Roles of the membrane-reentrant β-hairpin-like loop of RseP protease in selective substrate cleavage
Figures
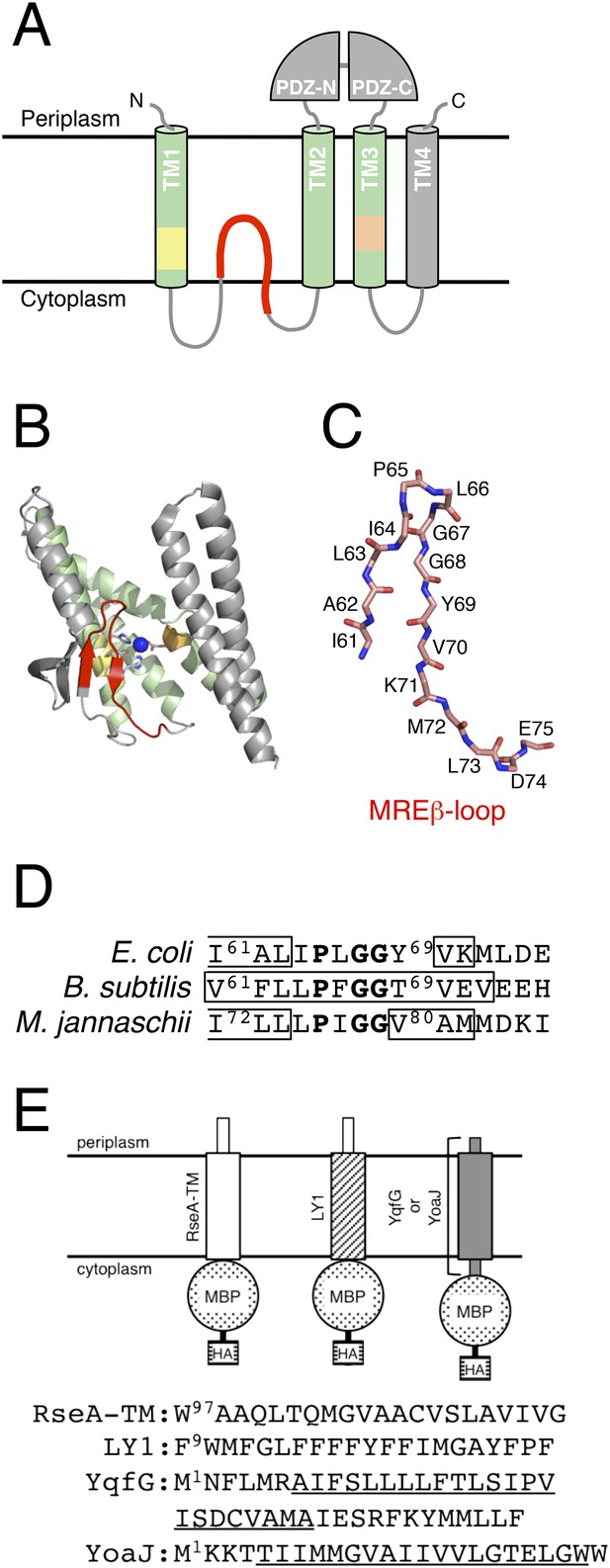
Structures of RseP and the model substrates used in this study.
(A) Schematic representation of RseP. (B) Crystal structure of mjS2P (PDB code: 3B4R A). The secondary structure was analysed using STRIDE (Frishman and Argos, 1995), and the image was generated using PyMOL (https://www.pymol.org) (Schrödinger, 2010). In A and B, the 3 TM helices composing the core membrane domain are shown in light green; the MRE β-loop, HExxH motif and LDG motif are shown in red, yellow and orange, respectively, and the coordinated zinc atom is shown in blue. (C) The structure of the mjS2P MRE β-loop. The polypeptide backbone is extracted from the model in Figure 1B and shown as sticks. C, O and N atoms are shown in pink, red and blue, respectively. The amino acid residues of the corresponding region of RseP are assigned on the model. (D) Sequences of the MRE β-loop of RseP (E. coli), SpoIVFB (B. subtilis) and mjS2P (M. jannaschii). The regions predicted to form β-strands are boxed. The secondary structure is assigned based on the analysis of amino acid sequences by PsiPred (http://bioinf.cs.ucl.ac.uk/psipred) (for RseP and SpoIVFB) or the analysis of the crystal structure by STRIDE (for mjS2P). The sequence alignment and secondary structure prediction suggest that this region of RseP also assumes the β-hairpin-like structure. The conserved PxGG motif is boldfaced. (E) Schematic representation of the model substrates and their amino acid sequences. RseA- and LacY-TM1 (LY1)-derived regions are shown as white and hatched rectangles, respectively. YqfG- or YoaJ-derived region is shown as a gray rectangle. Amino acid sequences of RseA and LY1 TMs and the entire region of YqfG and YoaJ are shown. The possible TMs of YqfG and YoaJ are underlined.
