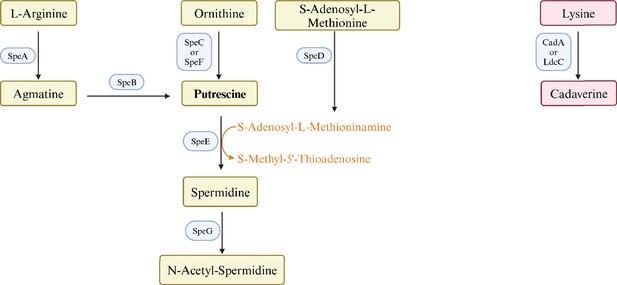
Control of pili synthesis and putrescine homeostasis in Escherichia coli
Figures
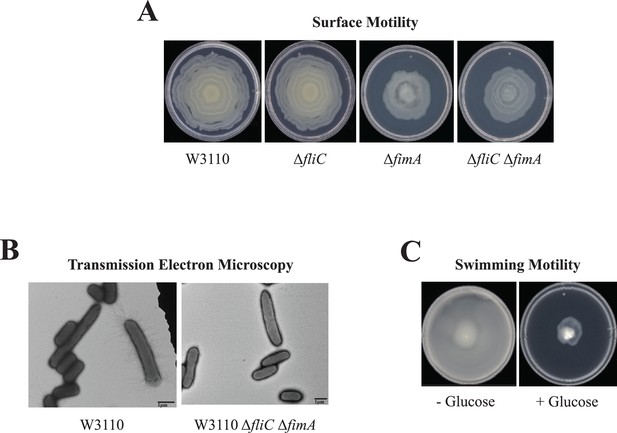
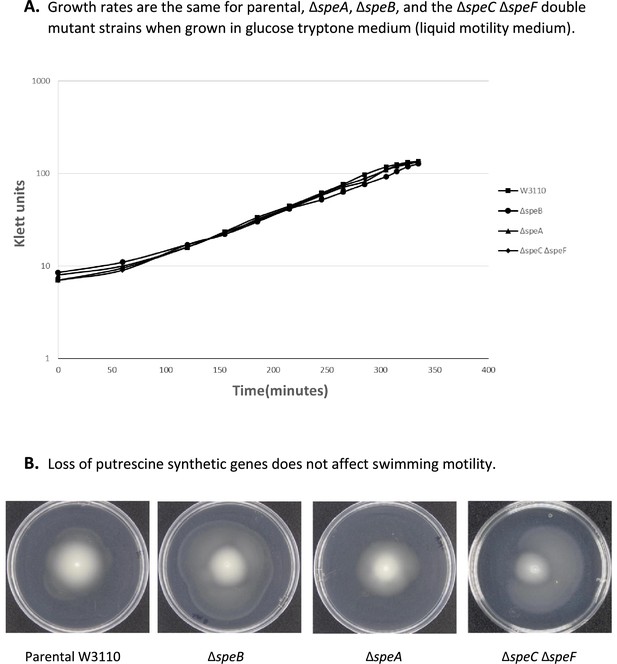
Surface motility of W3110.
(A) Surface motility of parental and derivative strains lacking the major subunits of the flagella (FliC), pili (FimA), or both. (B) Transmission electron microscopy (TEM) images of cells of W3110 and W3110 ΔfliC ΔfimA were taken from the movement’s edge directly from surface motility plates. (C) Swimming motility with and without 0.5% glucose.
