Complex transcriptional regulation and independent evolution of fungal-like traits in a relative of animals
Figures
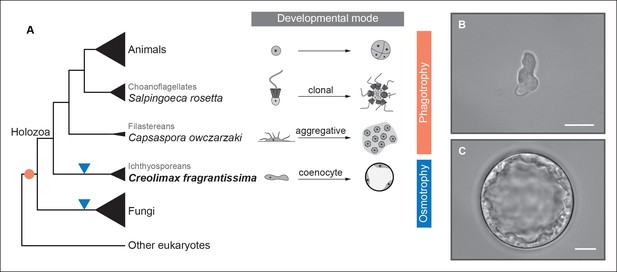
Evolution of developmental and feeding modes across holozoans.
(A) The cladogram represents known phylogenetic relationships among holozoans (Torruella et al., 2012; Torruella et al., 2015). Each lineage is represented by the species proposed as a model system with a schema of its developmental mode on the right. The evolution of specialized osmotrophy is shown as a blue triangle in the cladogram, while the putative ancestral phagotrophic feeding mode of opisthokonta is shown as an orange circle (Cavalier-Smith, 2012). Divergence times of the lineages shown in this figure range between 700 Mya (considered the latest estimates of animal origins) and 1200 Mya (earliest estimates of Opisthokont origins) (Sharpe* et al., 2015). Micrographs depicting the (B) amoeboid stage and (C) multinucleate stage of Creolimax fragrantissima are shown. Scale bars = 10 μm. Choanoflagellate adapted from Mark Dayel (CC BY-SA 3.0) www.dayel.com/blog/2010/10/07/ choanoflagellate-illustration.
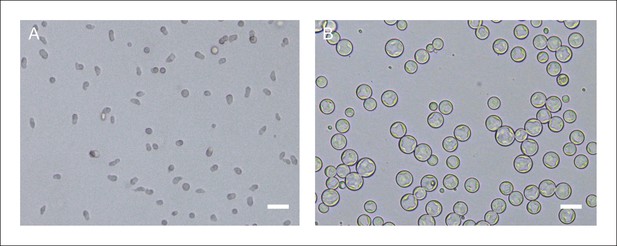
Creolimax synchronized stages.
(A) Culture after 5 μm filtering. (B) Culture grown for 24 hr after 5 μm filtering. Scale bars = 20 μm.
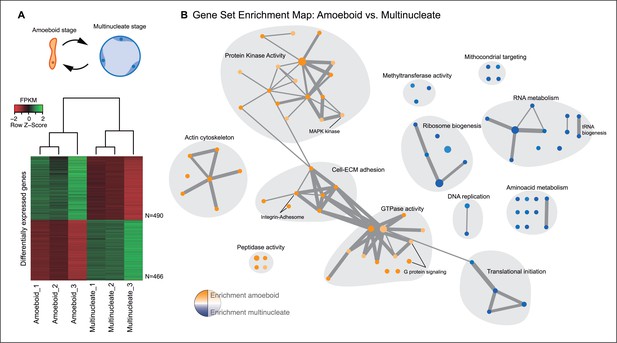
Differential gene expression in Creolimax.
(A) Diagram of the amoeboid and multinucleate stages, and heatmap showing the significantly differentially expressed genes across biological replicates in the pair-wise stage comparison. (B) Gene set enrichment analysis for the two stages. Orange represents enrichment in the amoeboid stage and blue represents enrichment in the multinucleate stage, color intensity depicts level of significance (p value). Node size represents the total number of genes in each GO, and edge width represents the total number of genes shared between each enriched GO category. Functionally related GOs are manually circled in gray shade according to functional and genic redundancy established by network connectivity. Complete list of GOs and inclusive groupings are found in Figure 2—source data 1. GOs, Gene Ontologies.
-
Figure 2—source data 1
GOs enrichments and groupings from Figure 2B.
- https://doi.org/10.7554/eLife.08904.006
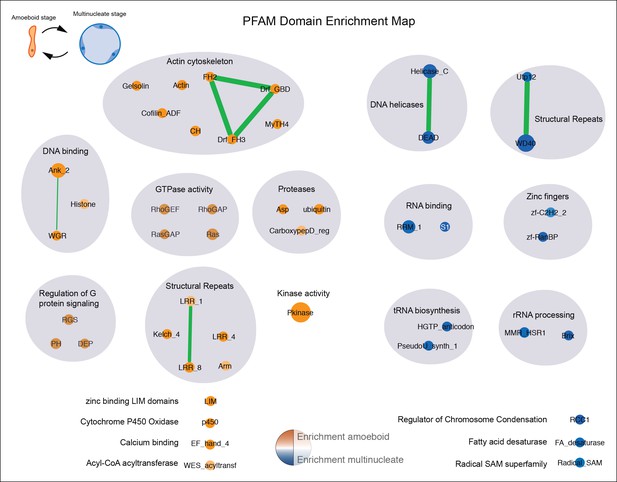
Pfam domain set enrichments in differentially expressed genes.
