) FRAP of GFP::EFA-6 (juEx6160) before and after axon injury; regions of interest indicated by red circles; green circles were used to calibrate baseline fluorescence intensity. (B) Normalized average fluorescence intensity after FRAP. (C) PLM termination defects in efa-6(lf) mutants and EFA-6 overexpressing transgenic animals. n = 40 for each bar. See Figure 2—figure supplement 1 for definitions of PLM overshooting and undershooting. (D) Normalized axon regrowth of efa-6(lf) mutants and EFA-6 overexpressors. n ≥ 10. (E) Quantitation of EBP::GFP dynamics in intact axons from wt, efa-6(tm3124) and transgenic animals expressing different EFA-6 fragments under Pmec-4 promoter. n ≥ 10. Statistics, one-way ANOVA with Bonferroni post test; ***p < 0.001; **p < 0.01; *p < 0.05; ns, not significant.
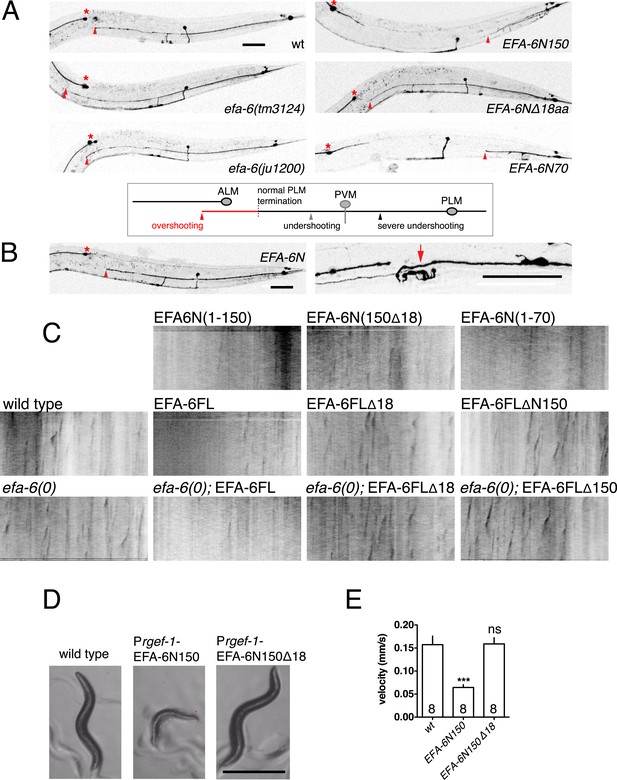
EFA-6 relocalization correlates with protein function in axon termination.
(A) Representative images of PLM development (see quantitation in Figure 2D and strain information in Supplementary file 1C). Asterisk indicates ALM soma. A normal PLM axon usually terminates posterior to ALM soma. The terminus (indicated by a red triangle) of an overshooting PLM axon is anterior to the ALM soma. Scale, 50 μm. (B) Transgenic animals with relatively normal PLM morphology (left panel) were used for axon regeneration analysis (right panel). Arrow: injury site. (C) Representative kymographs of EBP-GFP (juIs338) in wild type, efa-6(tm3124) and in transgenic strains expressing EFA-6 fragments. See quantitation in Figure 2F and strain genotypes in Supplementary file 1C. (D) Locomotor defects due to pan-neural overexpression of EFA-6N150 or EFA-6N150Δ18aa. Overexpression of EFA-6N150 in all neurons results in small, uncoordinated animals; deletion of the 18 aa motif abolishes this effect. (E) The locomotion velocity of animals overexpressing EFA-6N150 (rgef-1 promoter) is reduced compared to wild type, whereas EFA-6NΔ18aa-overexpressing animals display normal locomotion; WormTracker analysis. Statistics: One-way ANOVA with Bonferroni post test. ***p < 0.001.