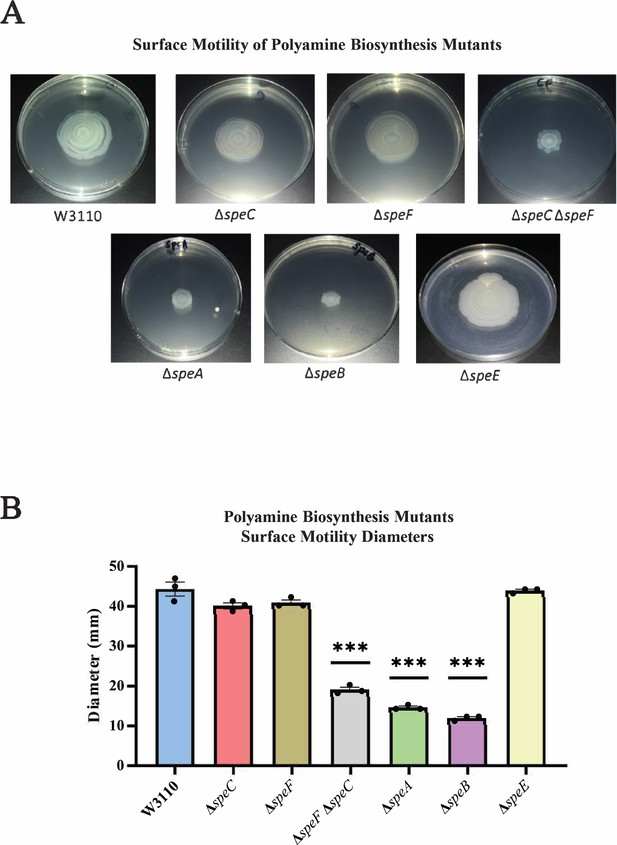
Genetics of surface motility.
(A) Pili-dependent surface motility (PDSM) of mutants with defects in polyamine anabolic genes. All assays were performed in triplicate, and representative images are shown. (B) Diameter of surface movement of polyamine mutants after 36 hr. Error bars represent standard deviations for three independent replicates. Statistical analysis was performed using the Dunnett test of significance: *p<0.05; **p<0.01; ***p<0.001. In this figure the ΔspeB mutant was IM26.
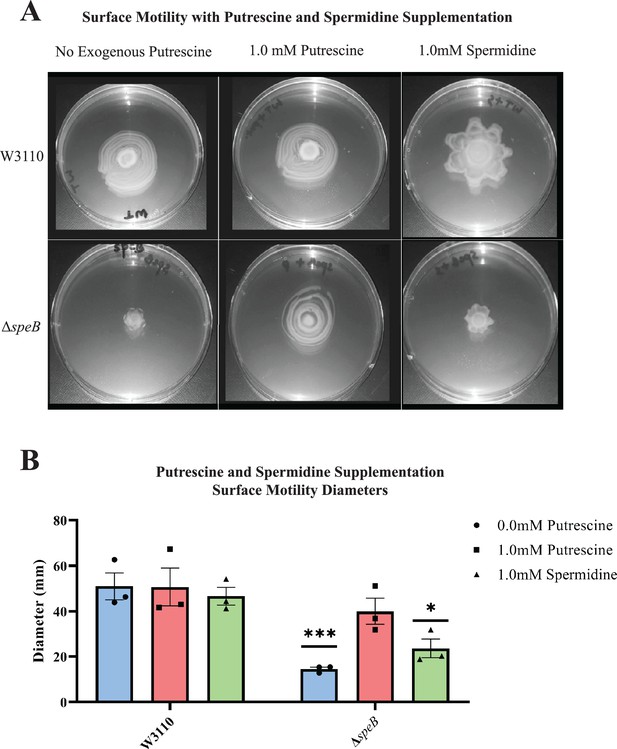
Nutritional supplementation of ΔspeB mutant.
(A) Putrescine and spermidine supplementation of wild-type and ΔspeB strains. (B) Diameter of surface movement of polyamine mutants after 36 hr. Error bars represent standard deviations for three independent replicates. Statistical analysis was performed using the Sadik test of significance: *p<0.05; **p<0.01; ***p<0.001. All assays were performed in triplicates and representative images are shown. In this figure the ΔspeB mutant was IM26.