Orange represents domains enriched in the amoeboid stage and blue represents domains enriched in the multinucleate stage, color intensity depicts level of significance (p value, Fisher’s exact test). Node size represents the total number of genes containing a Pfam domain and edge width represents the total number of genes sharing two distinct Pfam domains. Functionally related Pfams are manually circled in gray shade, primarily based on the information gathered in the Pfam database for each domain (including Pfam2Go annotations). Additional criteria to include a given domain in a functionally related category included: checking the list of GOs of the statistically differentially expressed domain-containing genes in a given stage and using a network connectivity redundancy between GO and Pfam categories in a mixed network (including both Pfam and GO annotation) done with Enrichment Map plugin in Cytoscape (Merico et al., 2010). GO, Gene Ontologies.
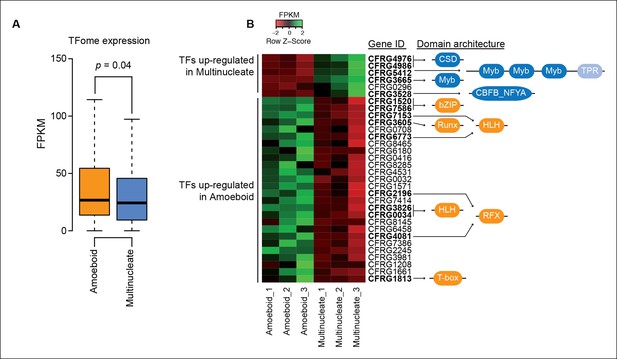
Differential TFome expression.
(A) Distribution of expression values (FPKM) for all TFs in the genome in the amoeboid and the multinucleate stages (p value, Wilcoxon signed rank test). (B) Heatmap showing the expression levels of all the TFs with a two-fold change in expression level between stages. Those with the gene ID in bold have statistically significant differential expression according to at least three different differential expression pipelines (see Material and methods). To the right, the domain architectures of the TFs show the DNA binding domain in dark blue and orange according to the stage where they are up-regulated. In the multinucleate stage: CSD (PF00313), Myb (PF00249), CBFB_NFYA (PF02045) and TPR (PF13414). In the amoeboid stage: bZIP_1 (PF00170), HLH (PF00010), T-box (PF00907), RFX_DNA_binding (PF02257), Runt (PF00853). CSD: Cold shock protein; FPKM, fragments per kilobase of exon per million fragments mapped; HLH: helix-loop-helix (protein structure); TFs, transcription factors
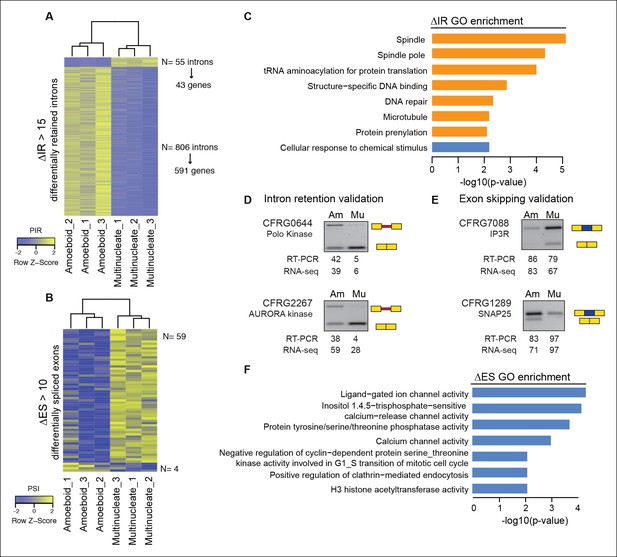
Regulated alternative splicing modes in Creolimax.
