A vocabulary of ancient peptides at the origin of folded proteins
Figures
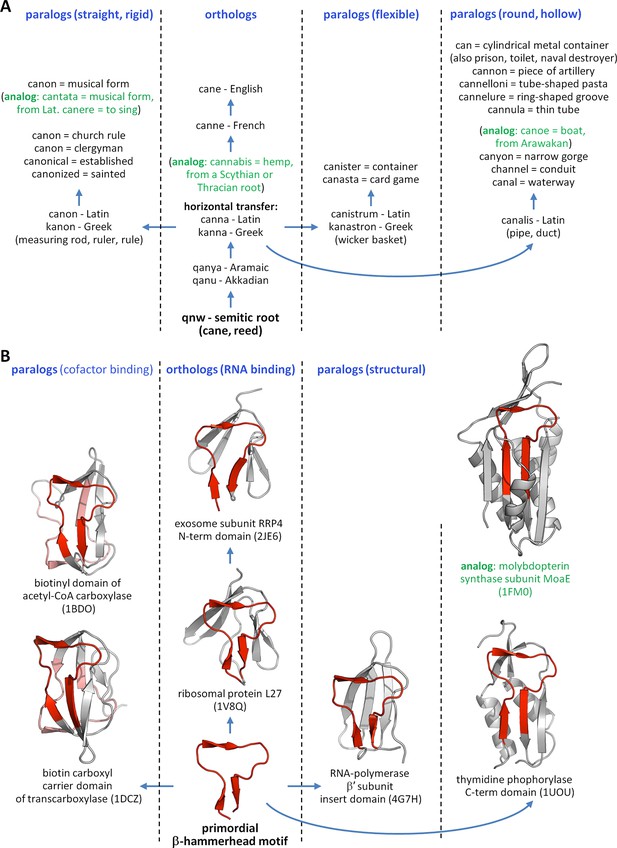
The evolution of words and proteins shows many parallels.
(A) The Semitic root qnw (*qanaw-), meaning reed, is the ancestor of hundreds of words in many different languages, following the same mechanisms as already known from biological evolution. Here we track the descendants of this root in the English language, arisen through the intermediary of Latin and Greek. In addition to the orthologous Greek word kanna (reed), paralogous cognates arose in antiquity based on certain attributes of reed, e.g., the levelling rule kanon (taking the straight and rigid attribute of reed), the wicker basket kanastron (flexible), and the Latin water duct canalis (round and hollow). A few examples of analogous words, which appear to be related to the descendants of qnw but have different evolutionary origins, are shown in green. (B) The primordial β-hammerhead motif (shown in red) is seen in four different folds, which cover a wide array of functions. Following our hypothesis of an origin in the RNA world, we propose that RNA binding is the orthologous function of this peptide, seen today in ribosomal protein L27 and exosome subunit RRP4. Paralogous functions arose around the time of the Last Universal Common Ancestor from its ability to form a biotin-binding domain by duplication, yielding the biotin-dependant enzymes of the barrel-sandwich hybrid fold, and to serve as a structural element in domains formed by accretion, yielding a domain of RNA-polymerase β’ subunit, as well as a range of enzymes with an α/β-hammerhead fold. By our analysis, enzymes classified in the α/β-hammerhead fold superfamily d.41.5, such as MoaE, are analogous to the other superfamilies in this fold, due to a lack of detectable sequence similarity, but nevertheless contain a supersecondary structure resembling the β-hammerhead.
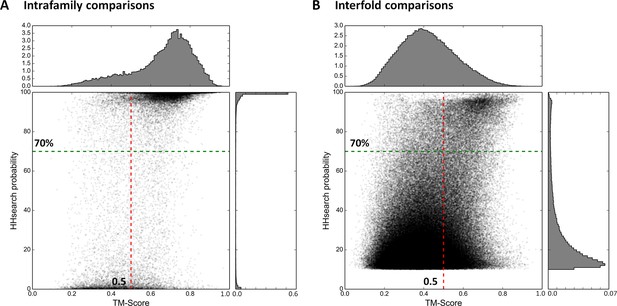
Estimation of cut-offs for HHsearch probability and TM-score.