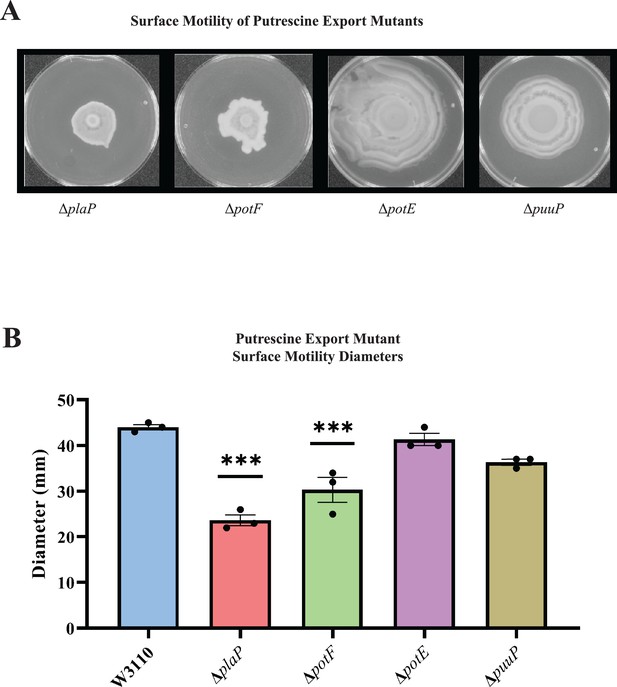
Putrescine transport and surface motility.
(A) Representative images of surface motility for strains defective in putrescine transport genes. (B) Diameter of surface movement of polyamine mutants after 36 hr. Error bars represent standard deviations for three independent replicates. Statistical analysis was performed using the Dunnett test of significance: *p<0.05; **p<0.01; ***p<0.001. All assays were performed in triplicate and representative images are shown.
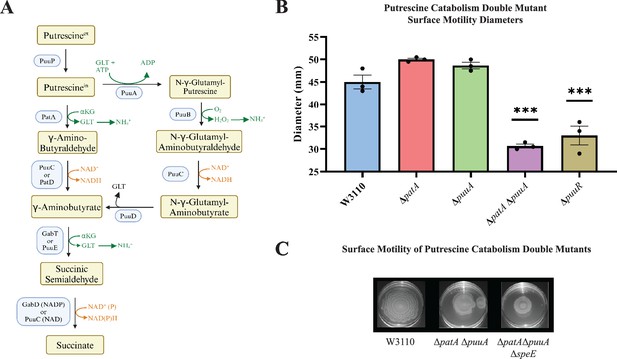
Putrescine catabolism and surface motility.
(A) Pathways and enzymes of putrescine catabolism. (B) Motility diameter of putrescine catabolic mutants after 36 hr. Error bars represent standard deviations for three independent replicates. Statistical analysis was performed using the Dunnett test of significance: *p<0.05; **p<0.01; ***p<0.001. All assays were performed in triplicates and representative images are shown. (C) Effect of loss of speE on the patA puuA catabolic double mutant.
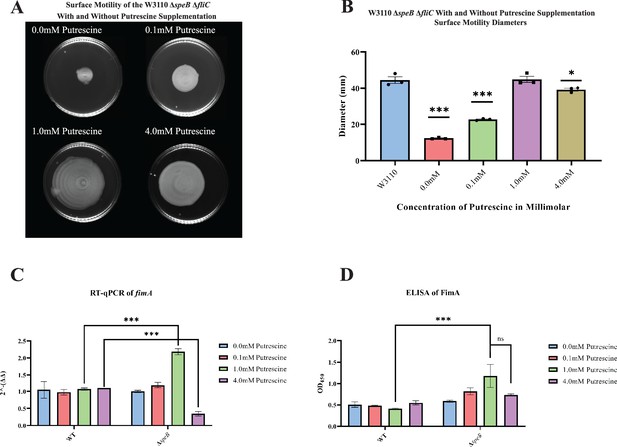
Surface motility and pili expression in W3110 and ΔspeB.
(A) W3110 ΔspeB surface motility with 0.0, 0.1, 1.0, and 4.0 mM exogenous putrescine. All assays were performed in triplicate. Representative images are shown. (B) Average diameter of three separate surface motility plates. The parental strain without putrescine is shown for reference. Significance was determined by comparing the diameters of the ΔspeB mutants in the different concentrations of putrescine compared to the parental W3110. One-way ANOVA was used with Dunnett hypothesis testing to determine p values, ***p<0.001. (C) Reverse transcriptase-quantitative PCR using primers targeting the fimA gene from cells grown in 0.0, 0.1, 1.0, and 4.0 mM of supplemented putrescine. Double deltas were generated by normalizing the parental and the ΔspeB mutant RNA libraries to rpoD t hen comparing fimA expression. Two-way ANOVA was used to determine significance followed by Sadik hypothesis testing. ***p<0.001. (D) Enzyme linked immunosorbent assays using antibodies targeting pili (FimA) from cells grown with 0.0, 0.1, 1.0, and 4.0 mM of supplemented putrescine. Two-way ANOVA was used to determine significance followed by FDR adjusting. ***p<0.001. In this figure the ΔspeB mutant was J15.
