Coordinated Tbx3/Tbx5 transcriptional control of the adult ventricular conduction system
Figures
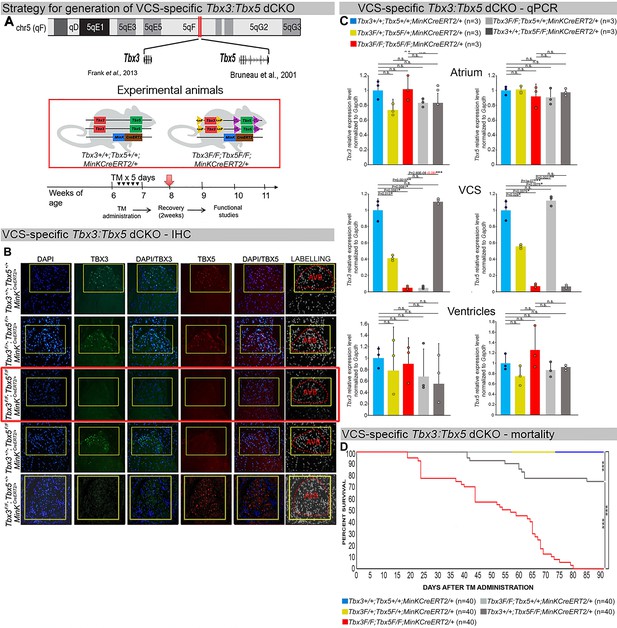
Generation of VCS-specific Tbx3:Tbx5 double-conditional knockout mice.
(A) Strategy to generate VCS-specific Tbx3:Tbx5 double-conditional knockout mouse line. A new Tbx3:Tbx5 double-conditional knockout mouse line (Tbx3fl/fl;Tbx5fl/fl) was generated using the CRISPR–Cas9 system (Gurumurthy et al., 2021; Dow et al., 2015) which allowed for the targeting of Tbx5 in the background of the previously validated Tbx3 floxed allele (Frank et al., 2011). A newly engineered Tbx5 floxed allele has been developed to mirror a previously published allele (Bruneau et al., 2001). This design has enabled the utilization of the previously published individual Tbx3 floxed allele (Frank et al., 2011) and individual Tbx5 floxed allele (Bruneau et al., 2001) as controls. To conditionally delete Tbx3 and Tbx5 genes specifically from the adult VCS and generate the experimental animals, the Tbx3:Tbx5 double-floxed mouse line (Tbx3fl/fl;Tbx5fl/fl) was combined with a VCS-specific tamoxifen inducible Cre transgenic mouse line (MinKCreERT2 [Tg(RP23-276I20-MinKCreERT2) Arnolds and Moskowitz, 2011b]). All allelic combinations were generated and evaluated as littermates in a mixed genetic background. The experimental mice employed in all studies were administered tamoxifen at 6 weeks of age and subsequently evaluated at 9 weeks of age (3 weeks post-tamoxifen administration). The loss of Tbx3 and Tbx5 expression, on both the protein and mRNA levels, assessed by immunohistochemistry (B) and qRT-PCR (C), respectively, was observed in the VCS of adult Tbx3:Tbx5 double-conditional mutant mice (Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+), but not in their littermate controls (Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+) (B, C). (C) qRT-PCR analysis showed a partial loss of Tbx3 and Tbx5 expression in the adult VCS of Tbx3:Tbx5 double-conditional heterozygous mice (Tbx3fl/+;Tbx5fl/+;R26ReYFP/+;MinKCreERT2/+) compared to their littermate controls (Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+). Additionally, qRT-PCR analysis confirmed the specificity of the Tbx3:Tbx5 double knockout for the VCS by assessing Tbx3 and Tbx5 expression levels in the atria and ventricles of tamoxifen-treated experimental mice. Consistent with the VCS selectivity of Cre activity in the MinKCreERT2 mice (Arnolds and Moskowitz, 2011b), Tbx3 and Tbx5 expression remained similar in the atrial and ventricular myocardium across all allelic combinations, including Tbx3:Tbx5 double-conditional knockout mice (Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+). (D) Conducted longitudinal studies revealed a significantly increased mortality rate in VCS-specific Tbx3:Tbx5-deficient mice compared to their littermate controls (***p < 0.0001, log-rank test, GraphPad Prism), suggesting a requirement for both Tbx3 and Tbx5 in the mature VCS. All allelic combinations of experimental and control mice (n = 40 biological replicates/genotype) were followed longitudinally after tamoxifen administration at 6 weeks of age. Tbx3:Tbx5 double-conditional knockout mice began to die suddenly at 3–4 weeks post-tamoxifen administration. Within the 3 months post-tamoxifen administration, all tamoxifen-treated Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+ mice had died suddenly (n = 40) without previous signs of illness. In contrast, no mortality was observed among the tamoxifen-treated Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+ and Tbx3fl/+;Tbx5fl/+;R26ReYFP/+;MinKCreERT2/+ littermates (each cohort n = 40) during this period. TBX3 and TBX5 protein expression was evaluated by immunohistochemistry (green and red signals, respectively) on serial sections from hearts of all allelic combinations (n = 3 biological replicates/genotype). Nuclei were stained with DAPI (blue signal). IHC original magnification: ×40. qRT-PCR data are presented as mean ± SD normalized to Gapdh and relative to Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+ mice (set as 1). N = 3 biological replicates/genotype (VCS cardiomyocytes pooled from 30 mice per biological replicate); multiple testing correction using Benjamini and Hochberg procedure. Significance was assessed by Welch t-test (*FDR <0.05; **FDR <0.005; ***FDR <0.001) and confirmed by one-tailed Wilcoxon test (p value in parentheses) when normally distribution was rejected. Abbreviations: AVB, atrioventricular bundle (also known as bundle of His); FDR, false discovery rate; VCS, ventricular conduction system.
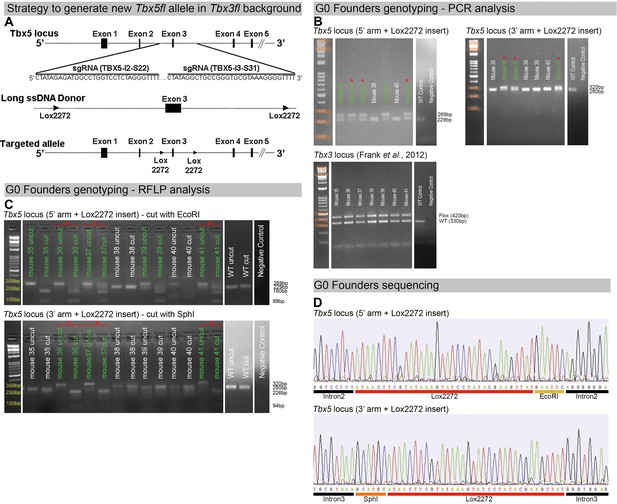
Generation of a novel Tbx5 floxed allele in aTbx3 floxed background.