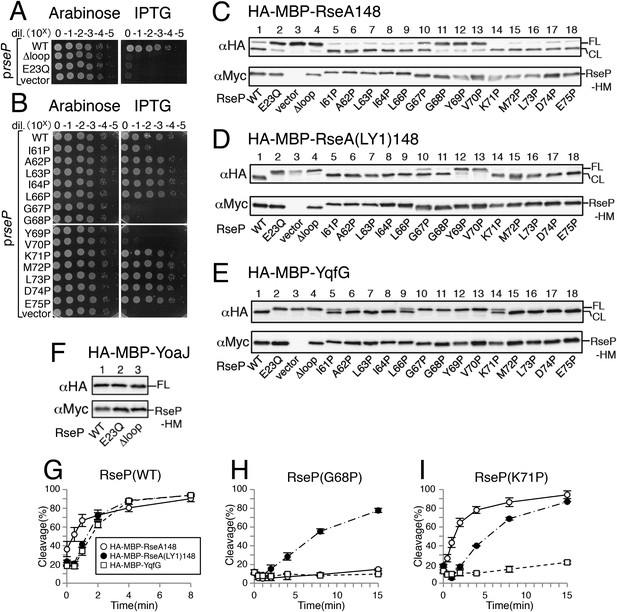
Effects of Pro substitutions in the MRE β-loop on RseP function.
(A and B) Complementation assay. Strain KK31 [rseP::kan/pKK6 (Para-rseP)] harbouring a plasmid encoding the indicated mutant form of RseP-His6-Myc (RseP-HM) under the lac promoter or the corresponding vector was grown in L medium containing 0.02% arabinose. The cultures were serially diluted with saline, and 4 μl of the diluted cultures were spotted on L agar plates containing 0.02% arabinose or 1 mM IPTG. The plates were incubated at 30°C for 20 hr. (C–F) Immunoblotting analysis of substrate cleavage. KA306 (ΔrseA/ΔrseP/ΔclpP) cells harbouring a plasmid encoding the indicated model substrate was transformed with plasmids encoding the indicated mutant forms of RseP-HM and were grown at 30°C in M9-based medium containing 1 mM IPTG and 1 mM cAMP for 3 hr. Proteins were precipitated using TCA, solubilised in 1% SDS and analysed by 10% Laemmli–SDS-PAGE and immunoblotting with anti-HA or anti-Myc antibody. FL and CL indicate full-length and RseP-cleaved forms, respectively, of each model substrate. (G–I) Pulse-chase analysis of substrate cleavage. KA306 cells harbouring an appropriate combination of plasmids encoding the indicated RseP-HM mutant and the model substrate were grown in M9-based medium containing 1 mM IPTG at 30°C. The cells were labelled with [35S]-methionine for 30 s and were chased with unlabelled methionine as indicated. Proteins were immunoprecipitated using agarose-conjugated anti-HA antibody and were analysed by 10% Laemmli–SDS-PAGE. Cleavage (%) was calculated using the following equation: cleavage (%) = 100 × (CL)/[(FL) + (CL)], where FL and CL are the intensities of the respective bands that were corrected for methionine content. Two independent experiments were performed, and mean values are shown along with standard deviations. See Figure 2—source data 1 for gel images and quantitated band intensities data for Figure 2G–I. See Figure 2—figure supplement 1 for integral association of HA-MBP-YqfG and HA-MBP-YoaJ with membrane. See Figure 2—figure supplement 2 for complementation and model substrate cleavage activity of the RseP derivatives with a Cys substitution in the MRE β-loop.
-
Figure 2—source data 1
Zip file containing gel images and quantified band intensity data for the pulse-chase experiments.