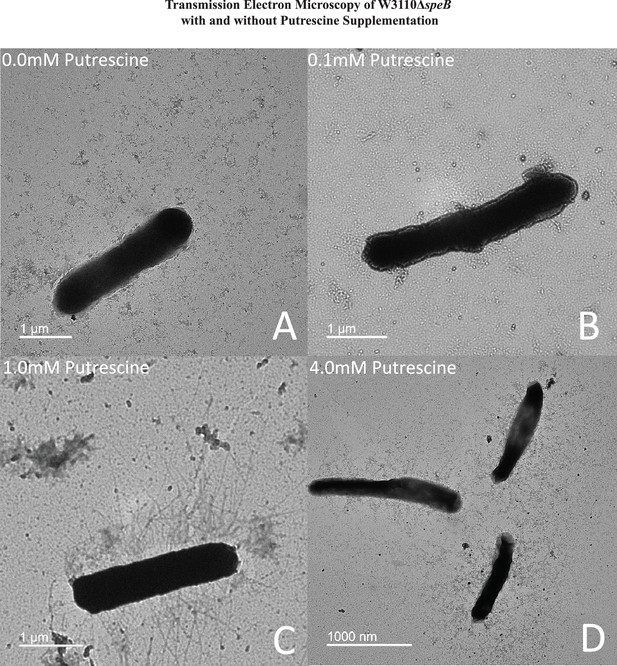
Transmission electron micrographs of ΔspeB mutant cells after surface motility.
Representative images are shown and cells were removed from motility plates with the following exogenous putrescine concentrations: (A) none, (B) 0.1 mM, (C) 1.0 mM, and (D) 4.0 mM. No pili were observed with 0 and 0.1 mM putrescine. Optimal pili production was observed with 1.0 mM putrescine. The bar represents one micron. In this figure the ΔspeB mutant was J15.
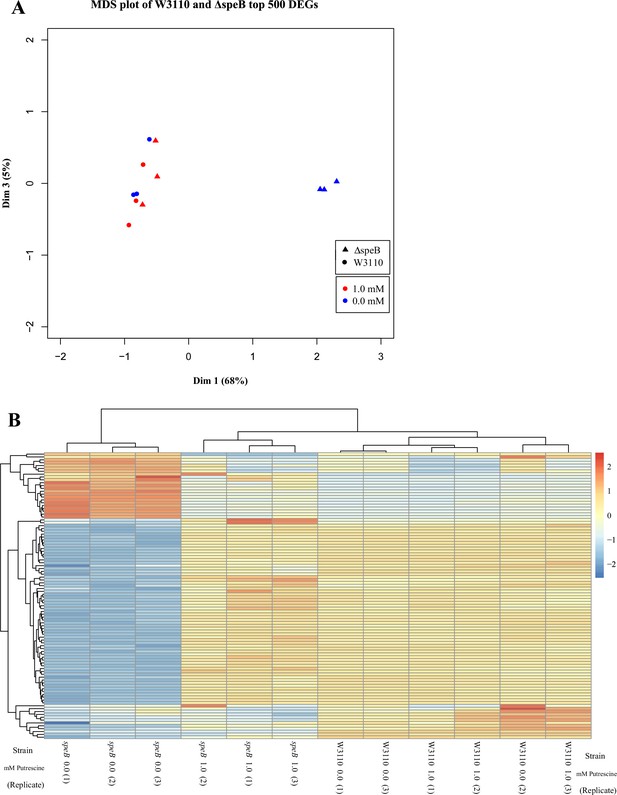
Visual representation of the results from transcriptomic sequencing of W3110 and the ΔspeB mutant’s gene expression in media with and without 1.0 mM putrescine.