(A) Strategy to generate a new Tbx5 floxed allele using the CRISPR/Cas9 system with long single-strand DNA donor (lssDNA donor) composed of the targeted exon 3 flanked by two lox2272 sites. The schematic shows the Cas9/sgRNA-targeting sites in the second and third introns of murine Tbx5, the lssDNA donor, and the targeted allele. (B) PCR analysis of Tbx5 and Tbx3 loci in G0 founders. Mice #35 to #41 are shown. Upper left panel: PCR genotyping of the Tbx5 locus using primers specific for the 5′ arm of the lssDNA donor with the lox2272 insertion. Upper right panel: PCR genotyping of the Tbx5 locus using primers specific for the 3′ arm of the lssDNA donor with the lox2272 insertion. Green labels indicate founders positive for lox2272 insertion in one arm. Red asterisks indicate founders positive for lox2272 insertion in both arms. Expected fragment sizes: for the 5′ arm with lox2272 insertion, positive band = 269 bp, WT = 229 bp; for the 3′ arm with insertion, positive band = 320 bp, WT = 280 bp. Lower panel: PCR genotyping of the Tbx3 locus using primers indicating Tbx3 floxed allele. Expected fragment sizes: Tbx3 floxed allele = 420 bp, Tbx3 WT allele = 330 bp. (C) RFLP analysis of the Tbx5 locus in G0 founders. Mice #35 to #41 are shown. Upper panel: PCR products amplified using primers specific for the 5′ arm of the lssDNA donor were digested with EcoRI to detect the insertion of the 5′ lox2272. WT amplicon = 229 bp; lox2272 positive amplicon = 269 bp. The WT amplicon lacks the EcoRI site. With the 5′ lox2272 insertion, the digestion products are 180 and 89 bp. Lower panel: PCR products amplified using primers specific for the 3′ arm of the lssDNA donor were digested with SphI to detect the insertion of the 3′ lox2272. WT amplicon = 280 bp; lox2272 positive amplicon = 320 bp. The WT amplicon lacks the SphI site. With the 3′ lox2272 insertion, the digestion products are 226 and 94 bp. WT controls are shown in the last lanes. Green labels indicate founders positive for lox2272 insertion in one arm. Red asterisks indicate founders positive for lox2272 insertion in both arms. (D) Example of Sanger sequencing results from a representative G0 founder previously identified as positive for 5′ and 3′ lox2272 sites insertion by PCR and RFLP analysis. The sequencing confirmed the precise integration of lox2272 sites in both arms, resulting in exon 3 being flanked by two lox2272 sites.
-
Figure 1—figure supplement 1—source data 1
File containing original gels corresponding to Figure 1—figure supplement 1B.
The black rectangle indicates the region included in the final figure panel.
- https://cdn.elifesciences.org/articles/102027/elife-102027-fig1-figsupp1-data1-v1.pdf
-
Figure 1—figure supplement 1—source data 2
Zipped folder containing original gels corresponding to Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/102027/elife-102027-fig1-figsupp1-data2-v1.pdf
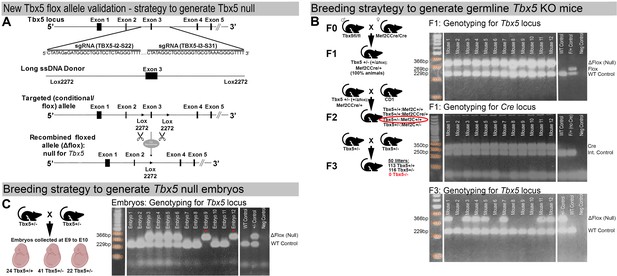
Validation of the newly generated Tbx5 floxed allele.
(A) Strategy to generate a Tbx5 null allele through recombination of the newly generated Tbx5 floxed allele in the presence of the Cre recombinase. (B) Strategy to generate germline Tbx5 null mice in presence of Mef2CCre allele. To verify the ability of the newly generated Tbx5 floxed allele to recombine into the Tbx5 KO (null) allele, three rounds of breeding were conducted (left panel), and a PCR assay (right panels) was utilized to distinguish the recombined Tbx5 null allele (Tbx5−) from the unrecombined floxed allele (Tbx5fl). Initially, homozygous Tbx5 floxed males (Tbx5fl/fl) were bred with homozygous Mef2CCre/Cre females, which express Cre recombinase at the zygote stage, to produce germline Tbx5+/−:Mef2Cre/+ double heterozygous mice. PCR analysis confirmed that 100% of the offspring were Tbx5+/−:Mef2CCre/+ double heterozygous mice (right upper and middle panels), demonstrating complete recombination of the Tbx5 floxed allele into the Tbx5 null allele in the presence of the Mef2CCre allele. Subsequently, Tbx5+/−:Mef2CCre/+ double heterozygous mice were backcrossed with wild-type CD1 IGS mice to obtain a germline Tbx5+/− heterozygous mouse line. Tbx5+/− mice were then bred to generate germline Tbx5 null mice (Tbx5−/−). Among 230 offspring (from 50 litters), 113 wild-type pups (Tbx5+/+) and 116 Tbx5 heterozygous pups (Tbx5+/−) were received, but no null pups (Tbx5−/−) were obtained. (Right lower panel) Example of genotyping results showing absence of Tbx5 null mice in newborn offspring generated by Tbx5+/− mice. Expected fragment sizes: Tbx5 Δflox (null) allele = 366 bp, Tbx5 flox allele = 269 bp, Tbx5 WT allele = 229 bp, Cre allele = 350 bp, Internal control allele = 250 bp. (C) Breeding strategy to generate E9.0–E10.0 germlineTbx5 null embryos. (Left panel) To generate germline Tbx5 null embryos (Tbx5−/−), Tbx5 heterozygous mice (Tbx5+/−) were bred, and embryos were collected at E9–E10. Genotyping analysis of 89 collected embryos from 10 timed pregnancies revealed the presence of 24 wild-type (Tbx5+/+), 42 heterozygous (Tbx5+/−), and 22 null (Tbx5−/−) embryos. These results confirmed that the newly generated Tbx5 floxed allele could recombine to form the null allele, mimicking the embryonic lethality observed with the previously generated allele (Bruneau et al., 2001). (Right panel) Example of genotyping results showing the presence of E9.0 Tbx5 null embryos collected from Tbx5+/− timed breedings. Expected fragment sizes: Tbx5 Δflox (null) allele = 366 bp, Tbx5 WT allele = 229 bp.