- https://doi.org/10.7554/eLife.08928.005
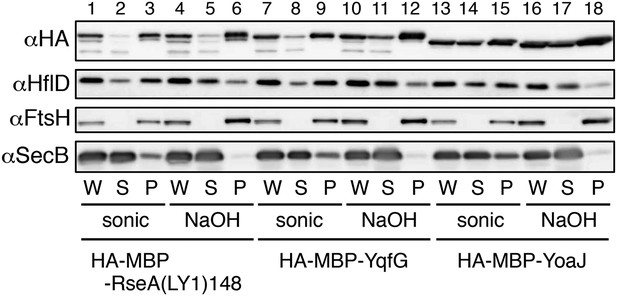
Membrane localization of the model substrates.
Strain KK374/pKH441(hflD) was further transformed with the plasmids encoding the indicated model substrate. Cells were grown in the M9-based medium containing 1 mM IPTG and 1 mM cAMP at 30°C. An aliquot of the cells were sonically disrupted and fractionated into the soluble (S) and membrane (P) fractions (‘sonic’ samples). Another aliquot was treated with 0.1 N NaOH and fractionated into the supernatant (S) and pellet (P) fractions (‘NaOH’ samples). W indicates unfractionated whole cell samples. Proteins were analyzed by 12.5% SDS-PAGE and immunoblotting using anti-HA, anti-HflD, anti-FtsH, and anti-SecB antibodies. HflD, FtsH and SecB are peripheral membrane, integral membrane and cytosolic proteins, respectively, and served as controls for the fractionation. Because the chromosomally-encoded HflD is expressed at a very low level and hard to detect, we expressed HflD from a multicopy plasmid. The majority of the model substrates, HA-MBP-RseA(LY1)148, HA-MBP-YqfG and HA-MBP-YoaJ, was associated with the membrane both after sonical disruption and after alkali extraction, although higher proportions of HA-MBP-YoaJ was recovered in the soluble fractions as compared with the other two model substrates, indicating that most of the SMP-derived model proteins are integrally associated with the membrane.
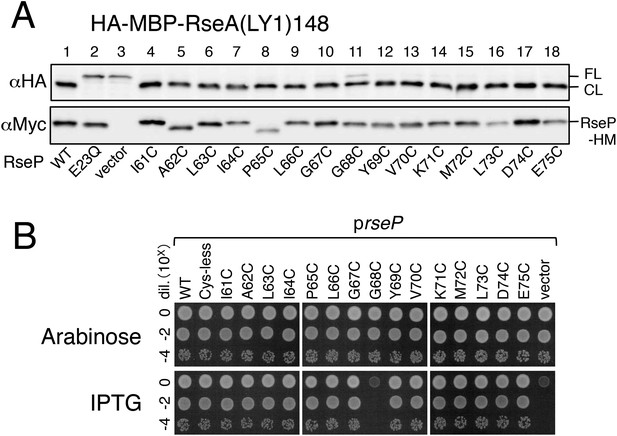
Effects of the MRE β-loop Cys substitutions on the RseP function.
(A) Assay of substrate cleavage by immunoblotting. Strain KA306 (ΔrseA/ΔrseP/ΔclpP) harboring pYH124 (HA-MBP-RseA(LY1)148) was further transformed with a plasmid encoding the indicated single Cys mutant form of RseP-HM. Cells were grown at 30°C in M9-based medium containing 1 mM IPTG and 1 mM cAMP for 3 hr. Proteins were acid-precipitated, solubilized in 1% SDS and analyzed by 10% SDS-PAGE and anti-HA or anti-Myc immunoblotting as indicated. (B) Complementation assay. Strain KK31 (rseP::kan/pKK6 (Para-rseP)) was further transformed with a plasmid encoding the indicated mutant form of RseP-HM under the lac promoter, or the corresponding vector. Cells were grown in L-0.02% arabinose, serially diluted with saline and a 4 µl each of the diluted culture was spotted on an L-0.02% arabinose or L-1 mM IPTG agar plate. The plates were incubated at 30°C for 20 hr.
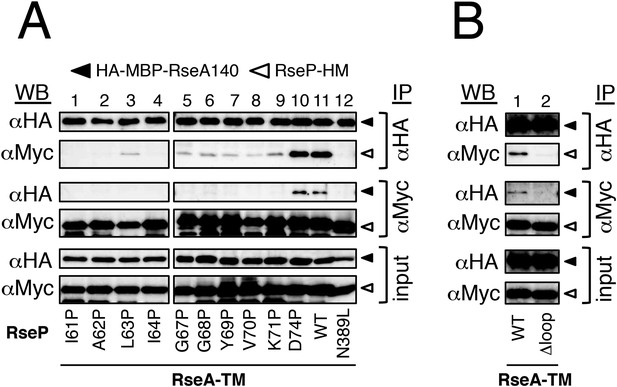
Effects of MRE β-loop mutations on RseP–substrate interaction.