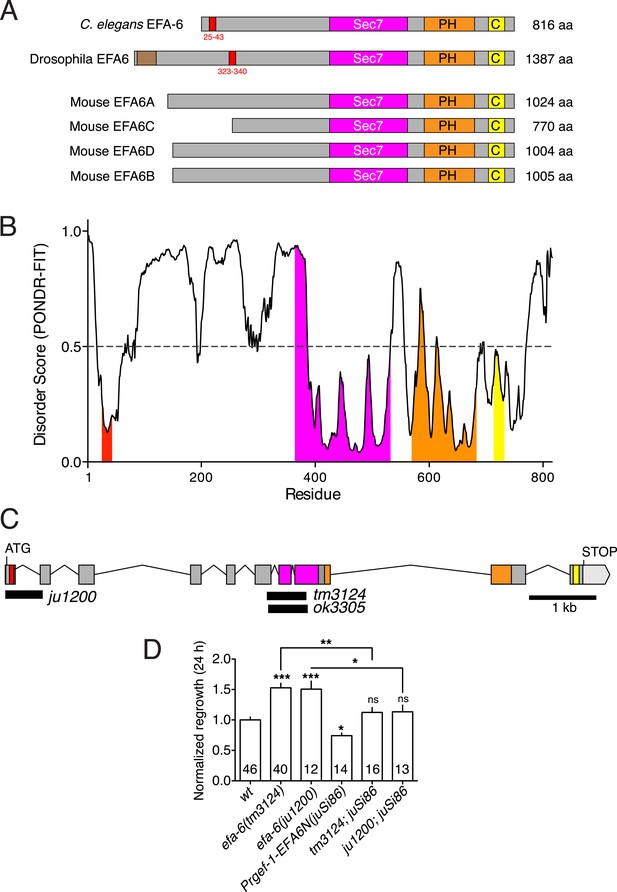
The conserved 18-aa motif in the EFA-6 N-terminus is a region of local protein order.
(A) Cartoons of EFA6 protein family domains, based on sequences from NCBI. The N-termini of EFA6 family members overall have low sequence complexity and lack recognizable domains, with the exception of a predicted PDZ domain in Drosophila EFA6. A conserved 18-aa motif (red box) is found in the N termini of Caenorhabditis elegans and Drosophila EFA6 proteins. Drosophila EFA6 has an N-terminal PDZ domain (brown box). (B) Plot of intrinsic protein disorder score for C. elegans EFA-6 using the metapredictor PONDR-FIT (disprot.org) (Xue et al., 2010). The Sec7, PH, and CC domains show low disorder probability, consistent with their defined tertiary structures. The EFA-6 N terminus has an overall high disorder probability except for the 18-aa motif. (C) C. elegans efa-6 intron-exon structure, with deletion mutations indicated as black boxes. Protein domains are colored as in (A). (D) Pan-neural (Prgef-1) expression of the EFA-6 N terminus from a single copy insertion transgene juSi86 rescues the enhanced regrowth of efa-6(ju1200) and efa-6(tm3124). Statistics: One-way ANOVA with Bonferroni post test. ***p < 0.001, **p < 0.01, *p < 0.05.
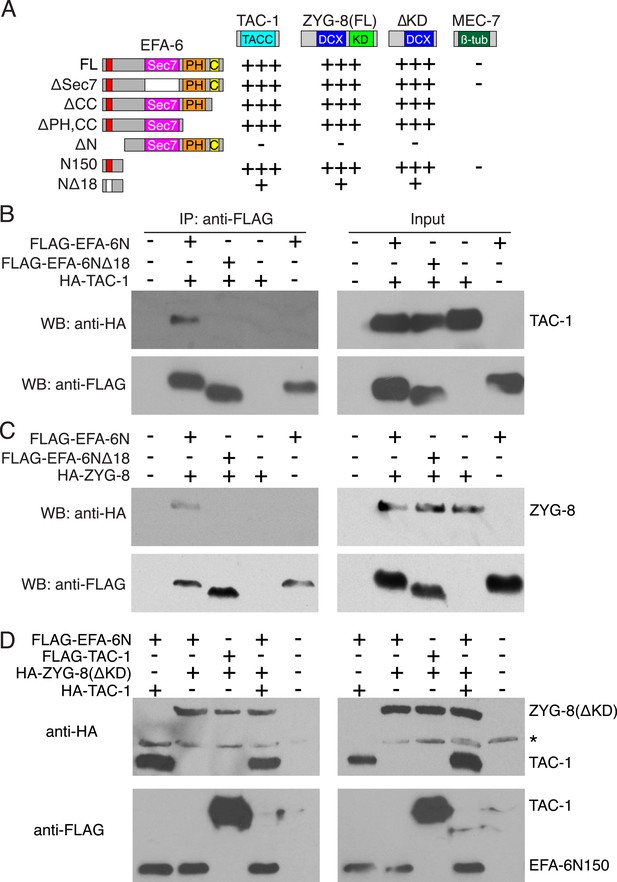
EFA-6 interacts with the MT-associated proteins (MAPs) TAC-1 and ZYG-8.
(A) Summary of two-hybrid analyses. The N-terminus of EFA-6 (N150) is necessary and sufficient for its interaction with TAC-1 and ZYG-8. Deletion of the 18-aa motif from the N-terminus severely impairs binding to TAC-1 and ZYG-8. The interaction between EFA-6 and ZYG-8 does not require the ZYG-8 kinase domain. EFA-6 did not interact with MEC-7/β-tubulin in the two-hybrid assay. ‘+++’, ‘+’, and ‘−’ indicate strong, weak, or undetectable interaction, respectively. (B–D) Co-immunoprecipitation (Co-IP) of EFA-6 and interactors in HEK293 cells. Indicated constructs were co-transfected into HEK293 cells at a 1:1 ratio. M2-FLAG conjugated magnetic beads were used for IP, and rabbit anti FLAG or anti HA antibodies used for western blotting (WB).
