Self-establishing communities enable cooperative metabolite exchange in a eukaryote
Figures
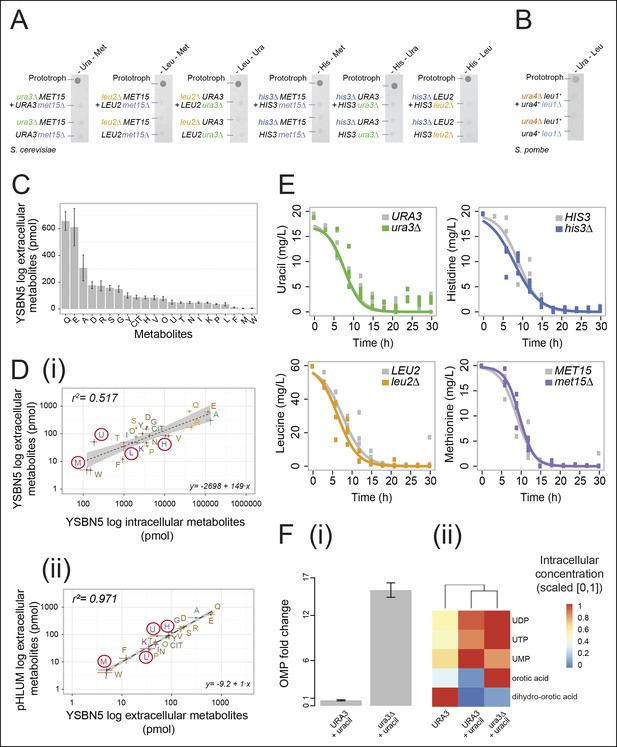
Yeast auxotrophs do not compensate for metabolic deficiencies upon co-culturing, yet export the relevant metabolites and prefer metabolite uptake over self-synthesis.
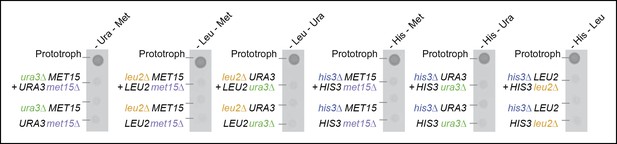
(A) Complementary pairs of Saccharomyces cerevisiae auxotrophs do not overcome metabolic deficiencies upon co-culturing. his3∆, leu2∆, met15∆,and ura3∆ yeasts were combined in complementary pairs and spotted on corresponding selective media. No pairs exhibited co-growth together. (B) A complementary pair of Schizosaccharomyces pombe auxotrophs does not overcome metabolic deficiencies upon co-culturing. leu1Δ and ura4Δ yeasts were combined in a complementary pair and spotted on corresponding selective media. No co-growth occurred. (C) The concentration of metabolites in the S. cerevisiae colony exometabolome obtained from 1.3e08 YSBN5 cells grown in a colony on synthetic minimal agar media (SM) and quantified by LC-MS/MS. Abbreviations: single letter IUPAC amino acid codes, O = ornithine, CIT = citrulline. n = 3, error bars = ± SD. (D) (i) Metabolites quantified as in (C), comparing intracellular (total cell extracts) and extracellular metabolite concentrations in YSBN5. n = 3, error bars = ± SD. Dashed line: linear regression fit, grey band shows 95% confidence region. (ii) Metabolites quantified as in (C), comparing extracellular metabolite concentrations of YSBN5 and BY4741-pHLUM yeast colonies grown on minimal media. H, L, U, and M are highlighted in red circles. n = 3, error bars = ± SD. Dashed line: linear regression fit, grey band shows 95% confidence region. Abbreviations IUPAC codes; H = histidine, L = leucine, U = uracil, M = methionine. (E) Consumption of uracil, histidine, leucine, and methionine in yeast batch cultures in synthetic complete (SC) media as measured by LC-MS/MS. Uracil, histidine, leucine, and methionine prototrophic cells consume these metabolites at rates and quantities comparable to the corresponding auxotrophic strains. (F) (i) Deletion of URA3 (Orotidine-5'-phosphate decarboxylase) causes accumulation of the Ura3p substrate orotidine-5'-phosphate (OMP), when cells are supplemented with 20 mg/L uracil (fold change of OMP abundance, relative to URA3 without uracil supplementation), as determined by LC-MS/MS. Error bars = ± SD. (ii) Uracil supplementation of wild-type cells alters their metabolite profile to resemble ura3∆ cells, which obtain uracil solely from the growth media. Heatmap scaling ([0,1] and min, max per metabolite) was based on median concentration. The dendrogram was constructed by comparing euclidean distance (dissimilarity) between samples.