We compared all domains in the SCOPe30 set in sequence space using HHsearch and subsequently in structure space using TM-align (see ‘Materials and methods’), and plotted the obtained scores. Separate plots for comparisons of domains within families (A) and between folds (B) were generated. Scores would have been expected at high HHsearch probabilities and TM-scores for intrafamily comparisons (presumed homologs, Panel A) and low HHsearch probabilities and TM-scores for interfold comparisons (presumed analogs, Panel B), but the score distributions were in fact bimodal, as also illustrated by the histograms top and right in each panel, which are plotted as probability density functions. In the comparison of domains of different fold, matches with an HHsearch probability of < 10% are not plotted.
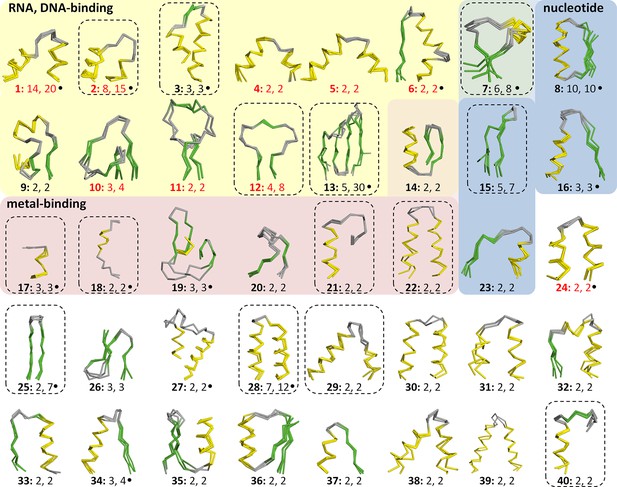
Vocabulary of primordial peptides that gave rise to folded proteins.
The 40 peptides we detected are shown as ensembles in backbone representation; α-helices are coloured in yellow, β-strands in green, and loops in gray. Detailed information on each fragment is provided in Table 1 and Figure 3—source data 1. The fragments are numbered sequentially and their occurrence in different folds and superfamilies of SCOPe is given. Fragments reported individually before are indicated by a dot. Nucleic-acid binding, nucleotide-binding, and metal-binding motifs are highlighted in yellow, blue, and red, respectively. Fragments found in ribosomal proteins are indicated by red font colour. Fragments that form folds by repetition are boxed.
-
Figure 3—source data 1
Multiple sequence alignments and accession details for the 40 primordial fragments shown in Figure 3 and the 5 B-set fragments shown in Figure 3—figure supplement 1.
Multiple homologous copies of a fragment found within the same domain are indicated by a *. Fragments reported individually before are indicated by a +.
- https://doi.org/10.7554/eLife.09410.006
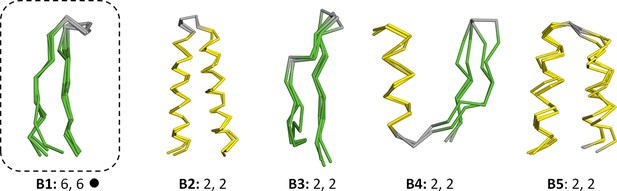
Vocabulary of primordial peptides (B-set).
In addition to the 40 fragments described in the main text, we detected five further fragments after relaxing the sequence similarity requirement to an HHsearch probability of 60%. One of these fragments, the Asp box (B1), has been previously described and it forms folds by repetition (emphasized by dotted boxes). Detailed information is provided in Figure 3—source data 1.
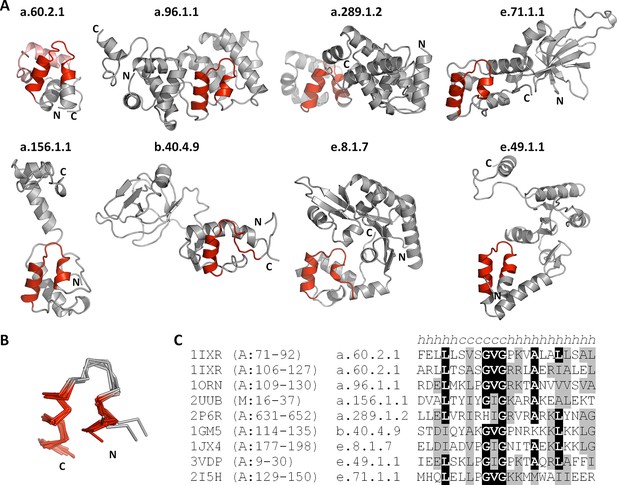
The nucleic-acid binding helix-hairpin-helix motif is found in 8 different folds comprising 15 superfamilies.