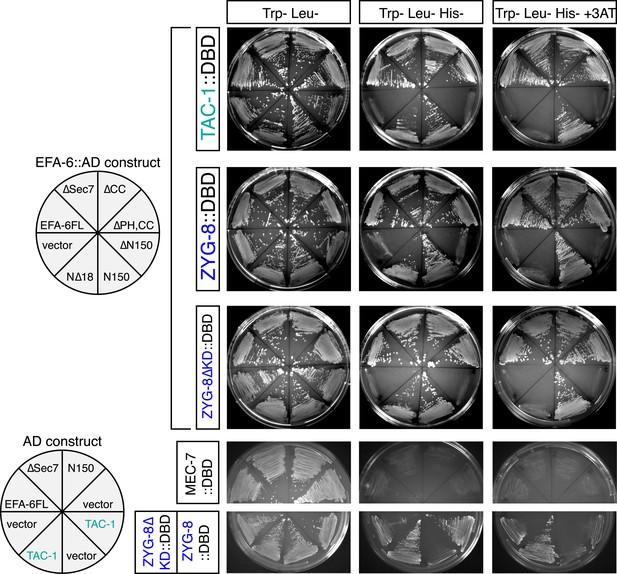
Two-hybrid analysis of the interactions between EFA-6 and TAC-1 or ZYG-8.
Pairs of constructs encoding an activation domain (AD) fusion protein and a DNA binding domain (DBD) fusion protein, as indicated, were co-transformed into yeast strain L40. See Supplementary file 1B for details of plasmids. Transformed yeasts were grown on agar plates with SD medium (synthetic minimal medium) lacking leucine and tryptophan. Interactions were examined on agar plates with SD medium lacking leucine, tryptophan, and histidine (KUWLH), with or without 1 mM 3-AT.
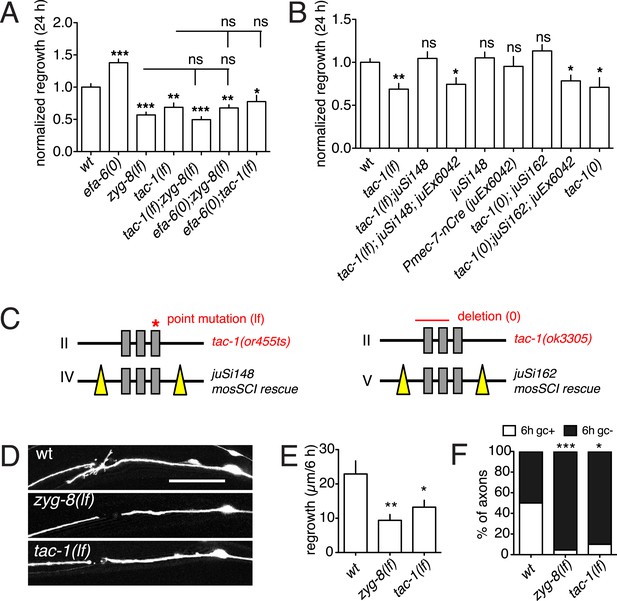
TAC-1 and ZYG-8 promote axon regrowth downstream of EFA-6.
(A) Normalized PLM axon regrowth at 24 hr. Strains were maintained at 15°C, shifted to 25°C 20 hr before axotomy, and kept at 25°C after axotomy for all experiments with ts (lf) alleles. (B) Normalized PLM axon regrowth at 24 hr post axotomy. Loss of TAC-1 impairs axon regrowth in a cell-autonomous manner. (C) Strategy for neuron-specific deletion of tac-1 mutants with Mos-SCI single copy transgene of floxed tac-1. (D) Representative images of axon regrowth at 6 hr post axotomy. WT regrowing axons usually displayed a regenerative growth cone (arrow) at 6 hr post-axotomy whereas zyg-8(lf) and tac-1(lf) mutant axons rarely display growth cones. (E) Quantitation of initial axon regrowth at 6 hr. (F) Percentage of axons with regenerative growth cones 6 hr post axotomy. Statistics, one-way ANOVA with Bonferroni post test; ***p < 0.001; **p < 0.01; *p < 0.05; ns, not significant. n ≥ 10. Scale, 25 μm.