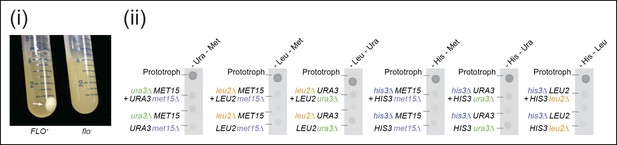
Flocculation does not enable Saccharomyces cerevisiae cells to establish viable co-cultures.
(i) (left) The FLO+ phenotype in yeast cells transforms their typical cell suspension (right) into a physiological state reminiscent of biofilms (left) (Smukalla et al., 2008). Cultures were grown to stationary phase in rich media (YPD) and flocculation was detected via an inability of cells to re-suspend following repeated tube inversion. (ii) Complementary pairs of flocculating S. cerevisiae auxotrophs do not overcome metabolic deficiencies upon co-culturing. his3∆, leu2∆, met15∆, and ura3∆ yeasts were combined in complementary pairs and spotted on corresponding selective media. No pairs exhibited co-growth together.
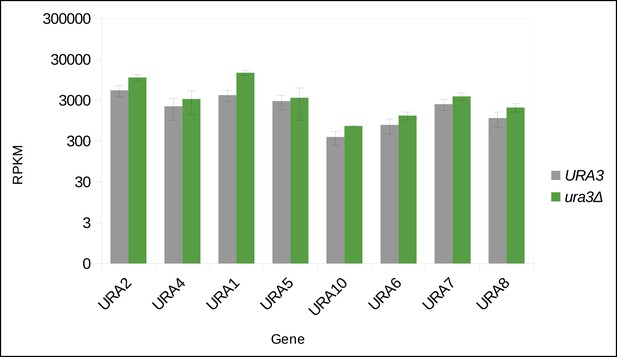
Uracil biosynthetic genes in the uracil prototroph (URA3) and auxotroph (ura3Δ) remain expressed in (uracil supplemented) SC media, as determined by RNA sequencing.
Abbreviations: RPKM = reads per kilobase per million. n = 3, error bars = ± SD.
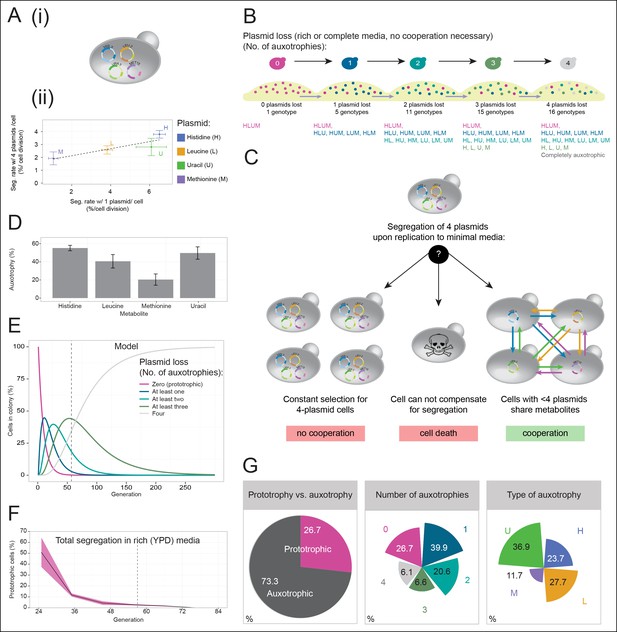
A self-establishing yeast community can cooperatively compensate for progressive loss of prototrophy on minimal media.