(A) Multidimensional scaling plot of the parental and ΔspeB mutant transcriptomes. When grown with 1.0 mM putrescine (red), the ΔspeB mutant transcriptome (triangles) are nearly identical to the parental transcriptomes when grown without and with putrescine (blue and red circles, respectively). When grown without putrescine, the ΔspeB mutant transcriptome (blue triangles) is greatly skewed from transcriptomes of the parental strain and the ΔspeB mutant grown with putrescine. (B) Heatmap of the top 100 most variable genes further demonstrates the distinctiveness of the ΔspeB mutant grown without putrescine and the similarities of the ΔspeB mutant transcriptome when grown with 1.0 mM putrescine supplementation and the parental grown with or without putrescine. In this figure the ΔspeB mutant was J15.
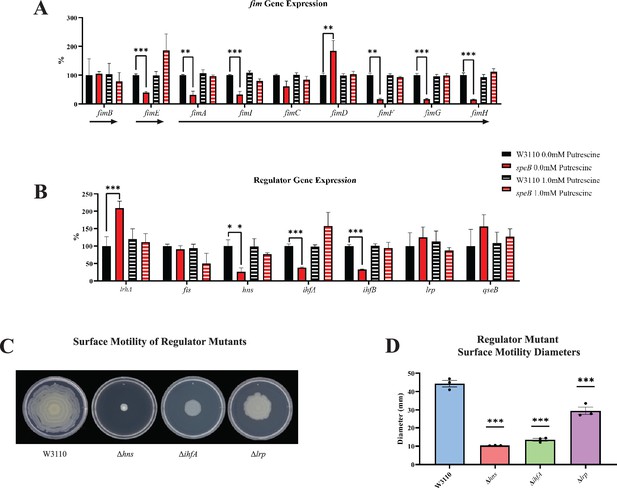
Transcriptomic sequencing of genes for the fim region and regulators that control fim gene expression.
(A) Expression of the fim genes in the parental W3110 and the speB mutant with and without putrescine supplementation. Values were calculated by dividing the individual replicates’ counts per million (CPM) value for each gene by the mean CPM of that gene in W3110 grown without putrescine and multiplying by 100 to yield the percent expression. Arrows below the genes signify the known operons: fimB and fimE belong to single gene operons, while fimAICDFGH belongs to one operon. One-way ANOVA was used to determine significance using Dunnett hypothesis testing. **p<0.01; ***p<0.001. (B) Expression of some regulators known to affect fim gene expression. Values were calculated as described in (A). ***p<0.001. (C) Surface motility of W3110 and three regulator mutants (Δhns, ΔihfA, and Δlrp). A gene found to be significantly different by this transcriptomic analysis (hns) was confirmed to be important in surface motility. In this figure the ΔspeB mutant was J15. (D) Diameter of surface movement of regulatory mutants after 36 hr. Error bars represent standard deviations for three independent replicates. Statistical analysis was performed using the Dunnett test of significance: *p<0.05; **p<0.01; ***p<0.001. All assays were performed in triplicate.

Phase variation in parental W3110, W3110 ΔspeB, W3110 Δhns, and MG1655.
(A) The diagram shows the genes for the FimB and FimE recombinases, the invertible fimS region which contains the promoter for the fim operon, and the fim operon which codes for the proteins of the type 1 pilus. Primer pairs 1–2 and 1–3 detect the fimS region in the phase OFF and ON orientations, respectively. The DNA sizes for phases OFF and ON are 884 and 394, respectively, for wild-type strains of E. coli, such as MG1655. Our lab strain of W3110 has an IS1 element insertion in fimE which increases the size of the amplified DNA fragment. MG1655 was analyzed as a control. Primer 1 is 5’-CCGCGATGCTTTCCTCTATG-3’; primer 2 is 5’-TAATGACGCCCTGAAATTGC-3’; and primer 3 is 5’-TGCTAACTGGAAAGGCGCTG-3’ (shown schematically). (B) Deletion of either speB or hns had no effect on fimS orientation. A possible explanation for the loss of pili or PDSM in the speB or hns mutants is locking the fimS switch in phase OFF. However, loss of either speB or hns had no effect on fimS orientation in W3110 (lanes 2–4), and putrescine did not alter fimS orientation of the W3110 ΔspeB mutant (lanes 6 and 7). We conclude that loss of speB in W3110 did not phase-lock fimS in phase OFF. Also note that W3110, which has an insertion in fimE, is not locked in phase ON.