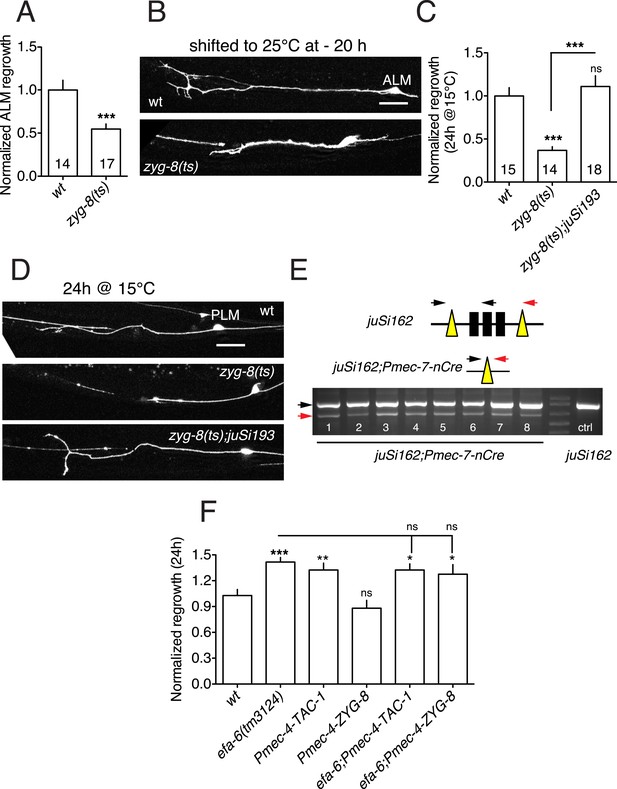
EFA-6 and its interactors regulate axon regeneration.
(A) zyg-8(or484ts) displays reduced ALM regrowth. Normalized ALM axon regrowth, 24 hr post axotomy. Strains were maintained at 15°C, shifted to 25°C 20 hr before axotomy, and kept at 25°C after axotomy. Statistics: Student's t-test. ***p < 0.001. (B) Representative images for panel (A); scale, 10 μm. (C) PLM axon regrowth defect of zyg-8(or484) at permissive temperature (15°C) and rescue by single copy insertion transgene Pmec-4-ZYG-8(juSi193). Statistics: one-way ANOVA with Bonferroni post test; ***p < 0.001. (D) Representative images for panel (C); Scale, 25 μm. (E) PCR assay for efficiency of the Cre-lox recombination in touch neurons. 8 animals of strain CZ20478 [tac-1(ok3305); juSi162(lox-tac-1-lox Mos-SCI) V; juEx6042(Pmec-7-nCre;ttx-3-rfp)] were tested for the excision product (red arrow on the gel image). Excision of the lox-flanked fragment occurred in 8/8 animals. No excision was seen in juSi162 animals lacking the Cre transgene. Individual animals expressing transgenic marker Pttx-3-RFP from the strain CZ20478 [tac-1(ok3305); juSi162(lox-tac-1-lox Mos-SCI); juEx6042(Pmec-7-nCre;Pttx-3-RFP)] were genotyped for the Cre-dependent deletion of tac-1 single copy insertion (juSi162). Animals not expressing Pttx-3-RFP, that is, tac-1(ok3305); juSi162(lox-tac-1-lox Mos-SCI), were used as negative control. The following primers (specific to juSi162 and not endogenous tac-1) were used in PCR: ttTi5605 homology arm: ACGCCCAGGAGAACACGTTAG (left black arrow) tac-1-5′ UTR: AGATCCACCCTCACCATCAC (middle black arrow) unc-119: TTCGCTGTCCTGTCACACTCG (red arrow). Without Cre-dependent deletion, this PCR will produce 3472 and 860 bp fragments. After deletion, the PCR product will be 642 bp. tac-1(ok3305) is a deletion of 812 bp with an insertion of 23 bp of random sequence at the deletion. We designed primers in the 23 bp insertion to detect ok3305, so that the tac-1(+) transgene will not interfere with genotyping for ok3305. (F) Overexpressing TAC-1 or ZYG-8 does not enhance axon regrowth of efa-6(lf), consistent with function in the same pathway. Normalized PLM axon regrowth of strains with indicated genotypes. Statistics: one-way ANOVA with Bonferroni post test; ***p < 0.001; **p < 0.01; *p < 0.05.