-
Figure 1—figure supplement 2—source data 1
File containing original gels corresponding to Figure 1—figure supplement 2B.
The black rectangle indicates the region included in the final figure panel.
- https://cdn.elifesciences.org/articles/102027/elife-102027-fig1-figsupp2-data1-v1.pdf
-
Figure 1—figure supplement 2—source data 2
Zipped folder containing original gels corresponding to Figure 1—figure supplement 2B.
- https://cdn.elifesciences.org/articles/102027/elife-102027-fig1-figsupp2-data2-v1.pdf

WT sequence of the targeted region at the mouse Tbx5 locus.
The partial intron 2 (black, lowercase letters), entire exon 3 (black, bold, uppercase letters), and partial intron 3 (black, lowercase letters) are shown. The sgRNA-targeting sequence at the 5′ of exon 3 (Tbx5-I2-S22) is labeled in bold, light green, while the sgRNA-targeting sequence at the 3′ of exon 3 (Tbx5-I2-S31) is labeled in bold, dark green. The cleavage sites at the sgRNA-targeting sequences are underlined. The protospacer-adjacent motif (PAM) sequences are labeled in bold, light blue.

Sequence of long ssDNA donor designed to target exon 3 of the mouse Tbx5 locus.
It is composed of targeted exon 3 flanked by two lox2272 sites, along with 100/150 bp 5′/3′ homology arms. The partial intron 2 (black, lowercase letters), entire exon 3 (bold, black, uppercase letters), partial intron 3 (black, lowercase letters), as well as 5′ (purple, lowercase letters) and 3′ (pink, lowercase letters) homology arms are displayed. The lox2272 sites, indicated in bold red, have been inserted at the cleavage sites of the sgRNA-binding sequences. Unique restriction enzyme sites used for RFLP analysis are shown as bold yellow for EcoRI and bold navy blue for SphI. T7 promoter is labeled in bold orange.

Sequence of mouse new Tbx5 floxed allele generated using CRISPR/Cas9 system with lssDNA composed of targeted exon 3 flanked by two lox2272 sites.
The partial intron 2 (black, lowercase letters), entire exon 3 (bold, black, uppercase letters), and partial intron 3 (black, lowercase letters) are displayed. The lox2272 sites, indicated in bold red, have been inserted at the cleavage sites of the sgRNA-binding sequences. Unique restriction enzyme sites used for RFLP analysis are shown as bold yellow for EcoRI and bold navy blue for SphI. PCR primers specific for the 5′ arm of the lssDNA donor with the lox2272 insertion (displayed as green box) as well as primers specific for the 3′ arm of the lssDNA donor with the lox2272 insertion (displayed as pink box) are shown.
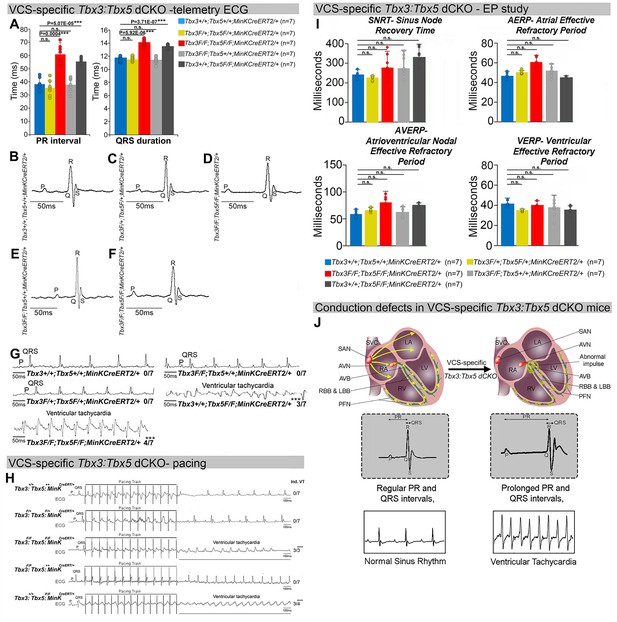
Arrhythmias and conduction abnormalities in mice with VCS-specific Tbx3:Tbx5 double-conditional knockout.
(A–F) VCS-specific Tbx3:Tbx5 double-conditional knockout causes significant VCS conduction slowing in adult Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+ mice. (A) PR (left graph) and QRS (right graph) intervals calculated from ambulatory telemetry electrocardiography (ECG) recordings in (B–F). Tbx3:Tbx5 double-conditional adult mice (Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+) displayed significant PR and QRS intervals prolongation compared to littermate controls (Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+) (A left and right graphs, respectively). Data are presented as mean ± SD. N = 7 biological replicates/genotype, multiple testing correction using Benjamini and Hochberg procedure; *Welch t-test p < 0.05 and FDR <0.05; **Welch t-test p < 0.005 and FDR <0.05; ***Welch t-test p < 0.001 and FDR <0.05. Representative ambulatory telemetry ECG of Tbx3+/+;Tbx5+/+; R26ReYFP/+;MinKCreERT2/+ (B), Tbx3fl/+;Tbx5fl/+;R26ReYFP/+;MinKCreERT2/+ (C), Tbx3fl/fl;Tbx5fl/fl; R26ReYFP/+;MinKCreERT2/+ (D), Tbx3fl/fl;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+ (E), Tbx3+/+;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+ (F) mice. (G) Simultaneous genetic removal of Tbx3 and Tbx5 from the adult VCS resulted in significantly increased episodes of spontaneous ventricular tachycardia. Episodes of spontaneous ventricular tachycardia were observed in four of seven Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+; MinKCreERT2/+ mice versus zero of seven littermate controls (Tbx3+/+;Tbx5+/+;R26ReYFP/+; MinKCreERT2/+) in ambulatory studies. N = 7 biological replicates/genotype; multiple testing correction using Benjamini and Hochberg procedure; *FDR of Welch t-test ≤0.05. (H) Tbx3:Tbx5 double-conditional knockout mice (Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+) showed significantly increased susceptibility to ventricular tachycardia following burst stimulation in invasive electrophysiology studies (three of three Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+; MinKCreERT2/+ mice vs. zero of seven control Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+ mice. Fisher’s exact test: *p < 0.05; n = 7 biological replicates/genotype). (I) Intracardiac electrophysiology detected no significant changes in SNRT, AERP, AVERP, and VERP recorded from experimental and control animals (n = 7 biological replicates/genotype; multiple testing correction using Benjamini and Hochberg procedure; *FDR of Welch t-test ≤0.05). (J) Graphical summary of conduction defects observed in adult, VCS-specific Tbx3:Tbx5-deficient mice. Simultaneous genetic deletion of Tbx3 and Tbx5 from the mature VCS results in conduction slowing, prolonged PR and QRS intervals, as well as ventricular tachycardia. Abbreviations: AVB, atrioventricular bundle (also known as bundle of His); AVN, atrioventricular node; FDR, false discovery rate; LA, left atrium; LBB, left bundle branches; LV, left ventricle; PFN, Purkinje fiber network; RA, right atrium; RBB, right bundle branches; RV, right ventricle; SAN, sinoatrial node; SVC, superior vena cava; VCS, ventricular conduction system.