(A and B) Co-immunoprecipitation assays of RseP–substrate interaction. IMVs were prepared from KK211 (ΔrseA/ΔrseP) cells harbouring a plasmid encoding the indicated mutant of RseP-HM and pSTD881 (HA-MBP-RseA140). The IMVs were solubilised with DDM and were subjected to immunoprecipitation with anti-HA or anti-Myc antibody. The immunoprecipitates and the DDM-solubilised proteins (input) were analysed by 12.5% Laemmli–SDS-PAGE and immunoblotting with anti-HA and anti-Myc antibodies. Proteins form approximately fourfold more number of cells were loaded on the gel for the immunoprecipitated samples as compared with the input samples. All the RseP-HM derivatives carried the E23Q mutation. See Figure 3—figure supplement 1 for interaction between RseP-HM and HA-MBP-YoaJ.
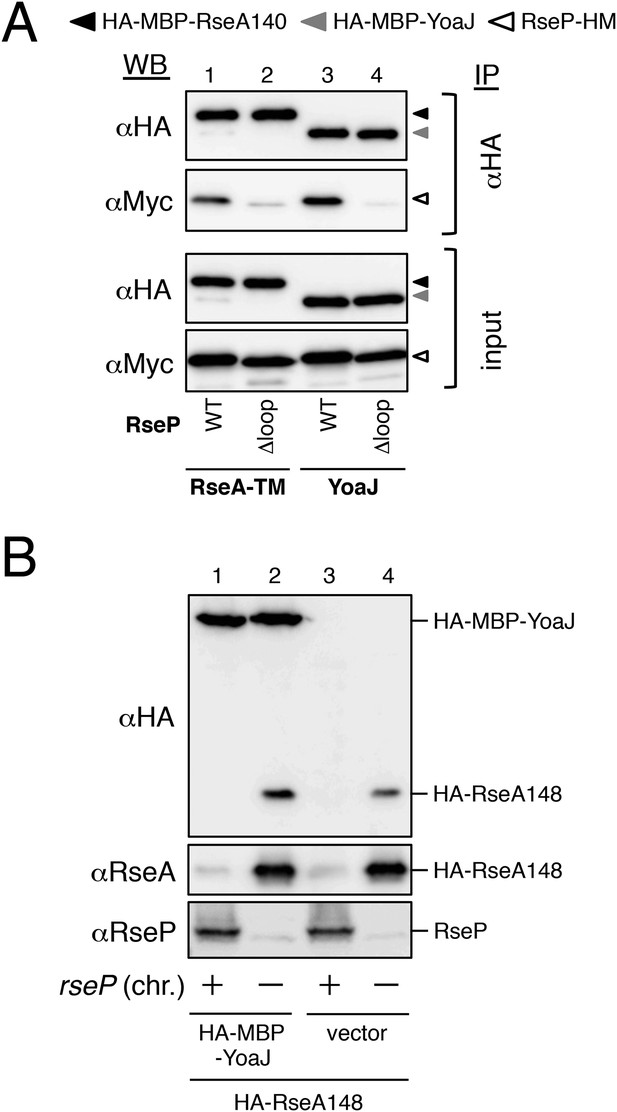
Analysis of RseP–YoaJ interaction.
(A) Co-immunoprecipitation assay of RseP–YoaJ interaction. IMVs were prepared from cells of KK211 (ΔrseA/ΔrseP) carrying a plasmid encoding the indicated mutant form of RseP-HM in addition to pSTD881 (HA-MBP-RseA140) or pKA210 (HA-MBP-YoaJ), solubilized with DDM and were subjected to immunoprecipitation with anti-HA. Immunoprecipitates and the DDM-solubilized proteins (input) were analyzed by 12.5% Laemmli–SDS-PAGE and anti-HA and anti-Myc immunoblotting. For the immunoprecipitated samples, proteins were loaded onto the gel in an amount corresponding to approximately fourfold more cells than were used for the input samples. All the RseP-HM derivatives carried the E23Q mutation. (B) Effect of YoaJ overproduction on cleavage of HA-RseA148. Cells of AD1811 (ΔrseA)/pKA195 (HA-RseA148)/pKA268 (HA-MBP-YoaJ) or KK211 (ΔrseA/ΔrseP)/pKA195/pKA268 were grown at 30°C in M9-based medium containing 1 mM IPTG and 1 mM cAMP for 3 hr. Proteins were acid-precipitated, solubilized in 1% SDS and analyzed by 10% Laemmli–SDS-PAGE and anti-HA, anti-RseA and anti-RseP immunoblotting as indicated.