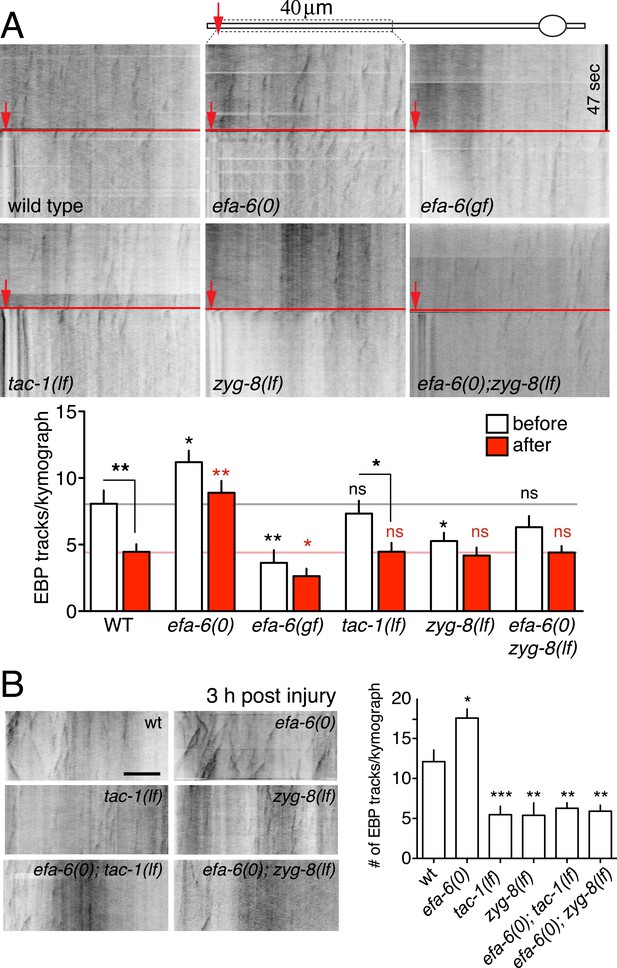
Injury triggers rapid down-regulation of MT dynamics dependent on EFA-6.
(A) MT dynamics (EBP-2::GFP) before and immediately after injury. Kymographs were created from videos of 400 frames (0.23 s/frame), 200 frames before and 200 frames after axotomy. Lower panel: Quantitation of EBP-2::GFP tracks in proximal axon before and immediately after injury. (B) MT dynamics 3 hr post injury. Kymographs were created from videos of 200 frames (0.23 s/frame). Quantitation of EBP-2::GFP tracks in 40 μm of the proximal axon for 47 s 3 hr post injury. Red line represents time of axotomy; arrow indicates injury site. Strains were maintained at 15°C, shifted to 25°C 20 hr before axotomy. Alleles: efa-6(tm3124), tac-1(or455ts), zyg-8(or484ts). Statistics, one-way ANOVA with Bonferroni post test; ***p < 0.001; **p < 0.01; *p < 0.05; ns, not significant. n ≥ 10 axons per condition.
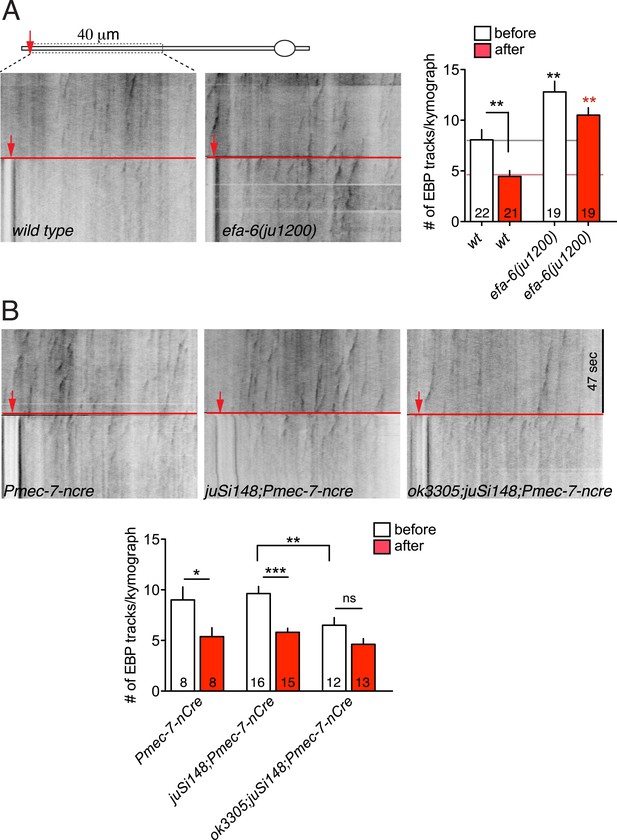
MT dynamics triggered by injury.
(A) MT dynamics (EBP-2::GFP) before and immediately after injury in wild type and efa-6(ju1200) mutant. A significant reduction in MT growth (defined as number of EBP-2::GFP tracks per kymograph) was seen in wild type animals, but not in efa-6(ju1200), similar to tm3124. (B) MT dynamics before and after injury in the tissue-specific tac-1 deletion mutant and in control strains. Statistics: one-way ANOVA with Bonferroni post test; ***p < 0.001; **p < 0.01; *p < 0.05.