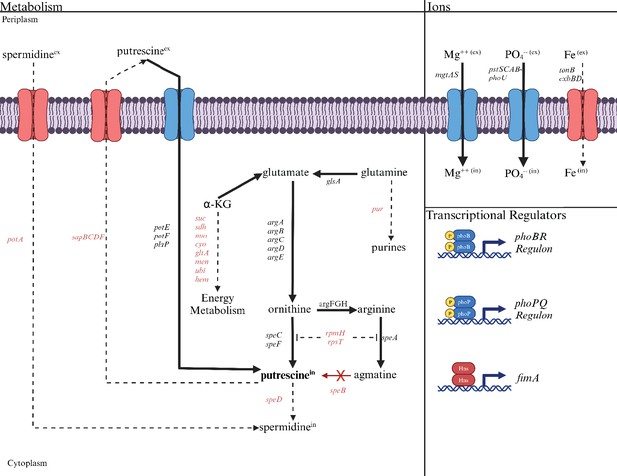
The deduced diversion of metabolism in a speB mutant away from energy metabolism toward putrescine synthesis compared to the parental strain.
The effects of low putrescine are shown. Genes and processes (transport and transcriptional regulators) in red have fewer transcripts, while those in black or blue have more. The dashed lines represent the proposed reduction in metabolic flux because of fewer transcripts from genes coding for the enzymes involved. Operons are shown except for the larger operons or regulons. For example, pur is meant to represent the unlinked genes that code for enzymes of purine synthesis. Table 2 or Supplementary file 2 should be consulted for specific genes and the quantitative change in transcripts.
Tables
Pairwise statistical comparisons of transcriptomes.
Abbreviation: putr is putrescine.
| Strain 1 | Strain 2 | R2 |
|---|---|---|
| Wild-type (1 mM putr) | wild-type (0 mM putr) | 0.950 |
| Wild-type (1 mM putr) | ΔspeB (1 mM putr) | 0.954 |
| ΔspeB (0 mM putr) | ΔspeB (1 mM putr) | 0.820 |
| ΔspeB (0 mM putr) | wild-type (0 mM putr) | 0.796 |
| ΔspeB (0 mM putr) | wild-type (1 mM putr) | 0.786 |
Differentially expressed genes that are proposed to contribute to putrescine homeostasis.
A positive number means more transcripts in the ΔspeB (lower putrescine) strain.
| Gene (function) | log2FC | FDR |
|---|---|---|
| Polyamine synthesis | ||
| speA (putrescine) | 0.99 | 0.003 |
| speC (putrescine) | 1.43 | 3E-4 |
| speD (spermidine) | –0.83 | 0.01 |
| speF (putrescine) | 1.05 | 0.007 |
| rpmH (inhibitor of SpeA and SpeC) | –2.77 | 2E-4 |
| rpsT (inhibitor of SpeA and SpeC) | –1.92 | 8E-4 |
| Polyamine transport | ||
| potABCD* (spermidine) | –2.17 | 3E-4 |
| potE (putrescine) | 2.07 | 2E-4 |
| potFGHI* (putrescine) | 1.41 | 4E-4 |
| plaP (putrescine) | 1.11 | 0.005 |
| sapBCDF* (putrescine export) | –1.18 | 8E-4 |
| Arginine synthesis | ||
| argA_2 (synthesis) | 1.77 | 0.001 |
| argB (synthesis) | 1.87 | 0.002 |
| argC (synthesis) | 2.86 | 0.005 |
| argD (synthesis) | 1.41 | 0.001 |
| argF (synthesis) | 1.63 | 0.01 |
| argG (synthesis) | 2.32 | 0.002 |
| argH (synthesis) | 0.97 | 0.001 |
| argI (synthesis) | 1.95 | 0.018 |
| argR (repressor of arginine regulon) | –1.35 | 4E-4 |
| Glutamate generation | ||
| glsA (glutamine degradation) | 2.16 | 5E-4 |
| glnG (regulation of ammonia assimilation) | –1.59 | 0.022 |
| TCA cycle/electron transport | ||
| acnA (TCA cycle) | –1.04 | 8E-4 |
| acnB (TCA cycle) | –1.96 | 2E-4 |
| fumA (TCA cycle) | –1.50 | 6E-4 |
| fumB (TCA cycle) | –1.49 | 0.003 |
| gltA (TCA cycle) | –1.64 | 2E-4 |
| icdA (TCA cycle) | –1.32 | 0.002 |
| sdhCDAB-sucABCD* (TCA cycle) | –3.88 | 5E-5 |
| nuo operon* (electron transport) | –0.82 | 0.004 |
| menFDHBCE* (electron transport) | –0.8 | 0.005 |
| ubiEJB* (electron transport) | –0.93 | 0.004 |
| cyoABCD* (electron transport) | –1.41 | 9E-4 |
| hemCD* (heme) | –2.05 | 2E-4 |
| Iron transport | ||