(A) Heatmap showing PIR inclusion levels of differentially retained introns. (B) Heatmap showing the PSI levels of differentially skipped exons. (C) GO enrichment activities of the genes showing differential IR. Bar length indicates the significance of the enrichment, orange indicates those with higher inclusion levels in the amoeboid stage and blue those with higher inclusion levels in the multinucleate stage. (D–E) RT-PCR validations of selected IR and ES events. The values correspond to relative intensity of the alternative isoform (retained intron or skipped exon) bands in the RT-PCR and the proportions observed for the inclusion values in the RNA-seq. (F) GOs enrichment of genes with differential ES, in blue those with higher exon inclusion levels in the multinucleate stage. ES, exon skipping; GO, Gene Ontology; IR, intron retention; PIR, percent intron retention; PSI, percent spliced in; RT-PCR, reverse transcription-polymerase chain reaction; RNA-seq, RNA sequencing.
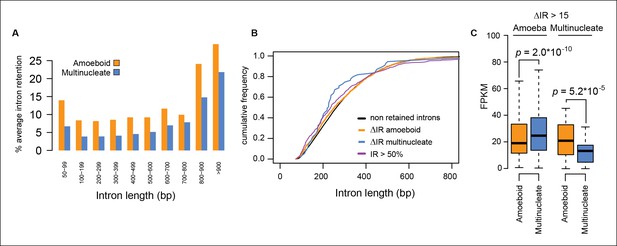
Intron size and transcriptional levels of differentially retained introns.
(A) Relationship between intron length and retention level. The barplot shows the percentage of retained introns (PSI >20) among the total number introns of a given size. (B) Cumulative frequency plot showing that the intron size distribution is the same for constitutive introns (PSI<2), highly retained introns (PSI >50 both stages) and differentially retained introns. (C) Distribution of expression values (FPKM) in the amoeboid and the multinucleate stage for genes differentially retained introns. FPKM, fragments per kilobase of exon per million fragments mapped; PSI, percent spliced in.
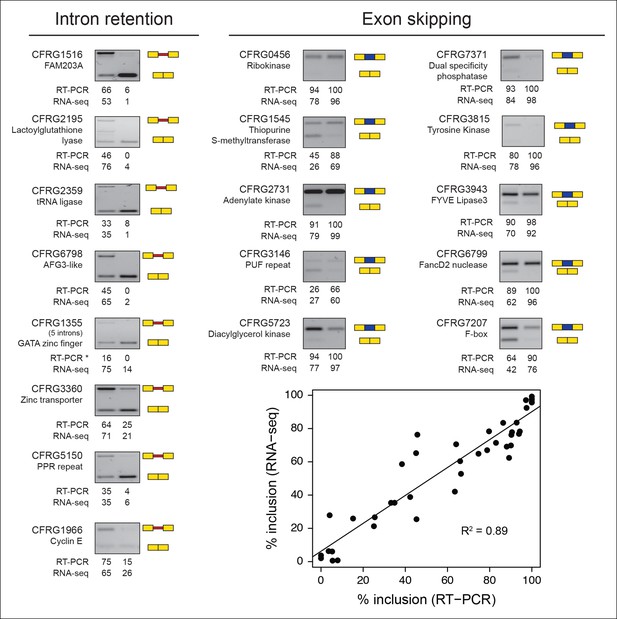
Validations of intron retention and ES events.
The RT-PCR gels show the different splice variants for each gene in the amoeboid stage (left) and multinucleate stage (right). RT-PCR values indicate the levels of inclusion of the alternative isoform (retained intron or skipped exon) compared to the canonically spliced form obtained with ImageJ; RNA-seq values are based on read coverage for each event. The scatter plot shows the differences between the RT-PCR measures and the RNA-seq-based values for all the validated examples, overall showing a high correlation. ES, exon skipping; RT-PCR, reverse transcription-polymerase chain reaction; RNA-seq, RNA sequencing
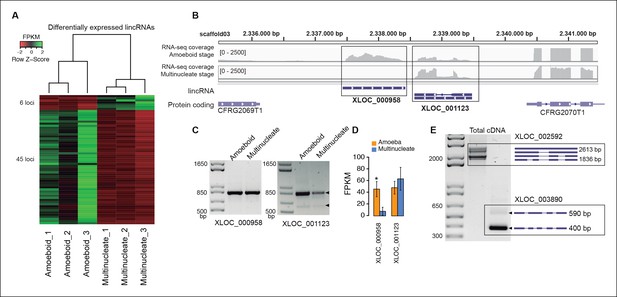
Transcriptional and post-transcriptional regulation of lincRNAs in Creolimax.