(A) Representative domains from the eight SCOPe folds. The motif is coloured in red and the remainder of the structure is shown in gray. The SCOPe family a.60.2.1 contains two copies of this motif, whereas the remaining folds contain one copy each. (B) Structural superimposition of the helix-hairpin-helix motifs displayed in panel A. (C) Sequence alignment of the motifs shown in panel A. Residues conserved in at least half of the aligned sequences are highlighted in black and similar residues are highlighted in gray.
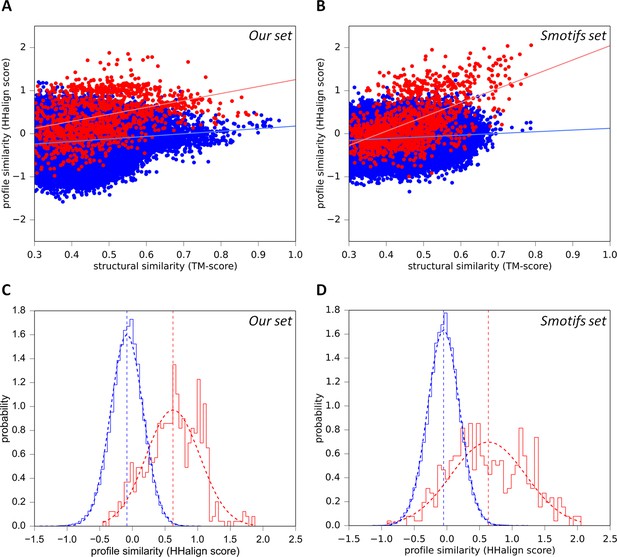
Sequence similarity of our fragments cannot be explained by structural constraints.
(A) For each occurrence of any of our 40 fragments, we searched for structural matches in SCOPe30 and plotted the TM-align score versus the profile-similarity score for the fixed alignment given by TM-align. The putatively homologous matches to occurrences of the same fragment in another superfamily are shown in red. Matches to fragments outside the list of folds in which the query fragment was found to occur (i.e. non-homologous matches) are blue. (B) Same as A, but using the Smotifs reference fragments as queries instead of our set. Matches within superfamilies (homologs) are shown in red, matches between fragments from different folds (analogs) are shown in blue. For both sets, sequence and structure similarity scores are significantly correlated for presumably homologous matches (our set: r=0.38; Smotifs: r=0.56, see linear regression lines) but not for analogous matches (our set: r=0.14; Smotifs: r=0.12). (C, D) Distribution of profile similarity scores for matches with a TM-score ≥ 0.5, for the homologous and analogous distribution in the plots (A) and (B), respectively. The means of the Gaussian fits are exactly the same in C and D.
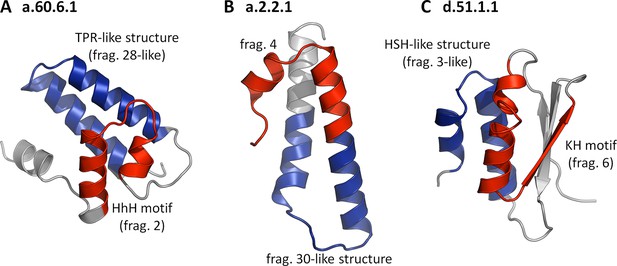
Folds showing two nonidentical fragments, one of which is which is not significant by our criteria.
No SCOP fold combines two of our fragments at the cutoffs used in this study (TM-score ≥ 0.5 and HHsearch probability ≥ 70%). If we however omit the sequence cutoff entirely for the second fragment, combinations become apparent. In these three examples, the 'significant' fragments are colored in red, the 'nonsignificant' ones in blue, and the remainder in gray. The structures are: (A) a.60.6.1, N-terminal domain of polymerase β (4KLI, A: 10-91), (B) a.2.2.1, 50S ribosomal protein L29 (1VQ8, V: 1-65), and (C) d.51.1.1, KH domain-like hypothetical protein APE0754 (1TUA, chain A: 1-84).

Amplification and accretion are key forces in the emergence of domains.
