Resolving dual binding conformations of cellulosome cohesin-dockerin complexes using single-molecule force spectroscopy
Figures
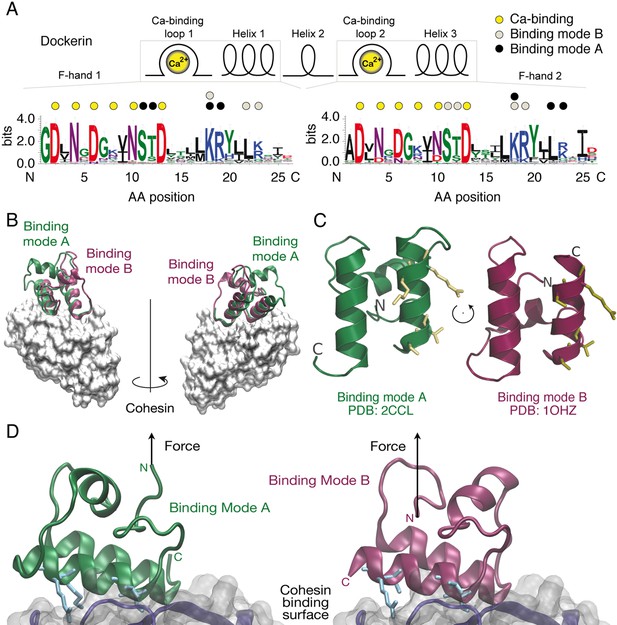
Cohesin:Dockerin dual binding modes.
(A) Secondary structure and consensus sequence logo (Crooks, 2004) assembled from 65 putative Ct type-I Doc variants. Dots above the amino acid codes indicate residues involved in: Ca2+ coordination (yellow), mode A binding (black), and mode B binding (gray). Letter colors represent chemical properties: Green, polar; purple, neutral; blue, basic; red, acidic; black, hydrophobic. Crucial Coh-binding residues are located at positions 11, 12, 18, 19, 22, and 23 in each F-hand motif. (B) Coh:Doc complex crystal structures showing overlaid Doc domains in the two binding modes. Images were generated by aligning the Coh domain (gray) from PDB 2CCL (green, binding mode (A) and 1OHZ (red, binding mode (B) using the VMD plugin MultiSeq (Humphrey et al., 1996; Roberts et al., 2006). (C) View of the Doc binding interface for each mode from the perspective of Coh. The conserved binding residues at positions 11, 12, 18, and 19 in the F-hand motif relevant for binding in the corresponding mode are depicted as stick models (yellow). (D) Close-up view of the interface for each binding mode with arrows indicating the location and direction of applied force. Binding residues 11, 12, 18, and 19 for binding mode A and 45, 46, 52, and 53 for binding mode B are shown as blue stick models. The Coh domain is oriented the exact same way in both views.
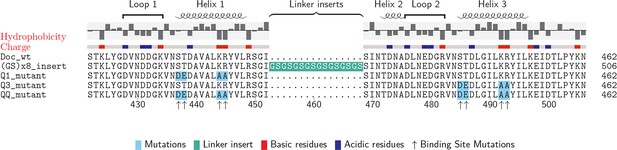
Doc sequences used in this study (N- to C-terminus).
Doc_wt: wild-type sequence; hydrophobicity and charge graphs are displayed for the wild-type-Doc (red: positively charged, blue: negatively charged); (GS)x8_insert: A (Gly-Ser)8 linker was incorporated between helix 1 and helix 2; Q1_mutant: Quadruple mutant in helix 1. Four point mutations (DE/AA) were incorporated into Doc helix 1 to knock out binding mode A; Q3_mutant: Quadruple mutant in helix 3. Four point mutations (DE/AA) were incorporated into Doc helix 3 to knock out binding mode B; QQ_mutant: Non-binding control with both binding modes knocked out. Numbers below indicate amino acid number of the fusion protein construct starting from the xylanase N-terminus.
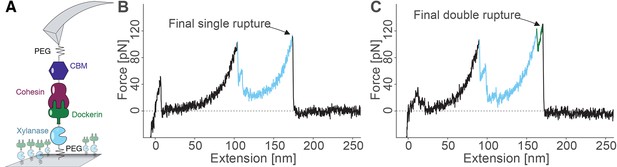
Overview of the experimental configuration and recorded single-molecule unfolding and unbinding traces.