(A) (i) Schematic illustration of BY4741 carrying four plasmids to complement its auxotrophies in histidine (his3Δ1), leucine (leu2Δ0), methionine (met15Δ0), and uracil (ura3Δ0). (ii) Plasmid segregation rates (probability of plasmid loss per cell division) of BY4741 carrying four plasmids encoding HIS3 (p423), LEU2 (pRS425), URA3 (p426), and MET15 (pRS411) (y-axis) compared to BY4741 carrying one plasmid at a time (x-axis). n = 3, error bars = ± SD. Dashed line: linear regression fit. (B) Schematic illustration of the segregate strain composition over time on rich or complete media where no cooperation is necessary for cells to survive. Sequential plasmid loss leads to an increase in auxotrophy, with loss of up to four plasmids leading to the formation of 16 cell types with varying metabolic capacity (metabotypes). (C) Three possible outcomes for BY4741 carrying four segregating plasmids, when establishing a colony on minimal media; (i) no cooperation, only cells carrying four plasmids grow, (ii) no cooperation but plasmid segregation is faster than the growth rate of cells carrying four plasmids leading to no growth capacity. Finally (iii), cells cooperate, wherein cells that have obtained auxotrophy continue growth by sharing metabolites with neighbouring cells in the colony. (D) Auxotrophy of BY4741 colonies carrying single plasmids encoding HIS3 (p423), LEU2 (pRS425), URA3 (p426), and MET15 (pRS411) on selective media after approximately 33 doublings. The number of plasmid-free cells (% auxotrophy abundance) was measured by replica plating. n = 3, error bars = ± SD. (E) Mathematical simulation of segregation over time, starting from 100% cells carrying four plasmids, based on the experimentally measured segregation rates. Highlighted is the situation after 57 doublings (achieved in dashed line) where >99.9% of cells have segregated >1 plasmid. (F) Segregation over time in a colony on rich media (no selection to maintain the plasmids); starting from a micro-colony of four-plasmid prototrophic cells on minimal media, cells were transferred to rich (YPD) media and established as a giant colony, segregation was followed by replica plating. Biomass gain is counted from the single cell. (G) Giant colonies established for 57 biomass doublings on minimal media are composed of (left) 73.3% auxotrophic cells, (centre) contain a mixed number of auxotrophies and (right) a non 1:1 ratio of auxotrophy types. (n = 542 genotyped cells). Colony growth is achieved, despite the majority of cells possessing one or more auxotrophies.
-
Figure 2—source data 1
Plasmid segregation rates.
- https://doi.org/10.7554/eLife.09943.007
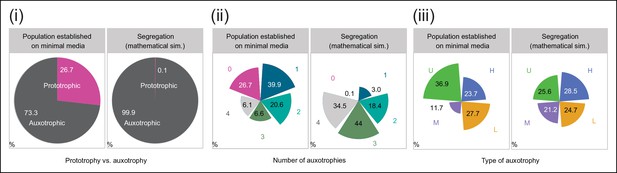
Experimentally obtained colony composition, compared to the composition expected if segregation continued without selective pressure to maintain cells able to synthesise leucine, uracil, methionine and histidine.
(i) (left) Colonies established for 57 biomass doublings on minimal media (SM) are composed of 73.3% auxotrophic cells (n = 542 genotyped cells). (right) Uninterrupted segregation would lead to 99.9% auxotrophic cells. (ii) (left) Auxotrophy number (0 to 4) for cells within the synthetic metabolically cooperating colony (SeMeCo). (right) Result when there is uninterrupted segregation (theoretical composition). (iii) (left) Composition of SeMeCo in terms of auxotrophy type (histidine, leucine, uracil and methionine). (right) Result when there is uninterrupted segregation (theoretical composition).
Schizosaccharomyces pombe, like Saccharomyces cerevisiae, are also able to establish SeMeCo colonies.
The four possible metabotypes resulting from a combination of uracil and leucine auxotrophies are found within S. pombe SeMeCos, despite growing colonies on selective media (n = 3). Separately established populations (>90 cells) were genotyped per replicate.
Complementary pairs of auxotrophs, re-isolated from established SeMeCo colonies, do not overcome metabolic deficiencies upon co-culturing, similar to the original strains .
his3∆, leu2∆, met15∆, and ura3∆ yeasts isolated from SeMeCo colonies were combined in complementary pairs and spotted on corresponding selective media. No pairs exhibited co-growth together, indicating that SeMeCo metabotypes did not acquire secondary mutations to overcome metabolic deficiencies, while establishing a cooperating community.
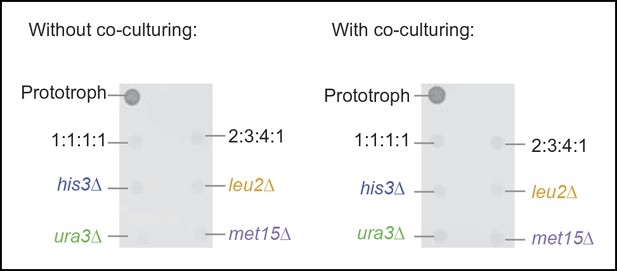
. Different auxotrophy combinations do not enable metabolic cooperation.