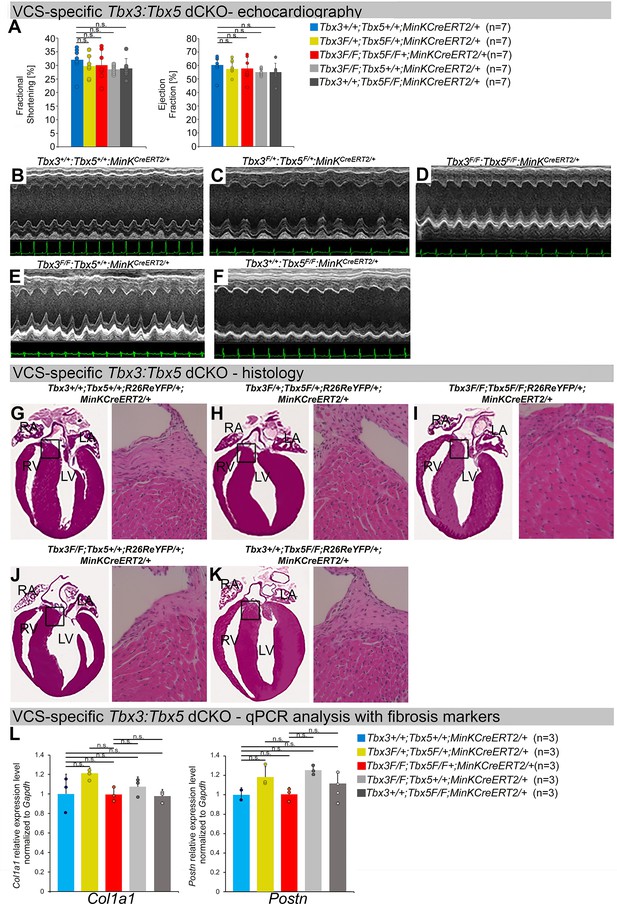
Cardiac function is preserved following double-conditional loss of Tbx3 and Tbx5 in the adult ventricular conduction system (VCS).
(A) Left ventricular (LV) fractional shortening (left graph) and LV ejection fraction (right graph) calculated from the M-mode electrocardiographies (ECGs) in (B–F) revealed no contractile dysfunction in VCS-specific Tbx3:Tbx5 double-conditional mutant mice (Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+; MinKCreERT2/+). Data are presented as mean ± SD. Welch t-test: ns, not significant (p > 0.05) ; n = 7 biological replicates/genotype. (B–F) Cardiac function, assessed by M-mode echocardiography from Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+ (B), Tbx3fl/+;Tbx5fl/+; R26ReYFP/+;MinKCreERT2/+ (C), Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+ (D), Tbx3fl/fl;Tbx5+/+; R26ReYFP/+;MinKCreERT2/+ (E), Tbx3+/+;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+ (F) mice shown above surface ECGs. No functional differences between mutant and control mice were detected. The most representative images for each genotype were utilized in the figure. n = 7 biological replicates/genotype. (G–K) Histological examination of all four chambers from Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+ (G), Tbx3fl/+;Tbx5fl/+;R26ReYFP/+; MinKCreERT2/+ (H), Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+ (I), Tbx3fl/fl;Tbx5+/+;R26ReYFP/+; MinKCreERT2/+ (J), Tbx3+/+;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+ (K) mice showed no histological abnormalities. The most representative images for each genotype were utilized in the figure. n = 3–4 biological replicates/genotype. Boxed areas in (G–K) have been shown at higher magnification at their right sides. (L) qRT-PCR analysis for fibrosis genes Col1a1 and Postn confirmed that there was no increase in expression of fibrosis markers in the VCS of Tbx3:Tbx5-deficient mice. Data are presented as mean ± SD normalized to Gapdh and relative to Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+ mice (set as 1). ns: FDR >0.05 in both Welch t-test and Wilcoxon test; n = 2–3 biological replicates/genotype (VCS cardiomyocytes pooled from 30 mice per biological replicate). Histological examination original magnification: ×2.5, boxed area showed at the higher magnification: ×40. Abbreviations: FDR, false discovery rate; LA, left atrium; RA, right atrium; LV, left ventricle; RV, right ventricle.
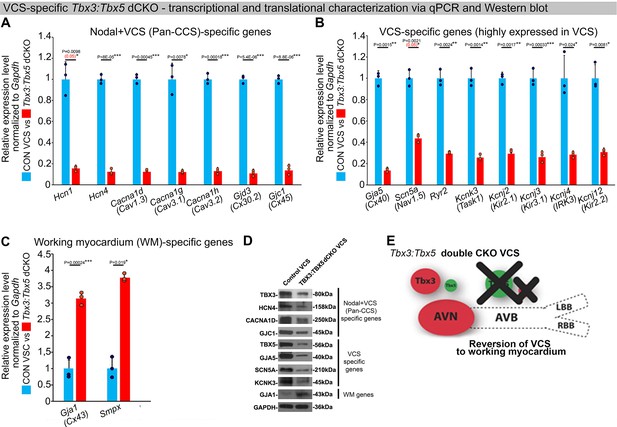
In the adult murine heart, Tbx3 and Tbx5 collectively promote ventricular conduction system (VCS) versus working myocardium (WM) phenotype.