(A) Heatmap showing transcriptional levels of significantly differentially expressed lincRNAs across biological replicates of amoeboid and multinucleate stages. (B) Example of genomic region where two lincRNA loci are found in tail-to-tail orientation surrounded by two protein-coding genes. (C) RT-PCR validations of the lincRNA loci. (D) Barplot depicting the average gene expression of those lincRNA in each stage. (E) Alternative splicing isoforms of lincRNAs showing various degrees of IR. IR, intron retention; lincRNA, long intergenic non-coding RNAs; RT-PCR, reverse transcription-polymerase chain reaction.
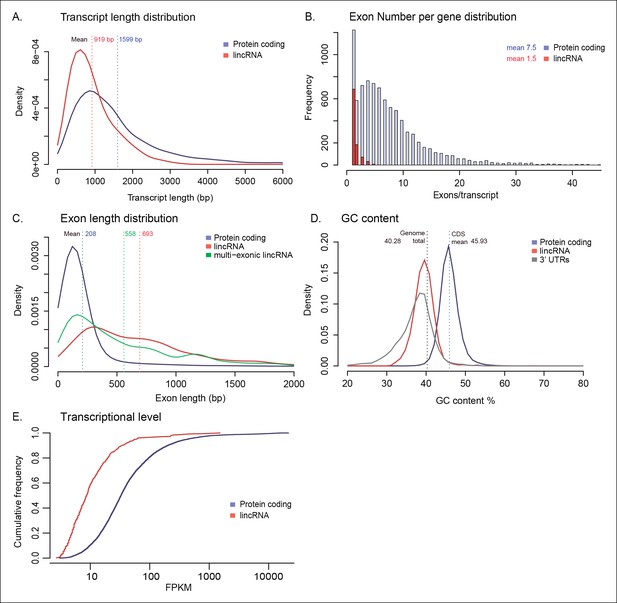
Genomic architecture of lincRNAs compared to protein-coding genes.
(A) Kernel density plot showing transcript length distribution. Protein-coding genes are shown in blue, lincRNA in red. (B) Density plot showing exon number distribution. (C) Kernel density plot showing exon length distribution; multi-exonic lincRNAs are shown in green. (D) Kernel density plot showing GC content distribution. The dashed line indicates the total genome GC content. (E) Cumulative frequency plot of expression levels obtained from log10 (FPKM). FPKM, fragments per kilobase of exon per million fragments mapped; lincRNA, long intergenic non-coding RNAs.
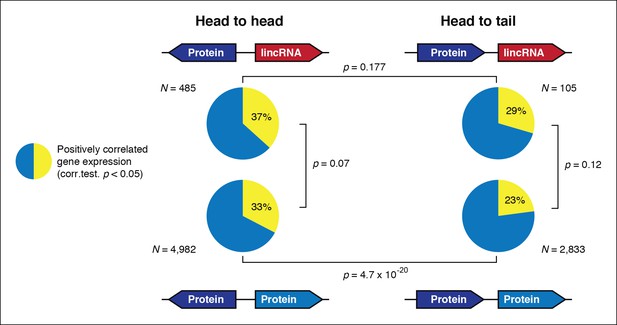
Gene orientation and transcriptional co-regulation of neighboring genes.
Distribution of Pearson correlation values between a gene and its upstream neighbor subdivided in four categories. Head-to-head oriented neighbors tend to be more co-expressed than head-to-tail genes, independently of coding potential.
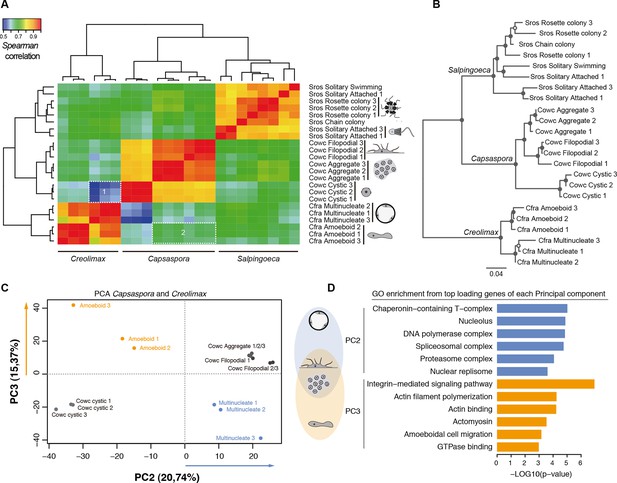
Holozoan cross-species comparison of transcriptional profiles.