(left) Quadruple mixed cultures of Saccharomyces cerevisiae auxotrophs do not overcome metabolic deficiencies upon co-culturing. his3Δ, leu2Δ, met15Δ,and ura3Δ yeasts were combined together in mixed ratios and spotted as a co-culture on corresponding selective media. (right) A similar outcome upon co-culturing the four genotypes over night in rich media prior to transferring to minimal media.
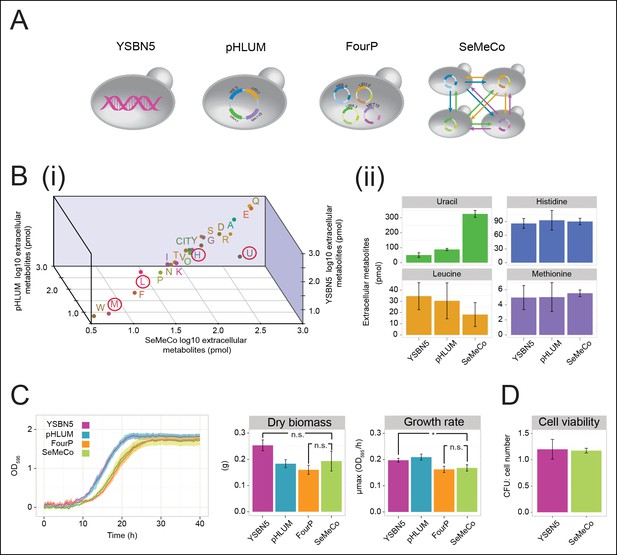
Growth and physiological parameters of the self-established metabolically cooperating yeast community 'SeMeCo'.
(A) Schematic illustration of colonies derived from the genomically prototrophic yeast strain YSBN5, the single-vector complemented BY4741-pHLUM ('pHLUM'), BY4741 complemented with four plasmids ('FourP'), and the self established yeast population (SeMeCo; self-established metabolically cooperating yeast community); (from left to right). (B) (i) Extracellular concentrations of metabolites in colonies of YSBN5, pHLUM and SeMeCo growing exponentially on minimal media as determined by LC-MS/MS, n = 3. Histidine (H), leucine (L), methionine (M), and uracil (U) are highlighted in red circles. (ii) Detailed extracellular concentration values of uracil, leucine, methionine, and histidine as determined by LC-MS/MS. n = 3, error bars = ± SD. (C) (left) Growth curve of YSBN5, pHLUM, FourP, and SeMeCo as determined by measuring optical density (OD595). n = 3, error area = ± SD. (centre) Dry biomass collected from 100 mL batch cultures after three days growth in minimal media, 30°C, n = 3, error bars = ± SD. (right) Maximum specific growth rate (µmax) as determined from OD595 growth curves using a model-richards fit (Kahm et al., 2010). n = 3, error bars = ± SD. (D) The ratio of colony-forming units (CFUs) to number of cells used for plating, for YSBN5 and SeMeCo. n = 3, error bars = ± SD.
-
Figure 3—source data 1
Absolute quantification of amino acids and uracil in yeast strains YSBN5, pHLUM and SeMeCo, absolute concentration values.
- https://doi.org/10.7554/eLife.09943.013
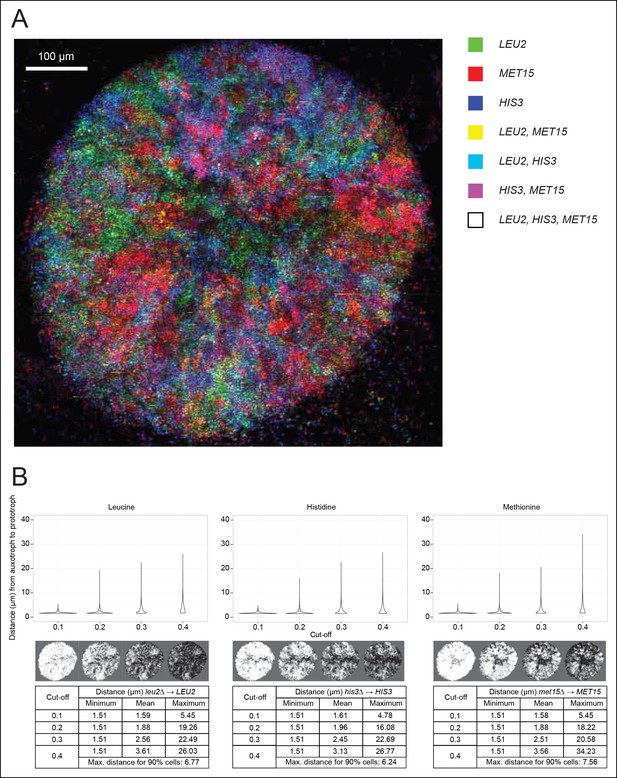
Spatial organisation of SeMeCo.