(A–C) qRT-PCR analysis of molecular changes driven by VCS-specific Tbx3:Tbx5 double-conditional knockout in adult mice. Transcriptional characterization of the adult VCS in Tbx3fl/fl;Tbx5fl/fl;R26ReYFP/+;MinKCreERT2/+ mutant mice, compared to their Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+ control littermates, was conducted using three distinct sets of molecular markers. (A) Genes expressed throughout the entire conduction system (Pan-CCS), implicated in the slow-conducting nodal phenotype. (B) Genes highly expressed in the fast-conducting VCS, critical for VCS function. (C) Markers specifically present in the working myocardium but absent in the CCS. VCS-specific Tbx3:Tbx5-deficient mice lost the VCS expression profile, including genes necessary for fast ventricular conduction (B) and those typically expressed in the entire CCS (Pan-CCS genes), essential for the slow-conducting nodal phenotype (A). In contrast, they acquired VCS expression of working myocardium-specific molecular markers important for working myocardial function (C). (D) Immunoblotting analysis confirmed transcriptional changes indicated by qRT-PCR analysis (A–C) in VCS-specific Tbx3:Tbx5 double-conditional knockout in adult mice. (E) Graphical summary of transcriptional changes observed in VCS of VCS-specific Tbx3:Tbx5-deficient mice. Simultaneous genetic deletion of Tbx3 and Tbx5 from the mature VCS resulted in a transcriptional profile resembling that of ventricular working myocardium. qRT-PCR data are presented as mean ± SD normalized to Gapdh and relative to Tbx3+/+;Tbx5+/+;R26ReYFP/+;MinKCreERT2/+ mice (set as 1). N = 3 biological replicates/genotype (VCS cardiomyocytes pooled from 30 mice per biological replicate); multiple testing correction using Benjamini and Hochberg procedure. Significance was assessed by Welch t-test (*FDR <0.05; **FDR <0.005; ***FDR <0.001) and confirmed by one-tailed Wilcoxon test (p value in parentheses) when normal distribution was rejected. Western blotting, n=2 biological replicates/ genotype (VCS cardiomyocytes pooled from 50 mice per each biological replicate). Abbreviations: AVB, atrioventricular bundle (also known as bundle of His); AVN, atrioventricular node; dCKO, double-conditional knockout; FDR, false discovery rate; LBB, left bundle branches; RBB, right bundle branches; VCS, ventricular conduction system.
-
Figure 4—source data 1
File containing original western blots corresponding to Figure 4D.
The black rectangle indicates the region included in the final figure panel.
- https://cdn.elifesciences.org/articles/102027/elife-102027-fig4-data1-v1.pdf
-
Figure 4—source data 2
Zipped folder containing original western blots files corresponding to Figure 4D.
- https://cdn.elifesciences.org/articles/102027/elife-102027-fig4-data2-v1.zip

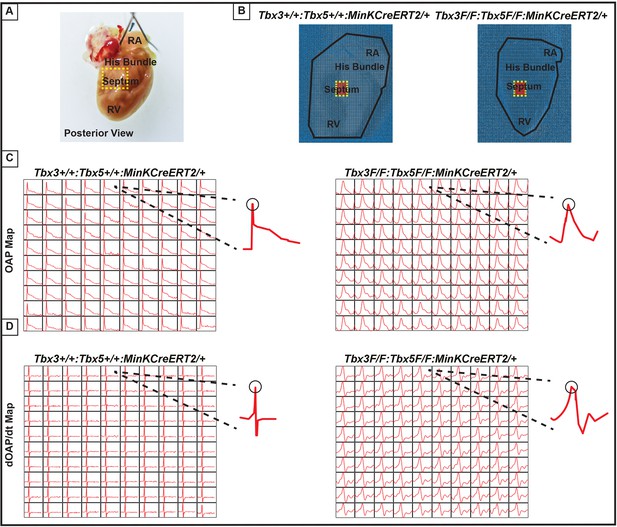
Loss of VCS optical action potential (OAP) morphology in Tbx3:Tbx5 double-conditional knockout mice.
(A) Schematic of the posterior view of a mouse heart with right ventricle (RV) free wall removed, highlighting the RV, septum, His bundle, and right atria (RA). (B) Representative 100 × 100 OAP map of OAP recorded during sinus rhythm from Tbx3:Tbx5 double-conditional knockout mice and control littermates with the free wall removed. The region of the His bundle is highlighted in red. (C) Representative 10 × 10 OAP map from the region of the His bundle. (D) Representative 10 × 10 map of the first derivative of the OAP from the region of the His bundle. (E) Representative ventricular OAP from whole heart intact preparation from Tbx3:Tbx5 double-conditional knockout mice (red) and control littermates (black). (F) Quantification of APD50 and APD80 at a basic cycle length of 125 ms. For (C-F), n=10 or 8 animals/ genotype.
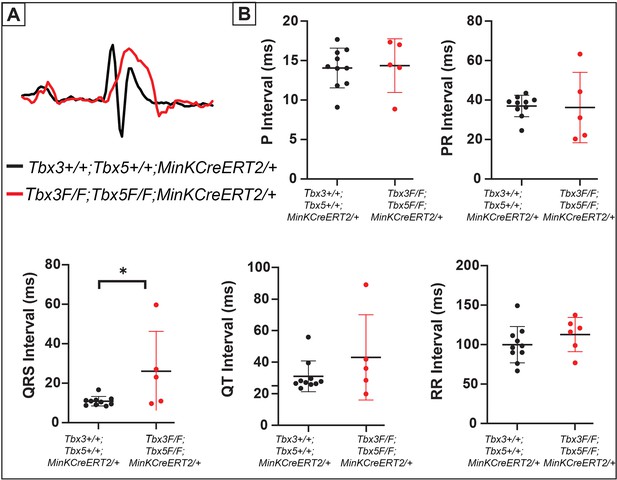
Tbx3:Tbx5 double-conditional knockout mice exhibit QRS prolongation.
(A) Representative electrocardiography (ECG) traces recorded from Tbx3:Tbx5 double-conditional knockout mice (red) and control littermates (black). (B) Quantification of P, PR, QRS, QT, and RR intervals. For (A and B), n=10 or 8 animals/ genotype.*p < 0.05 denotes significance.
Ventricular optical action potentials (OAPs) distal from His bundle have only one OAP upstroke.