(A) Symmetrical heatmap of the pair-wise Spearman correlation coefficients for the gene expression profiles of each cell stage. For each sample, log2(cRPKM+1) expression levels were obtained for 2177 one-to-one orthologs in the three species analyzed (see Materials and methods). Dashed-line squares highlight the direct comparisons for 1) Cfra multinucleate stage replicates against Cowc cystic stage replicates and 2) Cfra amoeboid stage replicates against Cowc aggregate and filopodial stage replicates. (B) Neighbor-joining tree of the species cell stages based on the aforementioned Spearman correlation distances matrix. Filled circles represent >95% bootstrap replicate nodal support. (C) The cell types plotted according to the values of the principal components 2 and 3 from a PCA of a dataset of 3030 1-to-1 orthologs between Capsaspora and Creolimax. (D) The significant GO enrichments for the top positive loading genes (>0.03) of the principal component 2 and 3. Cfra, Creolimax fragrantissima; Cowc, Capsaspora owczarzaki; GO, Gene Ontology; Sros, Salpingoeca rosetta; PCA, principal component analysis.
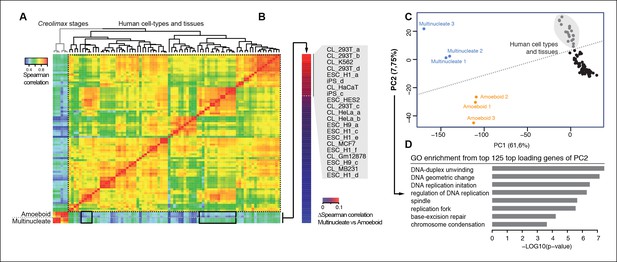
Comparison of human and Creolimax cell types and tissues.
(A) Symmetrical heatmap of the pair-wise Spearman correlation coefficients for the gene expression profiles of each cell type or tissue. For each sample log2(FPKM+1) expression levels were obtained for 2272 one-to-one orthologs between Creolimax and human (see Materials and methods). (B) The human cell types sorted by the difference of Spearman correlation between the amoeboid and the multinucleate cell stages. Highlighted in gray are those that displayed the major differences (>0.05). (C) The cell types plotted according to values of the principal components 1 and 2 from a PCA of the same transcriptional dataset of 2272 orthologs. The dots in gray are the human cell lines highlighted in the previous section. (D) The significant GO enrichments for the top positive loading genes of the principal component 2 (>0.04). Sampled human cell types described in Figure 6—source data 1. GO, Gene Ontology; PCA, principal component analysis.
-
Figure 6—source data 1
Human RNA-seq datasets used in this analysis.
- https://doi.org/10.7554/eLife.08904.017
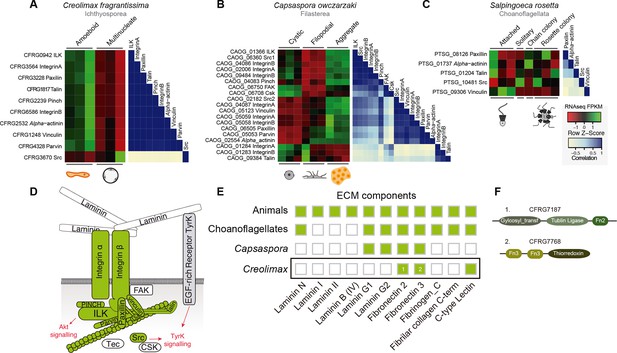
Co-regulation of the integrin adhesome in holozoans.
Heatmaps depicting expression levels of integrin adhesome orthologs (red–green) and their pair-wise Pearson correlation coefficients (white–blue) obtained from genome-wide FPKM transcriptional levels in the ichthyosporean Creolimax (A), the filasterean Capsaspora (B), and the choanoflagellate Salpingoeca (C). (D) Diagram of integrin adhesome components, those in green are found in Creolimax and those in white are absent. In gray, a tyrosine kinase receptor with extracellular EGF domains encoded Creolimax genome that could be interacting with an ECM component. (E) Repertoire of animal ECM domains in the three unicellular holozoan genomes; green = presence, white = absence. (F) Pfam domain architectures of fibronectin-domain containing genes in Creolimax. ECM, extracellular matrix; FPKM, fragments per kilobase of exon per million fragments mapped.