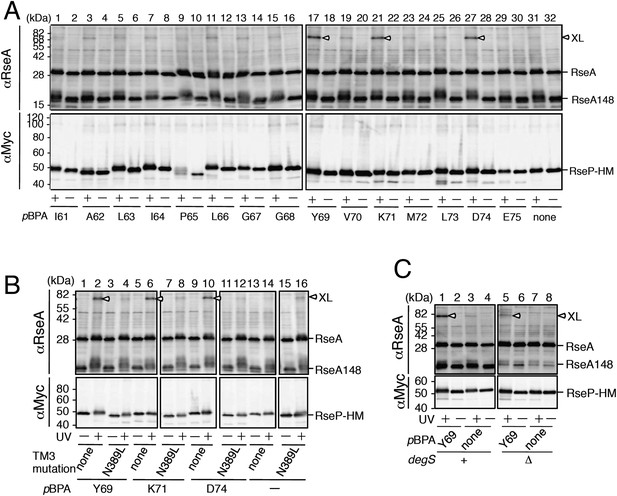
In vivo photo-crosslinking between RseP and RseA.
(A) Analysis of the MRE β-loop/RseA crosslinking. KA418 (ΔompA/ΔompC/ΔrseP)/pEVOL-pBpF cells were transformed with a plasmid encoding an RseP derivative with an amber mutation at the indicated position. The cells were grown at 30°C in M9-based medium containing 0.02% arabinose and 1 mM pBPA for 6 hr and were irradiated with UV light for 0 or 10 min. Proteins were precipitated using TCA and were analysed by SDS-PAGE with a 10% wide-range gel (Nacalai Tesque, Inc. Kyoto, Japan) containing 1% SDS and immunoblotting with anti-RseA antibody or by SDS-PAGE with a 10% Laemmli gel and immunoblotting with anti-Myc antibody. XL indicates crosslinked products. (B) Effect of the N389L mutation on photo-crosslinking. KA418/pEVOL-pBpF strain was transformed with a plasmid encoding an RseP derivative with an amber mutation at the indicated position with or without the N389L mutation. The cells were subjected to photo-crosslinking analysis, as indicated in A. (C) Effect of periplasmic truncation of RseA on photo-crosslinking. KA418/pEVOL-pBpF or KA438 (KA418, ΔdegS)/pEVOL-pBpF cells were transformed with a plasmid encoding an RseP derivative with an amber mutation at the indicated position. The cells were subjected to photo-crosslinking analysis, as indicated in A. All the RseP-HM derivatives in A–C carried the E23Q mutation. See Figure 4—figure supplement 1 for proteolytic activity of the pBPA-containing RseP derivatives and verification of RseP–RseA photo-crosslinking.
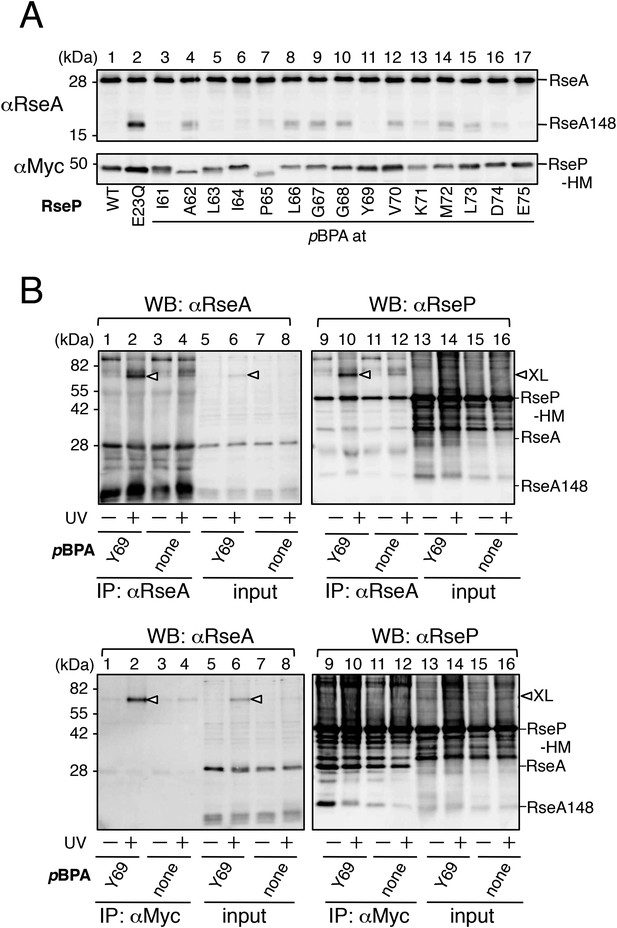
Functionality of the pBPA-containing RseP derivatives and characterization of the photo-crosslinked product.