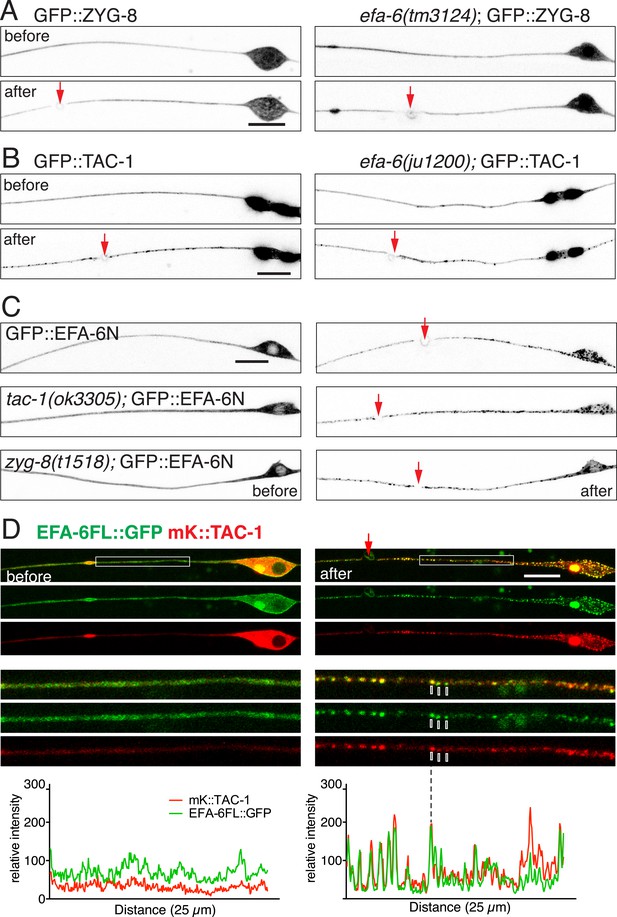
TAC-1 relocalizes in response to injury to become co-localized with EFA-6.
(A) Localization of GFP::ZYG-8 in touch neurons before and 2 min after axotomy in wild type and efa-6(tm3124). GFP::ZYG-8 localization is not affected by axon injury or loss of EFA-6. Transgene: Pmec-4-GFP::ZYG-8(juEx5932). (B) GFP::TAC-1 in PLM before and 2 min after axotomy at wild type and efa-6(ju1200) backgrounds. Injury triggered relocalization of TAC-1 was similar to EFA-6 and not dependent on EFA-6. Pmec-4-GFP::TAC-1(juEx5759). (C) GFP::EFA-6N150 (juEx3531) localization in wild-type, tac-1(ok3305) and a putative zyg-8 null allele zyg-8(t1518) (Gönczy et al., 2001). Relocalization of EFA-6N150 was not dependent on TAC-1 or ZYG-8. (D) Localization of EFA-6FL::GFP and mKate2::TAC-1 before and after axotomy in a touch neuron. Before axotomy, TAC-1 was diffuse in soma and along the axon, and concentrated in a large perinuclear dot. EFA-6 was predominantly localized to the plasma membrane and also in the perinuclear dot marked by TAC-1. After axotomy, both proteins became punctate and the puncta were partially co-localized; enlargements in small boxes below. Graphs of line scans along the axon are shown below the enlarged images. Arrow, injury site; scale, 10 μm.
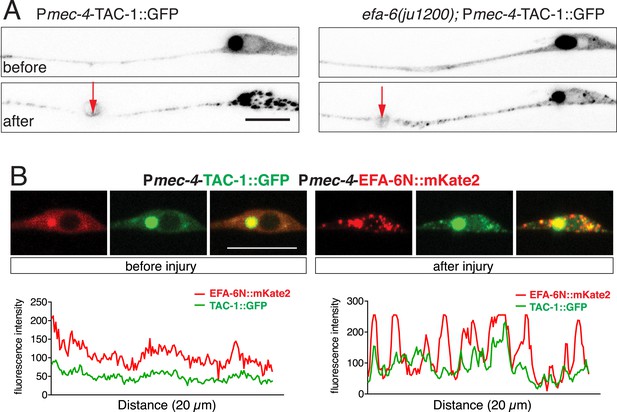
Injury triggered relocalization.
(A) Localization of TAC-1::GFP before and 2 min after injury; TAC-1 with a C-terminal GFP tag relocalized similarly to N-terminally tagged TAC-1. Localization of TAC-1::GFP in efa-6(ju1200) is similar to wild type background. Transgene: Pmec-4-TAC-1::GFP(juEx6362). (B) Co-localization of TAC-1::GFP and EFA-6N150::mKate2 in puncta after axotomy. Before axotomy, TAC-1 and EFA-6N150 were diffuse along the axon, and also localized in a perinuclear dot in the soma. After axotomy, both proteins were punctate and the puncta co-localized with each other. As the axonal signal was relatively dim, only soma images are shown. Line scan in soma below images. Arrow: injury site; scale, 10 μm.