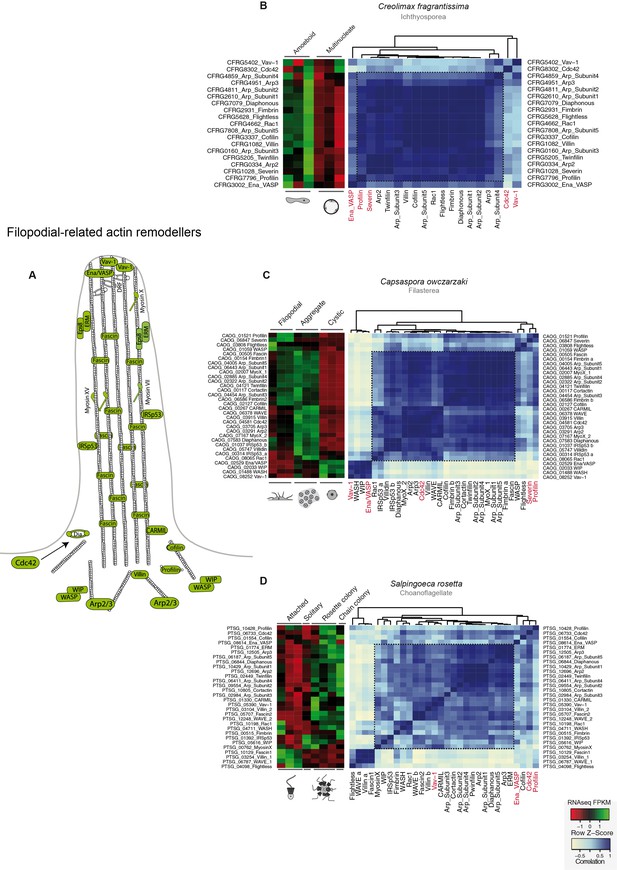
Co-regulation of the filopodial molecular toolkit genes in holozoans.
(A) Diagram of the filopodial machinery components as described in Sebé-Pedrós et al 2013 (Sebé-Pedrós et al., 2013); those in green are the genes found in unicellular holozoans. Heatmaps depicting expression levels of filopodial component orthologs (red–green) and their pair-wise Pearson correlation coefficients (white–blue) obtained from genome-wide FPKM transcriptional levels in the ichthyosporean Creolimax (B), the filasterean Capsaspora (C) and the choanoflagellate Salpingoeca (D). Highlighted in red are those genes that appear outside the module in at least two lineages. FPKM, fragments per kilobase of exon per million fragments mapped.
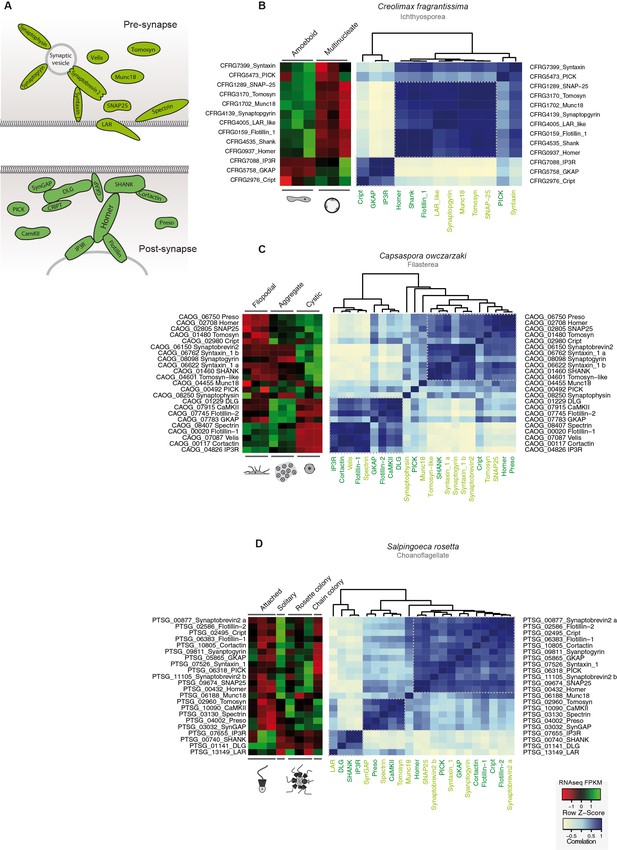
Co-regulation of the pre- and post- synaptic genes in holozoans.
(A) Diagram of the pre-synaptic (paler green) and post-synaptic (darker green) found in unicellular holozoans. Heatmaps depicting expression levels of pre- and post-synaptic orthologs (red–green) and their pair-wise Pearson correlation coefficients (white–blue) obtained from genome-wide FPKM transcriptional levels from the ichthyosporean Creolimax (B), the filasterean Capsaspora, (C) and the choanoflagellate Salpingoeca (D). Flotillin genes, despite not being directly related to the post-synpatic scaffold, have been shown to interact with Homer in choanoflagellates and animals (Burkhardt et al., 2014). FPKM, fragments per kilobase of exon per million fragments mapped.