Of the 40 fragments in our set, 14 form folds by repetition. The fragments are coloured in red in the shown structures. (A) The TPR element (Figure 3: 28) occurs repetitively in the TPR-like superfamily (a.118.8; 1ELW, shown on the left side) and singly in six other folds (e.g., a.7.16, 2CRB, right). (B) Outer membrane β-barrels comprise 4–12 homologous copies of a β-hairpin element (Figure 3: 25); examples include the eight-stranded OmpA (1QJP, left) and the twelve-stranded NalP (1UYN, right). The entire barrels are formed by repetition, but the strands of the hairpin split by the N- and C- termini are left gray. (C) The transcription factor AbrB (1YFB, left) is a homodimer and contains one copy of the β-α-β motif (Figure 3: 7) per subunit. MraZ has internal sequence symmetry and contains two homologous copies of the β-α-β motif (1N0E, right).
Tables
Data on the 40 primordial fragments.
| Fragment | Number of folds, superfamilies | Repetition in SCOPe foldor superfamily | Ribosomal (SCOPe id; protein) | Ligands | 9 most ancient and basal folds (SCOPe id) | Occurrence in the 10 folds with the largest number of superfamilies in SCOPe |
|---|---|---|---|---|---|---|
| 1+ | 14, 20 | a.4.5; S19e | DNA | a.4 | ||
| 2+ | 8, 15 | a.60.2 | a.156.1; S13 | DNA, RNA | a.60 | |
| 3+ | 3, 3 | a.174.1 | DNA | c.37.1.20 | ||
| 4 | 2, 2 | a.2.2; L29 | RNA | a.2 | ||
| 5 | 2, 2 | d.14.1; S5 | RNA | |||
| 6+ | 2, 2 | d.52.3; S3 | DNA, RNA | |||
| 7+ | 6, 8 | all folds containing it | DNA, CTP, SAM, FMN | |||
| 8+ | 10, 10 | FAD, NAD, NEA, NAJ, NAP, NAI, NDP, AMP, NMN, APR, LNC, A3D, ODP, UMA, SAH, SAM, COA, | c.2, c.66 | |||
| 9 | 2, 2 | DNA | ||||
| 10 | 3, 4 | d.66.1; S4 | RNA | |||
| 11 | 2, 2 | b.53.1; TL5 | RNA | |||
| 12 | 4, 8 | b.84.1 | b.84.4; L27 | RNA | ||
| 13+ | 5, 30 | all folds containing it | DNA | |||
| 14 | 2, 2 | ZN, DNA | ||||
| 15 | 5, 7 | d.37.1 | FMN, AMP, FAD | |||
| 16+ | 3, 3 | ATP, GTP, ADP, CTD, DGP, DCP, ANP, TMP, GDP | c.37 | |||
| 17+ | 3, 3 | a.138.1 | HEM, HEC | a.24 | ||
| 18+ | 2, 2 | a.1.2, d.58.1 | SF4(Fe4S4) | d.58.1 | d.58 | |
| 19+ | 3, 3 | ZN | ||||
| 20 | 2, 2 | FES | ||||
| 21 | 2, 2 | a.39.1, a.139.1 | CA | |||
| 22 | 2, 2 | a.25.1 | FEC, FE, FE2, HEM | |||
| 23 | 2, 2 | COA | ||||
| 24+ | 2, 2 | d.45.1; L7/12 | ||||
| 25+ | 2, 7 | f.4 | ||||
| 26 | 3, 3 | g.3 | ||||
| 27+ | 2, 2 | |||||
| 28+ | 7, 12 | a.118.8 | a.24, a.118 | |||
| 29 | 2, 2 | b.34.4 | b.34 | |||
| 30 | 2, 2 | a.2 | ||||
| 31 | 2, 2 | |||||
| 32 | 2, 2 | c.55.1 | ||||
| 33 | 2, 2 | |||||
| 34+ | 3, 4 | |||||
| 35 | 2, 2 | d.58.49 | d.58 | |||
| 36 | 2, 2 | c.55.5 | ||||
| 37 | 2, 2 | |||||
| 38 | 2, 2 | a.60 | ||||
| 39 | 2, 2 | a.118 | ||||
| 40 | 2, 2 | a.20.1 |
-
+previously reported fragments