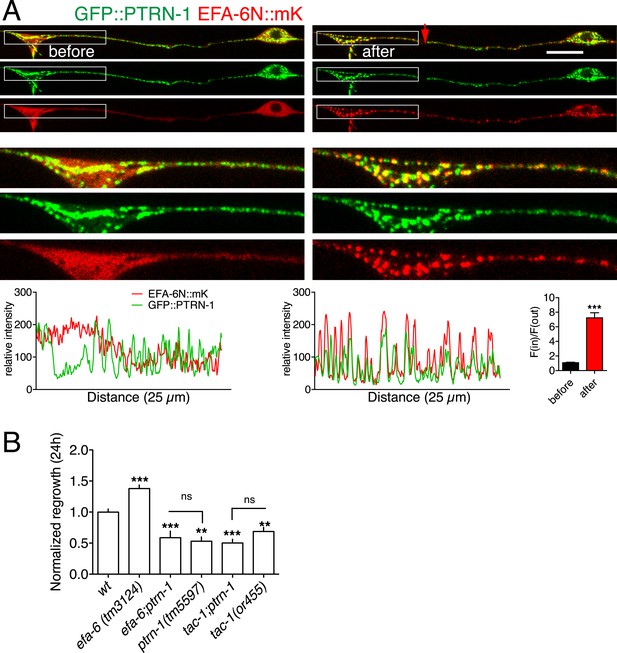
EFA-6 and TAC-1 re-localize to puncta overlapping with the MT minus end-binding protein Patronin/PTRN-1.
(A) Localization of PTRN-1 and EFA-6N150 in PLM before and after axotomy. EFA-6N150::mKate2 was diffuse in soma and axon before injury, and became punctate after injury and these puncta co-localized to GFP::PTRN-1. Enlarged images of the regions in boxes are shown below. Graphs of line scans along the axon and F(in)/F(out) ratio quantitation are shown below. Increased F(in)/F(out) ratio indicates higher degree of colocalization post axotomy; see Figure 7—figure supplement 1A and ‘Materials and methods’ for calculation of F(in)/F(out). Statistics: Student's t-test. ***p < 0.001. (B) Epistatic interactions between efa-6(0), ptrn-1(0), and tac-1(lf). Normalized PLM regrowth. Strains without or with temperature shift (cultured at 15°C and upshifted to 25°C 20 hr before axotomy and kept at 25°C for 24 hr after axotomy) were quantified separately. Statistics: one-way ANOVA with Bonferroni post test.
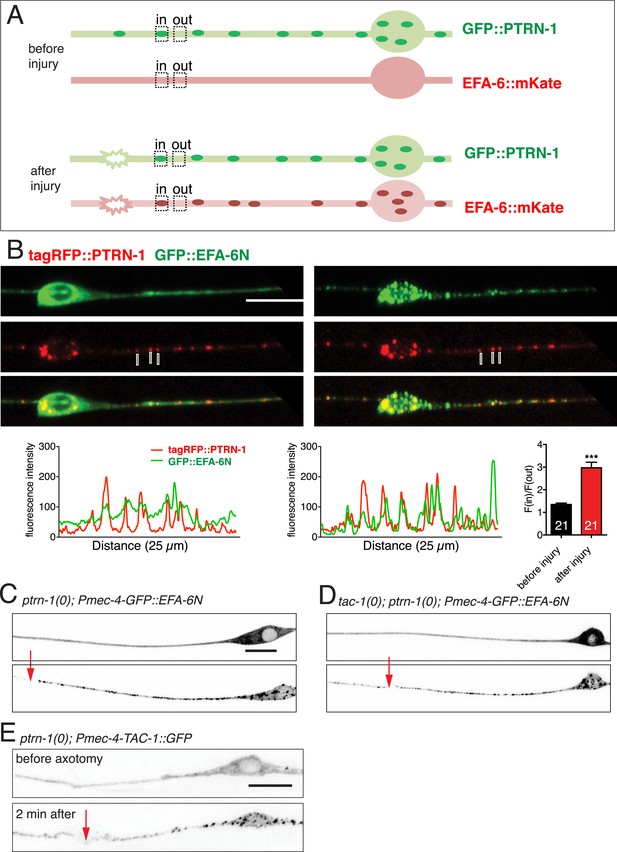
Co-localization of EFA-6 with PTRN-1 after injury.