(A) Schematic of the posterior view of mouse heart with right ventricle (RV) free wall removed. (B) Representative 100 × 100 pixel OAP map recorded during sinus rhythm from Tbx3:Tbx5 double-conditional knockout mice and control littermates with RV free wall removed. The region of the working ventricular myocardium distal from the His bundle is highlighted in red. (C) Representative 10 × 10 pixel OAP map from the region distal to the His bundle. (D) Representative 10 × 10 dOAP/dt map from the region distal to the His bundle. For (C and D), n=10 or 8 animals/ genotype.
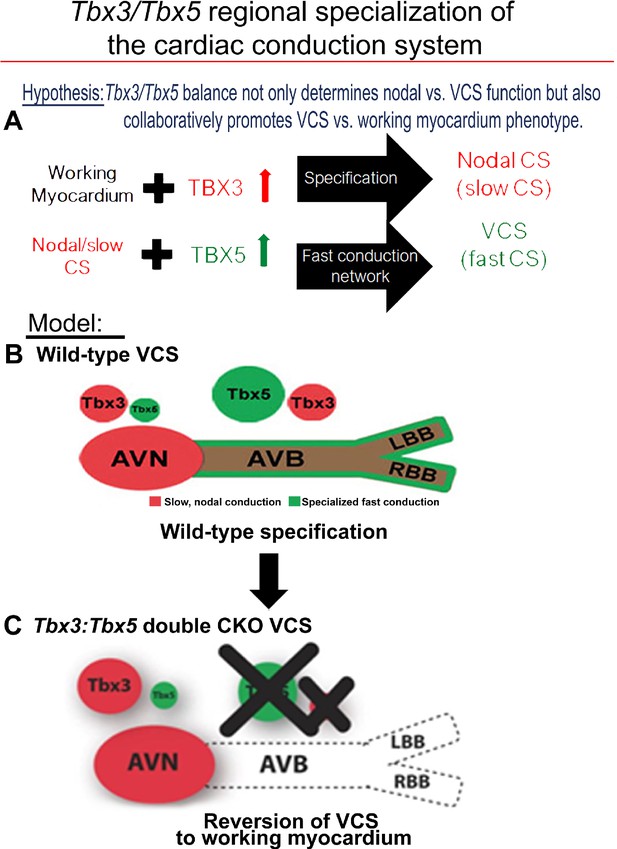
Tbx3 and Tbx5 play distinct roles in the adult VCS while collectively promoting CCS regional identity – a model elucidating our hypothesis for Tbx3/Tbx5 dose-dependent CCS regional specialization.
(A) The Tbx3/Tbx5 balance not only governs nodal versus ventricular conduction system (VCS) function but also collaboratively promotes the VCS versus working myocardium (WM) phenotype. Specifically, a high level of Tbx3 is linked to the specialization to the nodal conduction system, while an elevated Tbx5 level in nodal cells activates local expression of the Tbx5-dependent fast conduction network, resulting in the generation of VCS. (B) CCS regional specialization is driven by local expression of Tbx5-dependent fast conduction network in the VCS, which overlaps underlying Pan-CCS expression of nodal, slow conduction network. (C) VCS-specific simultaneous genetic removal of both the Tbx3 and Tbx5 transcription factors transforms the fast-conducting, adult VCS into cells resembling working myocardium, thereby shifting them from conduction to non-conduction myocytes. Therefore, within the adult CCS, the Tbx3 and Tbx5 expression levels are crucial not only for normal fast versus slow conduction system identity but also for maintaining the conduction versus contraction specialization of the VCS. AVB, atrioventricular bundle; AVN, atrioventricular node; CCS, cardiac conduction system; CKO, conditional knockout; LBB, left bundle branch; RBB, right bundle branch; VCS, ventricular conduction system.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Goat polyclonal anti-TBX3 | Santa Cruz Biotechnology | sc-31656 | IF (1:250) WB (1:250) |
| Antibody | Goat polyclonal anti-TBX5 | Santa Cruz Biotechnology | sc-17866 | IF (1:250) |
| Antibody | Donkey anti-goat IgG AlexaFluor-594 | Invitrogen | A-11058 | IF (1:250) |
| Antibody | Donkey anti-goat IgG AlexaFluor-488 | Invitrogen | A-11055 | IF (1:250) |
| Antibody | Rabbit polyclonal anti-HCN4 | Millipore | AB5808 | WB (1:500) |
| Antibody | Rabbit polyclonal anti-CAV1.3/CACNA1D | Alomone | ACC-005 | WB (1:200) |
| Antibody | Rabbit polyclonal anti-Cx45/GJC1 | Thermo Fisher | PA5-77357 | WB (1:250) |
| Antibody | Sheep polyclonal anti-TBX5 | R&D | AF5918 | WB (1:200) |
| Antibody | Rabbit polyclonal anti-CX40/GJA5 | Zymed/ Invitrogen | 36–4900 | WB (1:500) |
| Antibody | Rabbit polyclonal anti-NAV1.5/SCN5A | Alomone | ASC-005 | WB (1:200) |
| Antibody | Mouse monoclonal anti-KCNK3/TASK1 | Abcam | ab186352 | WB (1:1000) |
| Antibody | Rabbit polyclonal anti-CX43/GJA1 | Cell Signalling Technology | 3512 | WB (1:1000) |
| Antibody | Mouse monoclonal anti-GAPDH | Abcam | ab8245 | WB (1:1000) |
| Antibody | Rabbit anti-goat-HRP | Jackson Immuno Research | 305-035-003 | WB (1:10 000) |
| Antibody | Goat anti-rabbit-HRP | Jackson Immuno Research | 111-035-144 | WB (1:3000) |
| Antibody | Donkey anti-sheep-HRP | Abcam | ab6900 | WB (1:5000) |
| Antibody | Sheep anti-mouse-HRP | Amersham GE | NA931 | WB (1:2500) |
| Chemical compound, drug | BSA | Sigma-Aldrich | A9576 | |
| Chemical compound, drug | cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | 11836170001 | |
| Chemical compound, drug | DAPI | Invitrogen | 62248 | |
| Chemical compound, drug | Isoflurane | Sigma-Aldrich | 26675-46-7 | |
| Chemical compound, drug | POWER SYBR Green PCR Master Mix | Applied Biosystems | 43-676-59 | |
| Chemical compound, drug | ProLong Gold Antifade Mountant | Invitrogen | P10144 | |
| Chemical compound, drug | Propidium Iodide | Thermo Fisher Scientific | P3566 | |
| Chemical compound, drug | Tamoxifen | MP Biomedical | Dosage details look at Materials & Methods section | |
| Commercial assay or kit | MEGAshortscript T7 transcription kit | Invitrogen | AM1354 | |