(A) Spatial organisation of metabolically cooperating yeast micro-colony on minimal agar media (SM). SeMeCo was established with plasmids expressing fluorescent protein coupled to each auxotrophic marker; LEU2, MET15, and HIS3 cells are coloured green, red, and blue, respectively. Cells containing more than one marker are coloured as a product of the additive RGB colour model. Two–day-old live and growing micro-colony is visualised from underneath. (B) Minimum, mean and maximum distances between leucine, histidine, and methionine auxotrophs and their corresponding prototrophs in a SeMeCo colony. Maximum distance between auxotroph and prototroph for 90% of cells shows an average distance of 6.86 μm, using the highest cut-off. Despite the heterogeneous macroscopic colony composition, complementary auxotrophs are maintained in physical proximity to each other.
-
Figure 4—source data 1
Segregation rates of fluorescent protein plasmids from the yEp, pRS and p400 series.
- https://doi.org/10.7554/eLife.09943.015
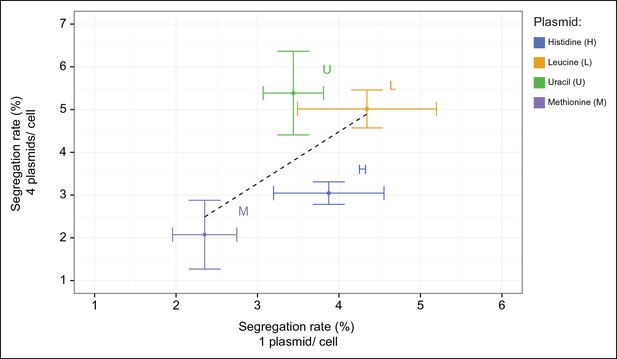
Plasmid segregation rates of fluorescent protein plasmids (%; probability of plasmid loss per cell division) of BY4741 carrying plasmids encoding HIS3 (yEpCFP_HIS), LEU2 (yEpSapphire_LEU), URA3 (yEpVenus_URA), and MET15 (pRS411-GPDpr-mCherry) respectively, compared to BY4741 carrying all four at the same time. n = 3, error bars = ± SD.
https://doi.org/10.7554/eLife.09943.016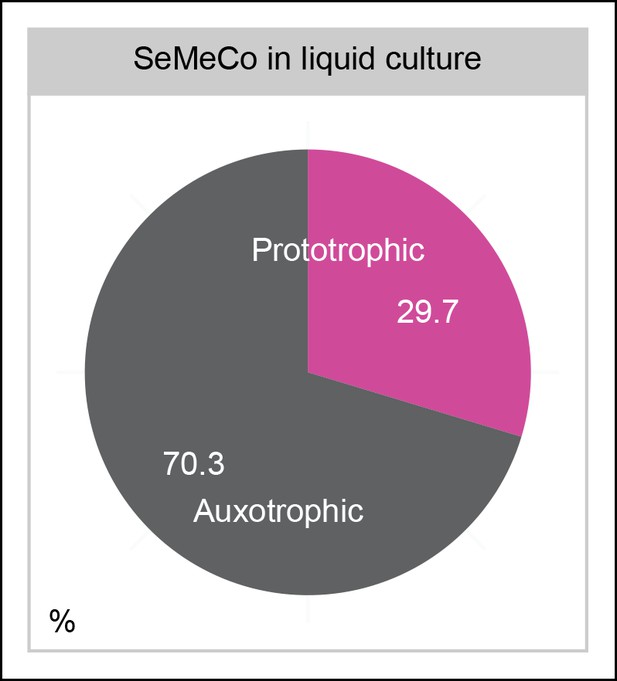
SeMeCos continue growth in minimal (SM) liquid culture.
As in the colony growth experiment, cells were transferred from a micro-colony on SM agar, and then grown for 7 days with re-diluting every 2 days, however, here cells were in shaking batch liquid culture (25 mL). Auxotrophy abundance is 70.3 ± 2.5% despite cells growing in minimal media.