(A) Cartoons illustrating quantitation of co-localization [F(in)/(out) ratio]; see ‘Materials and methods’ for details of the colocalization ratio calculation. (B) Localization of tagRFP::PTRN-1 and GFP::EFA-6N150 in PLM, before and after axotomy. GFP::EFA-6N150 became punctate after injury and co-localized with tagRFP::PTRN-1 puncta; 3 representative puncta are marked. Line scan and F(in)/F(out) ratio quantitation shown below the images. Statistics: Student's t-test. ***p < 0.001. (C) Localization of GFP::EFA-6N150 in ptrn-1(lt1), before and after axon injury. Localization of GFP::EFA-6N150 was not dependent on PTRN-1. ptrn-1(lt1) is a MosDel-induced deletion that removes the entire ptrn-1 coding sequence (Chuang et al., 2014). (D) Localization of GFP::EFA-6N150 in tac-1(lf) ptrn-1(0) double mutant is indistinguishable from wild type. (E) Localization of TAC-1::GFP in ptrn-1(0) before and after axon injury. Localization of TAC-1::GFP was not dependent on PTRN-1. Arrow: injury site; scale, 10 μm.
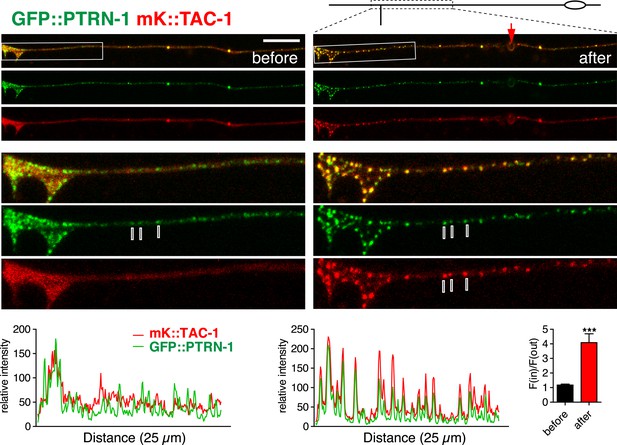
TAC-1 relocalizes to PTRN-1 puncta after injury.
Localization of GFP::PTRN-1 and mKate2::TAC-1 in distal PLM before and after axotomy; enlargements and graphs as in panel A. mKate2::TAC-1 was diffuse in the axon before injury, and became punctate after injury, colocalizing with GFP::PTRN-1 (3 representative puncta marked with lines). Arrow, injury site; Scale, 10 μm.
Videos
Injury-induced GFP::EFA-6 relocalization in neurons (ALM).
Transgene: Pmec-4-GFP::EFA-6(juEx6160). The video is 103 s, taken at 1 s/frame.
Tables
Localization and function of EFA-6 protein fragments in PLM neurons
| EFA-6 proteins | GFP fusion protein localization | Injury-induced re-localization | Overexpression effect on regrowth | Overexpression effect on axon development |
|---|---|---|---|---|
| Full length (FL) | Cortical membrane | yes | 51.2% ± 7.1% *** | 30% undershooting |
| FLΔN150 | Cortical membrane | no | 100.5% ± 6.6% ns | 22.5% overshooting |
| N150 | Cytosolic | yes | 28.5% ± 7.1% *** | 87.5% undershooting |
| 18 aa | Cytosolic + nuclear | no | 65.3% ± 4.7% * | 10% mild undershooting |
| N150Δ18aa | Cytosolic + nuclear | no | 55.0% ± 4.7% *** | 5% mild undershooting |
| N150 (33–38A) | Cytosolic + nuclear | no | 57.6% ± 10.4% *** | 5% mild undershooting |
| N150 (25–32A) | Cytosolic + nuclear | no | 59% ± 5.7% *** | 7.5% mild undershooting |
| N150 S33A, D35A | Cytosolic + nuclear | no | 63% ± 7.8% ** | 7.5% mild undershooting |
| FLΔ18aa | Cortical membrane | no | 77.9% ± 7.3% ns | wt |
| N100 | Cytosolic | yes | 26.6% ± 4.7% *** | 87.5% undershooting |
| N70 | Cytosolic | yes | 32.7% ± 4.5% *** | 85% undershooting |
| N42 | Cytosolic + nuclear | no | 66.2% ± 5.7% * | 12.5% undershooting |
| N24 | Cytosolic + nuclear | no | 80.7% ± 5.0% ns | wt |
-
Mild and severe undershooting are defined as PLM termination anterior or posterior to the PVM soma respectively; ‘undershooting’ includes both mild and severe undershooting. See Figure 2—figure supplement 1.
Additional files
-
Supplementary file 1
(A) Plasmids for C. elegans Transgenes. (B) Plasmids for Yeast Two-Hybrid and Co-immunoprecipitation. (C) C. elegans strains, transgenes, and clones.
- https://doi.org/10.7554/eLife.08695.023