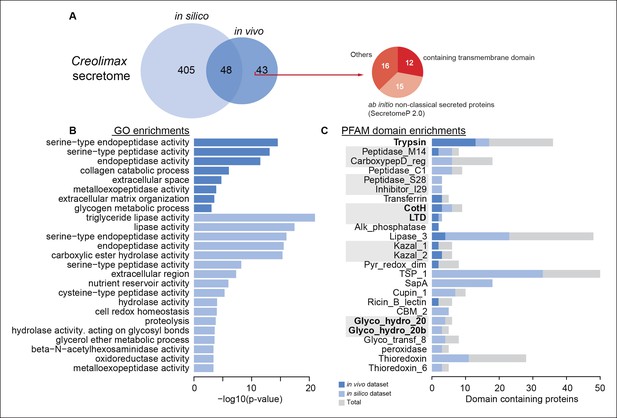
Functional enrichments of Creolimax secretome.
(A) Venn diagram showing the number of genes identified in the Creolimax secretome through an in silico approach (see Material and methods) and an in vivo proteomics approach. A circle diagram describes the features of genes only identified in the in vivo approach, lacking a signalP or having TM domains . (B) GO categories and (C) PFAM domains enriched in the secretome; in dark blue are those enriched in the in vivo dataset; in pale blue are those enriched in the in silico dataset; in gray are the total amount of PFAM-domain containing genes in the genome. GO, Gene Ontology.
-
Figure 8—source data 1
In vivo proteomics of Creolimax secretome.
- https://doi.org/10.7554/eLife.08904.022
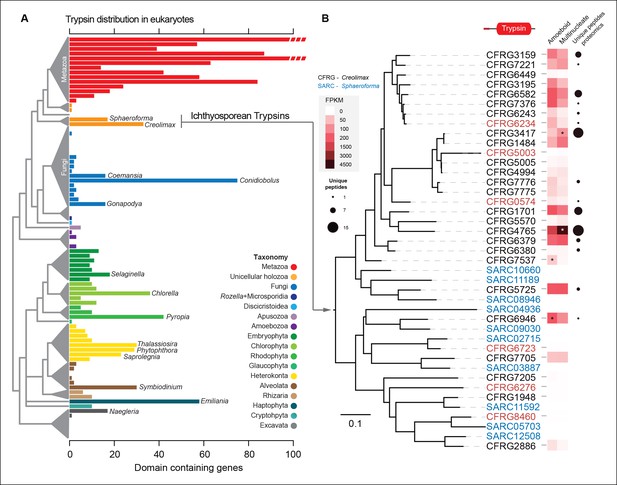
Trypsin evolution.
(A) Barplot showing the total number of Trypsin proteins (PF00089) found in the genomes of diverse eukaryotes. Branches are color coded according to the taxonomy shown in the legend. (B) Maximum-likelihood phylogenetic tree based on the amino acid sequence of the Trypsin domain from Creolimax fragrantissima and Sphaeroforma arctica. Expression levels obtained from genome-wide FPKM calculation are shown. Number of unique peptides obtained from the in vivo secretome proteomic dataset is also shown. In red are those genes that do not present a signal peptide according to SignalP. FPKM, fragments per kilobase of exon per million fragments mapped.
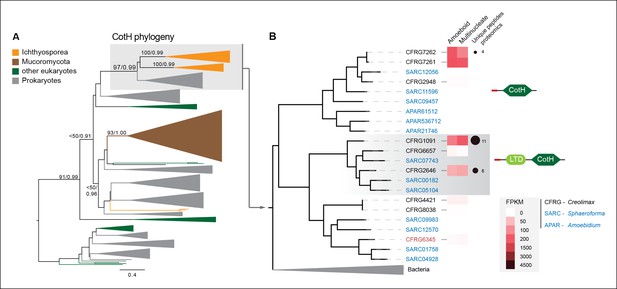
CotH evolution.
(A) Maximum-likelihood phylogenetic tree of the CotH domain (PF08757). Nodal support is shown in key branches (100 maximum likelihood replicates bootstrap values and Bayesian posterior probabilities). Color code indicates taxon distribution in each clade as depicted in the legend; for a detailed tree, see Figure 10—figure supplement 1. (B) Detail of the phylogenetic tree depicting ichthyosporean CotH sequences, covering Creolimax, Sphaeroforma arctica, and Amoebidium parasiticum. Expression levels obtained from genome-wide FPKM calculation and the number of unique peptides obtained from the in vivo secretome proteomic dataset are shown. Domain configurations obtained from a PfamScan analysis. Gene identifiers in red are those that do not present a signal peptide according to SignalP. FPKM, fragments per kilobase of exon per million fragments mapped.