| Commercial assay or kit | mMessage Machine T7 transcription kit | Invitrogen | AM1344 | |
| Commercial assay or kit | Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 | |
| Commercial assay or kit | Pierce ECL | Thermo Fisher Scientific | 32106 | |
| Commercial assay or kit | Pierce ECL Plus | Thermo Fisher Scientific | 32132 | |
| Commercial assay or kit | RNeasy Mini Kit | Qiagen | 74104 | |
| Commercial assay or kit | SuperScript III First-Strand Synthesis SuperMix | Invitrogen | 18080400 | |
| Genetic reagent (M. musculus) | Tbx3fl/fl;Tbx5fl/fl Mixed 129/SvJ:C57BL/6 J:CD-1 background | This paper | Generated using CRISPR–Cas9 technology (look at Experimental animals section) | |
| Genetic reagent (M. musculus) | Tbx3fl/fl;Tbx5fl/fl; R26ReYFP/+; MinKCreERT2/+ Mixed 129/SvJ:C57BL/6 J:CD-1 background | This paper | Generated using CRISPR–Cas9 technology (look at Experimental animals section) | |
| Genetic reagent (M. musculus) | Kcne1CreERT2 [Tg(RP23-276I20-MinKCreERT2)] – referred to as MinKCreERT2 in the manuscript for consistency with field-standard nomenclature and to avoid confusion. Mixed 129/SvJ:C57BL/6 J:CD-1 background | PMID: 21504046 | Dr. Ivan P. Moskowitz (University of Chicago) | |
| Genetic reagent (M. musculus) | Tbx3fl/fl Mixed 129/SvJ:C57BL/6 J:CD-1 background | PMID: 22203979 | Dr. Anne M Moon (University of Utah) | |
| Genetic reagent (M. musculus) | CD-1 | Charles Rivers Laboratories | https://www.criver.com/products-services/find-model/cd-1r-igs-mouse?region=3611 | |
| Genetic reagent (M. musculus) | C57Bl/6 J | Jackson Laboratory | https://www.jax.org/strain/000664 | |
| Other | Long ssDNA donor | This paper | Sequence details look at Figure 1—figure supplement 4 | |
| Other | SDS-PAGE on 4–20% TGX Gels | Bio-Rad | 4561096 | 4–20% Mini-PROTEAN TGX Precast Protein Gels |
| Other | Tbx5-I2-S22 | This paper | sgRNA | Sequence details look at Figure 1—figure supplement 3 |
| Other | Tbx5-I2-S31 | This paper | sgRNA | Sequence details look at Figure 1—figure supplement 3 |
| Other | VectaShield +DAPI | Vector Laboratories | https://vectorlabs.com/vectashield-mounting-medium-with-dapi.html | |
| Sequence-based reagent | Tbx5 locus: 5’arm-F | This paper | PCR primers | Sequence details look at Figure 1—figure supplement 5 |
| Sequence-based reagent | Tbx5 locus: 5’arm-R | This paper | PCR primers | Sequence details look at Figure 1—figure supplement 5 |
| Sequence-based reagent | Tbx5 locus: 3’arm-F | This paper | PCR primers | Sequence details look at Figure 1—figure supplement 5 |
| Sequence-based reagent | Tbx5 locus: 3’arm-R | This paper | PCR primers | Sequence details look at Figure 1—figure supplement 5 |
| Sequence-based reagent | Tbx3 locus: WT-F | PMID: 22203979 | PCR primers | Dr. Anne M Moon (University of Utah) |
| Sequence-based reagent | Tbx3 locus: WT-R | PMID: 22203979 | PCR primers | Dr. Anne M Moon (University of Utah) |
| Sequence-based reagent | Tbx3 locus: Flox-F | PMID: 22203979 | PCR primers | Dr. Anne M Moon (University of Utah) |
| Sequence-based reagent | Tbx3 locus: Flox-R | PMID: 22203979 | PCR primers | Dr. Anne M Moon (University of Utah) |
| Sequence-based reagent | qPCR_Tbx3-F | PMID: 32290757 | qRT-PCR primers | 5’-AGATCCGGTT ATCCCTGGGAC-3’ |
| Sequence-based reagent | qPCR_Tbx3-R | PMID: 32290757 | qRT-PCR primers | 5’-CAGCAGCCCC CACTAACTG-3’ |
| Sequence-based reagent | qPCR_Tbx5-F | PMID: 32290757 | qRT-PCR primers | 5’-GGCATGGAAG GAATCAAGGT-3’ |
| Sequence-based reagent | qPCR_Tbx5-R | PMID: 32290757 | qRT-PCR primers | 5’-CTAGGAAACATT CTCCTCCCTGC-3’ |
| Sequence-based reagent | qPCR_Gja1-F | PMID: 22086960 (Primer Bank ID:166091435 c3) | qRT-PCR primers | 5’-ACAGCGGTTG AGTCAGCTTG-3’ |
| Sequence-based reagent | qPCR_Gja1-R | PMID: 22086960 (Primer Bank ID:166091435 c3) | qRT-PCR primers | 5’-GAGAGATGGG GAAGGACTTGT-3’ |
| Sequence-based reagent | qPCR_Gja5-F | PMID: 32290757 | qRT-PCR primers | 5’-AGCTCCAGTC ACCCATCTTG-3’ |
| Sequence-based reagent | qPCR_Gja5-R | PMID: 32290757 | qRT-PCR primers | 5’-CAGTTGAACA GCAGCCAGAG-3’ |
| Sequence-based reagent | qPCR_Gjc1-F | PMID: 32290757 | qRT-PCR primers | 5’-AGATCCACAA CCATTCGACATTT-3’ |
| Sequence-based reagent | qPCR_Gjc1-R | PMID: 32290757 | qRT-PCR primers | 5’-TCCCAGGTAC ATCACAGAGGG-3’ |
| Sequence-based reagent | qPCR_Gjd3-F | PMID: 22086960 (Primer Bank ID: 30519904 a1) | qRT-PCR primers | 5’-TCATGCTGAT CTTCCGCATCC-3’ |