| exbBD* (transport of all iron chelates) | –2.33 | 4E-5 |
| tonB (transport of all iron chelates) | –1.9 | 4E-5 |
| entCEBA* (enterochelin iron) | –5.2 | 7E-6 |
| fecABCDE * (ferric citrate) | –4.4 | 3E-5 |
| fecIR (regulators, ferric citrate) | –4.4 | 6E-6 |
| feoABC* (ferrous iron) | –3.4 | 7E-4 |
| fur (regulator, iron assimilation) | –0.40 | 0.11 |
| Magnesium and phosphate transport | ||
| mgtA (magnesium) | 5.90 | 5E-6 |
| phoQ (regulator, magnesium) | 0.79 | 0.009 |
| pstSCAB* (phosphate) | 3.61 | 0.01 |
| phoBR* (regulator, phosphate assimilation) | 2.80 | 0.02 |
-
*
Values for transcripts of the first gene of the operon are given. Results for other genes of the operon are in Supplementary file 2.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Escherichia coli) | speB | PMID:16738554 | P1 transduction construction | |
| Gene (E. coli) | various, see strain list | PMID:16738554 | P1 transduction construction | |
| Strain, strain background (E. coli) | W3110 | other | Lab strain; history provided in Methods | |
| Genetic reagent (E. coli transducing virus) | P1 vir | other | Lab strain | |
| Antibody | anti-E. coli RpoD (rabbit polyclonal) | Cusabio | Cat #: CSB-PA360419XA01ENV RRID:AB_3678626 | 1:10,000 |
| Antibody | anti-E. coli FimA (rabbit polyclonal) | Cusabio | Cat #: CSB-PA361210ZA01ENV RRID:AB_3678627 | 1:10,000 |
| Antibody | Goat anti-rabbit IgG (H+L) HRP conjugated | Cusabio | Cat #: CSB-PA489724 RRID:AB_3678628 | 1:10,000 |
| Sequence-based reagent | FimA forward | This paper | PCR primers | ATGGTGGGACCGTTCACTTT |
| Sequence-based reagent | FimA reverse | This paper | PCR primers | GGCAACAGCGGCTTTAGATG |
| Sequence-based reagent | RpoD forward | This paper | PCR primers | TCGTGTTGAAGCAGAAGAAGCG |
| Sequence-based reagent | RpoD reverse | This paper | PCR primers | TCGTCATCGCCATCTTCTTCG |
| Sequence-based reagent | Phase variation test primer 1 | This paper | PCR primers | CCGCGATGCTTTCCTCTATG |
| Sequence-based reagent | Phase variation test primer 2 | This paper | PCR primers | TAATGACGCCCTGAAATTGC |
| Sequence-based reagent | Phase variation test primer 3 | This paper | PCR primers | TGCTAACTGGAAAGGCGCTG |
| Commercial assay or kit | TMB substrate kit | ThermoFisher | Cat #: 34021 | |
| Commercial assay or kit | Ligation sequencing DNA V14 | Oxford Nanopore Technologies | Cat #: SQK-LSK114 | |
| Commercial assay or kit | Rneasy Mini Kit | Qiagen | Cat #: 74104 | |
| Commercial assay or kit | RiboMinus Transcriptome Isolation Kit, bacteria | ThermoFisher | Cat #: K155004 | |
| Commercial assay or kit | RiboCop rRNA Depletion Kits for Bacteria | Lexogen | Cat #: 126.24 | |
| Commercial assay or kit | LunaScript RT Super | NEB | Cat #: E3010 | |
| Commercial assay or kit | PowerUp STBR Green Master Mix for qPCR | ThermoFisher | Cat #: A25777 | |
| Chemical compound, drug | Eiken Agar | Eiken Chemical Co., Ltd, Tokyo, Japan | Cat #: E-MJ00 | |
| Software, algorithm | bcl2fastq (v 4.2.4) | Illumina | RRID:SCR_015058 | |