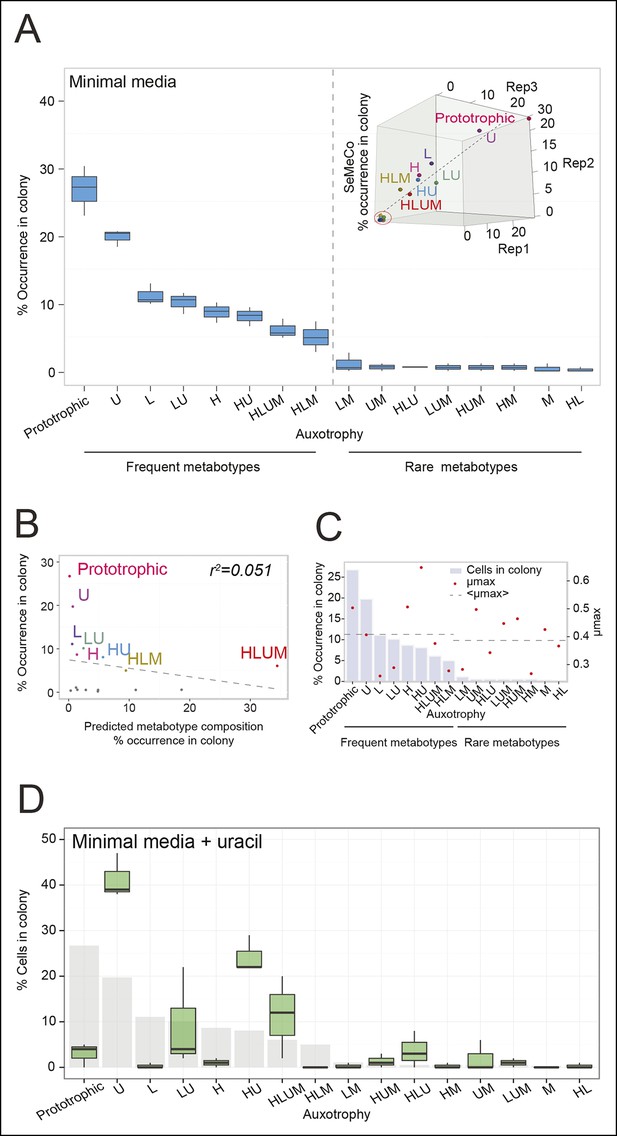
The community composition is distinct and dynamic.
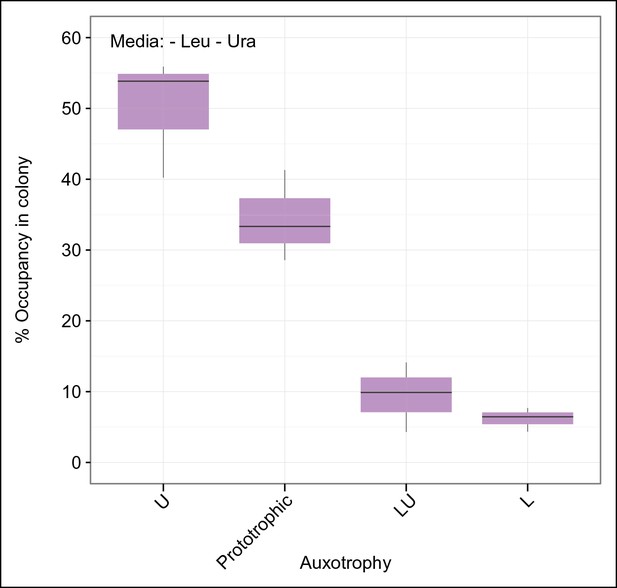
(A) Frequency of the 16 metabotypes that result from combination of histidine, leucine, methionine, and uracil auxotrophies as found within SeMeCo colonies. Separately established populations (n=3) were genotyped (>180 cells per colony) and eight metabotypes showed to dominate in the populations (inset). Frequency of the 16 metabotypes in independently established cell populations. The eight metabotypes of low frequency, which were depleted in all experiments, are highlighted with a red circle. (B) No correlation shown between the frequency of the 16 metabotypes in SeMeCo versus a segregation rate-predicted colony composition. Coloured points correspond to the experimentally observed eight frequent metabotypes. Dashed line: linear regression fit. (C) Maximum specific growth rate (µmax) in supplemented minimal media (red dots), of the 16 strains carrying HIS3, LEU2, URA3, and MET15 plasmids in all combinations obtained from (Mülleder et al., 2012), relative to the frequency of the specific metabotype in SeMeCo (light blue). Dashed line indicates average µmax for the eight most and least frequent metabotypes within SeMeCo colonies. (D) SeMeCo re-established on minimal media supplemented with uracil. After 7 days of growth (with re-spotting every two days), SeMeCo adapted with an entirely different composition of metabotypes (green box plot) compared to original SeMeCo colony composition (grey bars). Uracil producing cells decline, including the FourP genotype, so that 97% of cells are cooperating auxotrophs.