(A) Proteolytic activities of the RseP-HM derivatives having pBPA in the MRE β-loop. Strain KA418 (ΔrseP/ΔompA/ΔompC)/pEVOL-pBpF was further transformed with a plasmid encoding an RseP-derivative with an amber mutation at the indicated positions. Cells were grown at 30°C in the M9-based medium containing 0.02% arabinose and 1 mM pBPA for 6 hr. Proteins were acid-precipitated, solubilized in SDS sample buffer and analyzed by 10% SDS-PAGE and anti-RseA or anti-Myc immunoblotting as indicated. (B) Identification of the photo-crosslinked proteins by immunoprecipitation. The UV-irradiated, acid-precipitated proteins from cells of KA429 or KA436 carrying a plasmid encoding the indicated RseP-HM derivatives in addition to pEVOL-pBpF were solubilized in 1% SDS and subjected to immunoprecipitation with agarose-conjugated anti-RseA (upper panels) or anti-Myc (lower panels) antibodies. Immunocomplexes were dissolved in SDS-sample buffer and analyzed by 10% SDS-PAGE and anti-RseA or anti-Myc immunoblotting as indicated. For the immunoprecipitated samples, proteins were loaded onto the gel in an amount corresponding to approximately 70-fold more cells than was used for the input samples.
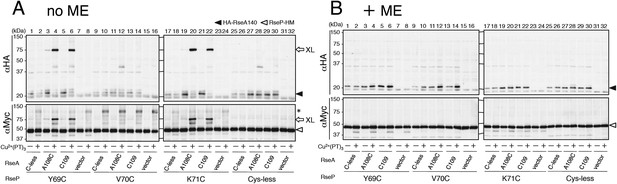
Disulfide crosslinking of RseP and RseA.
AD1840 (ΔrseA/ΔrseP/ΔdegS) cells harbouring a combination of plasmids encoding Cys-less or the indicated single Cys derivative of RseP-HM and HA-RseA140 were treated with Cu2+(phenanthroline (PT))3 at 37°C for 5 min. After quenching the oxidant, proteins were precipitated using TCA and free thiol groups were blocked by treatment with N-ethylmaleimide (NEM). The samples were analysed by 10% Laemmli–SDS-PAGE and immunoblotting with anti-HA (upper panels) or anti-Myc (lower panels) antibody, followed by treatment without (A) or with (B) 10% 2-mercaptoethanol (ME). All the RseP-HM derivatives carried the E23Q mutation. Asterisk indicates a possible dimer of RseP-HM (Koide et al., 2008) that was not characterised further.