| Software, algorithm | Guppy basecaller (v 6.5.7) | Oxford Nanopore Technologies | RRID:SCR_023196 | |
| Software, algorithm | Porechop (v 0.2.4) | Oxford Nanopore Technologies | RRID:SCR_016967 | |
| Software, algorithm | Flye (v 2.9.2) | PMID:27956617 | RRID:SCR_017016 | |
| Software, algorithm | Pilon (v 1.24) | https://doi.org.10.1371/journal.pone.0112963 | RRID:SCR_014731 | |
| Software, algorithm | Circulator (v 1.5.5) | PMID:26714481 | ||
| Software, algorithm | Prokka (v 1.14.6) | https://doi.org.10.1093/bioinformatics/btu153 | RRID:SCR_014732 | |
| Software, algorithm | QUAST (v 5.2.0) | PMID:23422339 | RRID:SCR_001228 | |
| Software, algorithm | CLC Genomics Workbench | Qiagen | RRID:SCR_011853 | |
| Software, algorithm | edgeR | PMID:19910308 | RRID:SCR_012802 |
Strains.
| Strain name | Genotype | Reference |
|---|---|---|
| BLS77 | W3110 ∆puuR::cat | Schneider and Reitzer, 2012 |
| BLS80 | W3110 ∆puuA::cat | Schneider and Reitzer, 2012 |
| BLS88 | W3110 ∆patA ∆puuA | Schneider and Reitzer, 2012 |
| CP2 | W3110 ∆patA | Schneider and Reitzer, 2012 |
| IM26 | W3110 ∆speB::kan fliC-lacZ | This study |
| IM27 | W3110 ∆speE::kan fliC-lacZ | This study |
| IM28 | W3110 ∆speF::cat fliC-lacZ | This study |
| IM29 | W3110 ∆speC::kan fliC-lacZ | This study |
| IM34 | W3110 ∆speC ∆speF::cat fliC-lacZ | This study |
| IM60 | W3110 ∆cadA::kan fliC-lacZ | This study |
| IM61 | W3110 ∆speA::kan | This study |
| IM62 | W3110 ∆patA ∆puuA ∆speE::kan | This study |
| IM63 | W3110 ∆potE::kan | This study |
| IM64 | W3110 ∆potF::kan | This study |
| IM65 | W3110 ∆plaP::kan | This study |
| IM66 | W3110 ∆puuP::kan | This study |
| J15 | W3110 ΔspeB ΔfliC::kan | This study |
| SA1 | W3110 ∆fliC::kan | This study |
| SA2 | W3110 ∆fimA::kan | This study |
| SA3 | W3110 ∆fliC ∆fimA | This study |
| SA4 | W3110 ∆hns::kan | This study |
| SA5 | W3110 ∆ihfA::kan | This study |
| SA6 | W3110 ∆lrp::kan | This study |
| W3110-LR referred to as W3110 | lacIqlacL8 | Lab strain |
Additional files
-
Supplementary file 1
Expression graphs comparing the logCPM of the transcriptomes of the average of the three replicates of the W3110 with and without 1 mM putrescine and the speB mutant with and without 1 mM putrescine.
A regression line was calculated and the correlation between each set of transcriptomes was noted on the graph. Higher R2 values indicate greater similarity between the transcriptomes.
- https://cdn.elifesciences.org/articles/102439/elife-102439-supp1-v1.pdf
-
Supplementary file 2
Differential gene expression analysis of W3110 and ΔspeB.
J15 refers to the ΔspeB mutant. In the tab labeled ‘W3110_0_vs_J15_1_ Putrescine’ W3110 was grown without putrescine, and J15 was grown with 1.0 mM putrescine; positive fold change values refer to increases in J15 gene expression.
- https://cdn.elifesciences.org/articles/102439/elife-102439-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/102439/elife-102439-mdarchecklist1-v1.docx