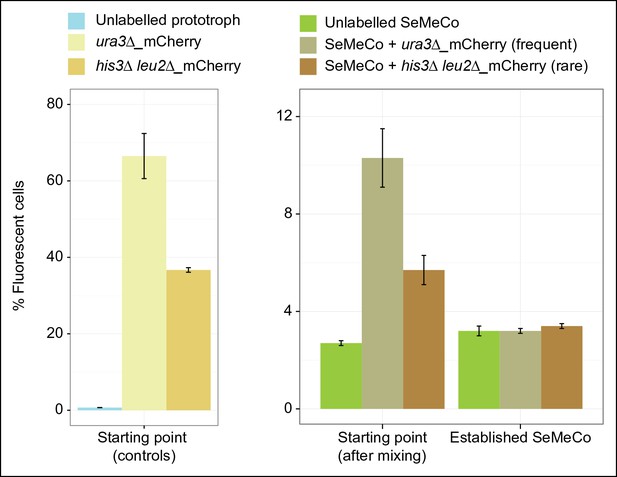
Abundance over time of a fluorescent labelled frequent (HIS3, LEU2, MET15, ura3∆) and rare (his3∆, leu2∆, URA3, MET15) genotype spiked into SeMeCo, as measured by FACS.
(Metabotype frequency determined from SeMeCo colony, Figure 5A). (left)% Fluorescence of the frequent and rare metabotype established individually as a colony shows frequent and rare abundance respectively (unlabelled prototroph control is BY4741-pHLUM).(right) Frequent and rare metabotypes spiked into pre-established SeMeCo shows depletion of both cell types after approx. 48 hr. n = 3, error bars = ± SD. FACS: Fluorescence-activated cell sorting
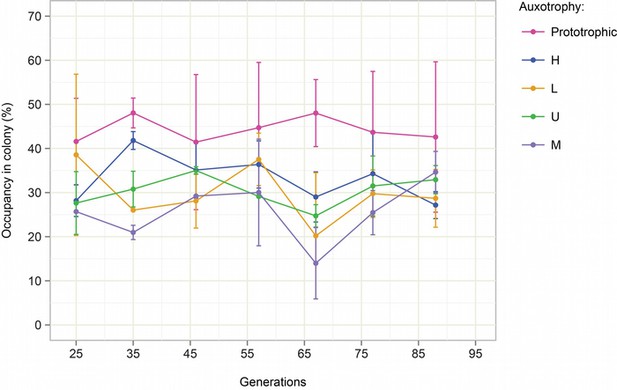
Stability of SeMeCo colony over time.
Starting from a SeMeCo micro-colony on minimal media, a giant colony was established and composition was followed by replica plating for 90 generations. Biomass gain is calculated starting from the single cell.
Tables
Strains and plasmids used in this study.