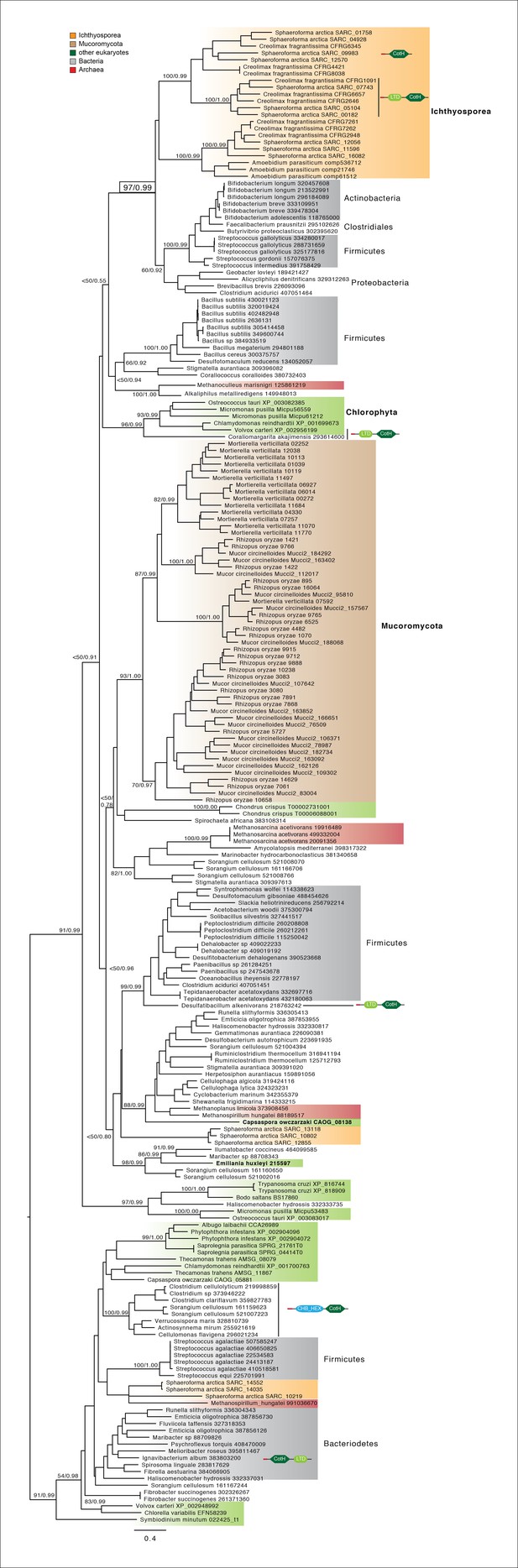
CotH extended phylogeny.
Maximum-likelihood phylogenetic tree of the CotH domain (PF08757). Nodal support is shown in key branches (100 maximum likelihood replicates bootstrap values and Bayesian posterior probabilities). Color code indicates taxon distribution in each clade as depicted in the legend. Domain configurations obtained from a PfamScan analysis.
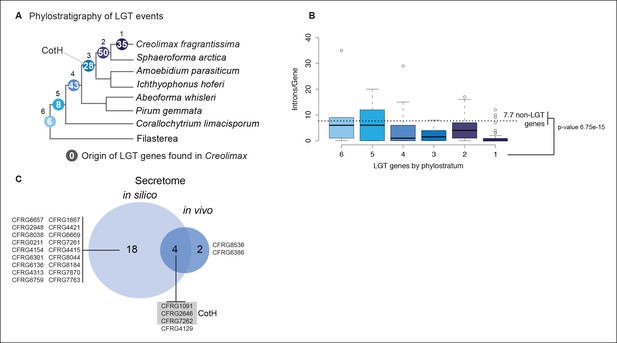
Features of prokaryotic LGT.
(A) Phylostratigraphy depicting origins of LGT events found in Creolimax. Each node represents the total number of horizontally acquired genes in Creolimax found in any of the remaining ichthyosporean transcriptomes/genomes. Phylogenetic relationships among ichthyosporeans and out-groups obtained from (Torruella et al., 2015). (B) Boxplot showing intron number distribution according to LGT phylostratigraphic age. (C) Venn diagram of horizontally acquired genes found in the in silico and the in vivo secretome datasets. LGT, lateral gene transfer.