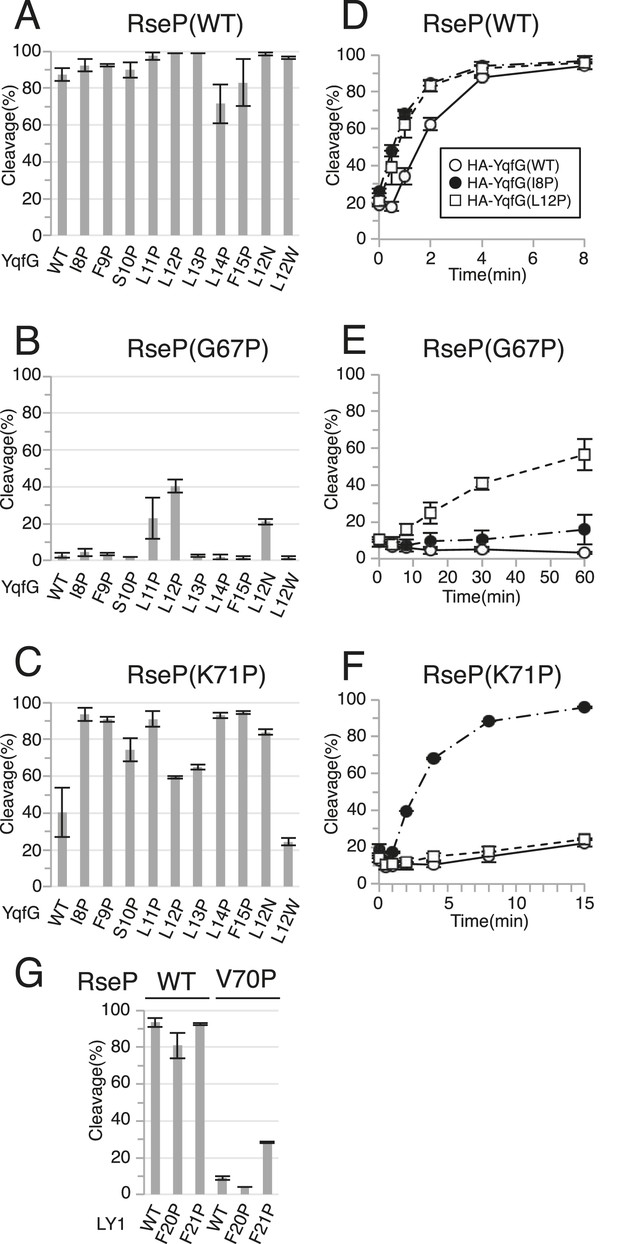
Suppression of the effect of RseP MRE β-loop mutations by destabilising substrate TM helix.
(A–C) Immunoblotting analysis of YqfG cleavage. KA306 cells carrying a combination of plasmids encoding the indicated RseP-HM mutants and HA-MBP-YqfG mutants were grown and were subjected to immunoblotting analysis, as described in Figure 2. (D–F) Pulse-chase analysis of YqfG cleavage. KA306 cells harbouring a combination of plasmids encoding the indicated RseP-HM mutants and HA-MBP-YqfG mutants were grown and subjected to pulse-chase analysis, as described in Figure 2. Two independent experiments were performed, and mean values are shown along with standard deviations. See Figure 6—source data 1 for gel images and quantitated band intensities data for Figure 6D–F. (G) Immunoblotting analysis of LY1 cleavage. KA306 cells carrying a combination of plasmids encoding the indicated RseP-HM mutants and HA-MBP-RseA(LY1)148 mutants were grown and were subjected to immunoblotting analysis, as above.
-
Figure 6—source data 1
Zip file containing gel images and quantified band intensity data for the pulse-chase experiments.
- https://doi.org/10.7554/eLife.08928.014
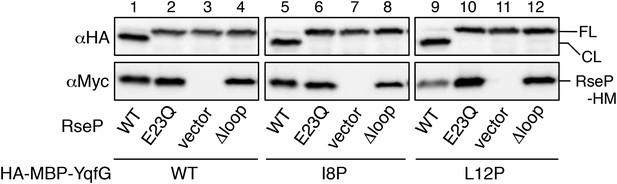
Cleavage of the I8P and L12P mutants of HA-MBP-YqfG depends on the MRE β-loop.
KA306 cells harbouring a combination of plasmids encoding the indicated RseP-HM mutants and HA-MBP-YqfG mutants were grown and total proteins were subjected to immunoblotting analysis, as described in Figure 2.

A model describing the function of the MRE β-loop.
(A) The periplasmic PDZ tandem of RseP acts as a size-exclusion filter to discriminate between substrates based on the size of their periplasmic region. (B) The binding of the MRE β-loop serves as an additional checkpoint for substrates with a small periplasmic region and contributes to selective substrate cleavage. (C) The MRE β-loop binds to substrate TMs with some specificity because of β-strand addition, stabilises its extended conformation and presents the substrate to the recessed proteolytic active site of RseP.
Additional files
-
Supplementary file 1
Table S1. Strains used in this study.
- https://doi.org/10.7554/eLife.08928.017
-
Supplementary file 2
Table S2. Plasmids used in this study.
- https://doi.org/10.7554/eLife.08928.018