| Name | Description | Reference |
|---|---|---|
| Strains | ||
| BY4741 | MATa, his3∆1 leu2∆0 met15∆0 ura3∆0 (ATCC 201388) | (Brachmann et al., 1998) |
| BY4741 FLO+ | Derived from tetrad dissection after crossing and sporulating a flocculating BY4741 strain derived from the knock out collection (∆tpo1)with BY4742 and isolating a FLO+ TPO1 wild-type progeny | This study |
| YSBN5 | MATa, FY3 ho::Ble | (Canelas et al., 2010) |
| ED666 | h+ ade6-M210 ura4-D18 leu1-32 | Bioneer Cat. No. M-3030H |
| Plasmids | ||
| p423GPD | 2 µ vector with HIS3 marker | (Mumberg et al., 1995) |
| pRS425 | 2 µ vector with LEU2 marker | (Christianson et al., 1992) |
| p426GPD | 2 µ vector with URA3 marker | (Mumberg et al., 1995) |
| pRS411 | Yeast centromeric vector with MET15 marker | (Brachmann et al., 1998) |
| pHLUM | Yeast centromeric vector with HIS3, URA3, LEU2 and MET15 markers (minichromosome). (Addgene number: 40276) | (Mülleder et al., 2012) |
| pFS118 | Yeast high-copy vector with endogenous promoter for ura4+ (Addgene number: 12378) | (Sivakumar et al., 2004) |
| pREP41-MCS+ | Yeast high-copy vector with endogenous promoter for LEU2. (Addgene number: 52690) | A gift from Michael Nick Boddy |
| p416GPD | Yeast centromeric vector with endogenous promoter for URA3 | (Mumberg et al., 1995) |
| pHS12-mCherry | Yeast vector with mCherry fluorescent tag and LEU2 marker. (Addgene number: 25444) | A gift from Benjamin Glick |
| p426-GPDpr-mCherry | Yeast 2 µ vector with endogenous promoter for URA3 marker and a GPD promoter for mCherry fluorescent tag | This study. Derived from p416GPD, pHS12-mCherry and p426GPD |
| pRS411-GPDpr-mCherry | Yeast 2 µ vector with endogenous promoter for MET15 marker and a GPD promoter for mCherry fluorescent tag | This study. Derived from p426-GPDpr-mCherry and pRS411 |
| yEpVenus_URA | Yeast 2 µ vector with TDH3-promoter-driven Venus (YFP) and URA3 marker | (Bilsland et al., 2013) |
| yEpCFP_HIS | Yeast 2 µ vector with TDH3-promoter-driven CFP and HIS3 marker | (Bilsland et al., 2013) |
| yEpSapphire_LEU | Yeast 2 µ vector with TDH3-promoter-driven Sapphire (a UV-excitable GFP) and LEU2 marker | (Bilsland et al., 2013) |
| pHLM-GPDpr-mCherry | Yeast centromeric vector with a GPD promoter for mCherry fluorescent tag | This study. Derived from pHLUM |
| pUM-GPDpr-mCherry | Yeast centromeric vector with a GPD promoter for mCherry fluorescent tag | This study. Derived from pHLUM |
Oligonucleotides used to create expression plasmid p426-GPDpr-mCherry.
| Name | Sequence |
|---|---|
| mCherry_Bam_Sac_fw | AAGAAGAGCTCAAAAGGATCCGGGATGGTGAGCAAGGGCGAGG |
| mCherry_Xho_rv | CCTTTTCTCGAGCTTGTACAGCTCGTCCATGC |
SRM transitions for quantification of amino acids and uracil.
| Compound name | Compound abbreviation | SRM transition | Fragmentor (V) | Collision energy (V) | Polarity |
|---|---|---|---|---|---|
| Uracil | U | 111.0 > 42.1 | 62 | 9 | -− |
| Phenylalanine | F | 166.1 > 120 | 100 | 9 | + |
| Leucine | L | 132.1 > 86 | 80 | 8 | + |
| Tryptophan | W | 205.1 > 188 | 85 | 5 | + |
| Isoleucine | I | 132.1 > 86 | 80 | 8 | + |
| Methionine | M | 150.1 > 104 | 40 | 8 | + |
| Taurine | Tau | 126 > 44.1 | 110 | 16 | + |
| Valine | V | 118.1 > 71.9 | 100 | 10 | + |
| Proline | P | 116.1 > 70.1 | 100 | 13 | + |
| Tyrosine | Y | 182 > 165 | 90 | 5 | + |
| Alanine | A | 90 > 44.1 | 50 | 8 | + |
| Threonine | T | 120.1 > 74 | 80 | 9 | + |
| Glycine | G | 76 > 30.1 | 50 | 5 | + |
| Glutamine | Q | 147.1 > 84 | 50 | 16 | + |
| Glutamate | E | 148.1 > 84.1 | 75 | 10 | + |
| Serine | S | 106 > 60 | 40 | 9 | + |
| Asparagine | N | 133.1 > 74 | 80 | 9 | + |
| Aspartate | D | 134.1 > 74 | 80 | 10 | + |
| Histidine | H | 156.1 > 110.2 | 80 | 12 | + |
| Arginine | R | 175.1 > 70 | 100 | 15 | + |
| Lysine | K | 147.1 > 84 | 50 | 16 | + |
| Citrulline | Cit | 176 > 159 | 60 | 4 | + |
| Ornithine | O | 133 > 70 | 90 | 10 | + |
Additional files
-
Supplementary file 1
Script used for the simulation of plasmid segregation over time, using R (r-project.org).
- https://doi.org/10.7554/eLife.09943.023