| Sequence-based reagent | qPCR_Gjd3-R | PMID: 22086960 (Primer Bank ID: 30519904 a1) | qRT-PCR primers | 5’-GAAGCGGTAG TGGGACACC-3’ |
| Sequence-based reagent | qPCR_Scn5a-F | PMID: 32290757 | qRT-PCR primers | 5’-CGCTCCTCCA GGTAGATGTC-3’ |
| Sequence-based reagent | qPCR_Scn5a-R | PMID: 32290757 | qRT-PCR primers | 5’-CTACCGCATA GTGGAGCACA-3’ |
| Sequence-based reagent | qPCR_Ryr2-F | PMID: 32290757 | qRT-PCR primers | 5’-CAAATCCTTC TGCTGCCAAG-3’ |
| Sequence-based reagent | qPCR_Ryr2-R | PMID: 32290757 | qRT-PCR primers | 5’-CGAGGATGAG ATCCAGTTCC-3’ |
| Sequence-based reagent | qPCR_Kcnk3-F | PMID: 32290757 | qRT-PCR primers | 5’-CTCCTTCTAC TTCGCCATCA-3’ |
| Sequence-based reagent | qPCR_Kcnk3-R | PMID: 32290757 | qRT-PCR primers | 5’-GAAGGTGTTG ATGCGTTCA-3’ |
| Sequence-based reagent | qPCR_Kcnj2-F | PMID: 32290757 | qRT-PCR primers | 5’-CGACTGCCAT GACAACTCAA-3’ |
| Sequence-based reagent | qPCR_Kcnj2-R | PMID: 32290757 | qRT-PCR primers | 5’-CATATCTCCG ATTCTCGCCT-3’ |
| Sequence-based reagent | qPCR_Kcnj3-F | PMID: 32290757 | qRT-PCR primers | 5’-GCTGGCAACT ACACTCCCTG-3’ |
| Sequence-based reagent | qPCR_Kcnj3-R | PMID: 32290757 | qRT-PCR primers | 5’-AACATGCAGC CGATGAGGAA-3’ |
| Sequence-based reagent | qPCR_Kcnj4-F | PMID: 32290757 | qRT-PCR primers | 5’-CACGTAAACG GCTTTTTGGGG-3’ |
| Sequence-based reagent | qPCR_Kcnj4-R | PMID: 32290757 | qRT-PCR primers | 5’-CCGTCTCGAA CGGAGATGAC-3’ |
| Sequence-based reagent | qPCR_Kcnj12-F | PMID: 32290757 | qRT-PCR primers | 5’-ACCCCTACAG CATCGTATCAT-3’ |
| Sequence-based reagent | qPCR_Kcnj12-R | PMID: 32290757 | qRT-PCR primers | 5’-GTTGCACTGA CCGTTCTTCTT-3’ |
| Sequence-based reagent | qPCR_Hcn1-F | PMID: 22086960 (Primer Bank ID: 6754168 a1) | qRT-PCR primers | 5’-CAAATTCTCC CTCCGCATGTT-3’ |
| Sequence-based reagent | qPCR_Hcn1-R | PMID: 22086960 (Primer Bank ID: 6754168 a1) | qRT-PCR primers | 5’-TGAAGAACGT GATTCCAACTGG-3’ |
| Sequence-based reagent | qPCR_Hcn4-F | PMID: 32290757 | qRT-PCR primers | 5’-GGCGGACACC GCTATCAAA-3’ |
| Sequence-based reagent | qPCR_Hcn4-R | PMID: 32290757 | qRT-PCR primers | 5’-TGCCGAACAT CCTTAGGGAGA-3’ |
| Sequence-based reagent | qPCR_Cacna1d-F | PMID: 22086960 (Primer Bank ID: 27413155 a1) | qRT-PCR primers | 5’-GACTGATGCC CGATATAAAGGC-3’ |
| Sequence-based reagent | qPCR_Cacna1d-R | PMID: 22086960 (Primer Bank ID: 27413155 a1) | qRT-PCR primers | 5’-CCTTCACCAGA AATAGGGAGTCT-3’ |
| Sequence-based reagent | qPCR_Cacna1g-F | PMID: 32290757 | qRT-PCR primers | 5’-TGTCTCCGCA CGGTCTGTAA-3’ |
| Sequence-based reagent | qPCR_Cacna1g-R | PMID: 32290757 | qRT-PCR primers | 5’-AGATACCCAA AGCGACCATCTT-3’ |
| Sequence-based reagent | qPCR_Cacna1h-F | PMID: 32290757 | qRT-PCR primers | 5’-GAACGTGGTT CTTTACAACGGC-3’ |
| Sequence-based reagent | qPCR_Cacna1h-R | PMID: 32290757 | qRT-PCR primers | 5’-GCACATAGTT CCCAAAGGTCA-3’ |
| Sequence-based reagent | qPCR_Smpx-F | PMID: 22086960 (Primer Bank ID: 14149752 a1) | qRT-PCR primers | 5’-ATGTCGAAGC AGCCAATTTCC-3’ |
| Sequence-based reagent | qPCR_Smpx-R | PMID: 22086960 (Primer Bank ID: 14149752 a1) | qRT-PCR primers | 5’-TCAGACAAGT TGACAACAGGTC-3’ |
| Sequence-based reagent | qPCR_Col1a1-F | PMID: 22086960 (Primer Bank ID: 34328108 a1) | qRT-PCR primers | 5’-GCTCCTCTTA GGGGCCACT-3’ |
| Sequence-based reagent | qPCR_Col1a1-R | PMID: 22086960 (Primer Bank ID: 34328108 a1) | qRT-PCR primers | 5’-CCACGTCTCA CCATTGGGG-3’ |
| Sequence-based reagent | qPCR_Postn-F | PMID: 22086960 (Primer Bank ID: 7657429 a1) | qRT-PCR primers | 5’-CCTGCCCTTA TATGCTCTGCT-3’ |
| Sequence-based reagent | qPCR_Postn-R | PMID: 22086960 (Primer Bank ID: 7657429 a1) | qRT-PCR primers | 5’-AAACATGGTCA ATAGGCATCACT-3’ |
Additional files
-
Source data 1
Expanded statistical analysis of the qRT-PCR data presented in Figures 1C, 3L and 4A–C.
Statistical analyses were performed in R (v4.2.0). Normality for each experimental group was assessed using the Shapiro-Wilk test. Group comparisons were conducted using Welch’s t-test, with multiple testing correction by the Benjamini & Hochberg method to control the false discovery rate (FDR) (74). If FDR < 0.05 but normality was rejected (Shapiro-Wilk P < 0.05), significance was confirmed using a one-tailed Wilcoxon rank-sum test (P ≤ 0.05).
- https://cdn.elifesciences.org/articles/102027/elife-102027-data1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/102027/elife-102027-mdarchecklist1-v1.docx