(A) Schematic depiction showing the pulling geometry with CBM-Coh on the AFM Cantilever and Xyn-Doc on the glass substrate. Each fusion protein is site-specifically and covalently immobilized on a PEG-coated surface. (B-C) Each force vs. extension trace shows PEG linker stretching (black), xylanase unfolding and subsequent stretching (blue), and Coh:Doc complex rupture. The Coh:Doc complex rupture occurred in two distinct event types: single (B) and double (C) ruptures. The 8-nm contour length increment separating the double peaks was assigned to Doc unfolding (C, green).
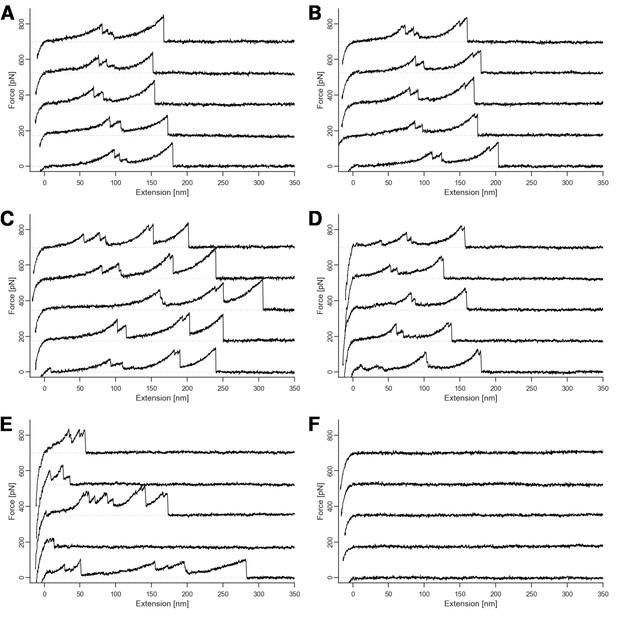
Representative sample of force traces.
Traces showing xylanase unfolding and (A) single-event rupture, (B) double-event rupture, (C) CBM unfolding prior to unbinding, (D) shielded double-event for Q3 mutants, (E) nonspecific multiple interactions, and (F) no interactions. Traces such as E and F were excluded from the analysis. Multiple traces are offset on the force axis for readability.
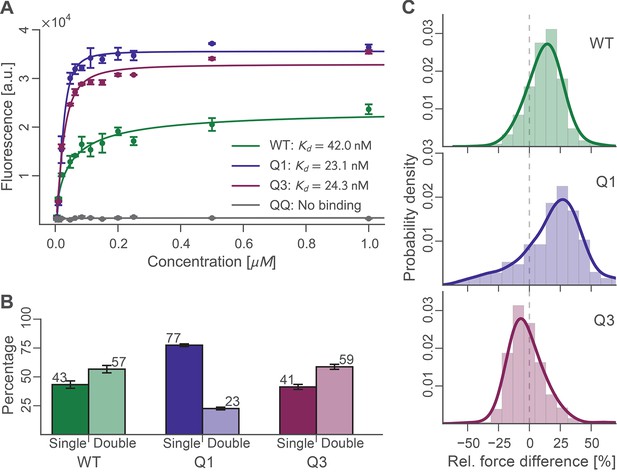
Bulk and single-molecule characterization of Doc mutants.
(A) Fluorescence binding curve showing binding of TagRFP-labelled Coh to wild-type and mutant Doc nonspecifically immobilized in a 96-well plate. Both Q1 and Q3 mutants bound TagRFP-Coh similarly to wild-type with dissociation constants (KD) in the low nM range. The negative control QQ mutant showed no binding. Solid lines are 4 parameter logistic nonlinear regression model fits to the data. Error bars represent the standard deviation of three independent samples. (B) Event probabilities for single (opaque colors) and double (translucent colors) Coh:Doc rupture peaks determined for Doc wild-type and DE/AA quadruple mutants. Data originate from 947, 4959, and 1998 force-extension traces from wild-type, Q1 and Q3 variants, respectively. Error bars represent 95% Clopper-Pearson confidence intervals based on the beta probability distribution. (C) Relative difference in double peak rupture forces for the different Doc variants. Positive values indicate a stronger final peak. Histograms represent concatenated data from various pulling speeds. Drawn lines are kernel density estimates calculated on the raw data.
-
Figure 4—source data 1
Probability Data.
Single/double peak counting statistics displayed in Figure 4B.
- https://doi.org/10.7554/eLife.10319.008
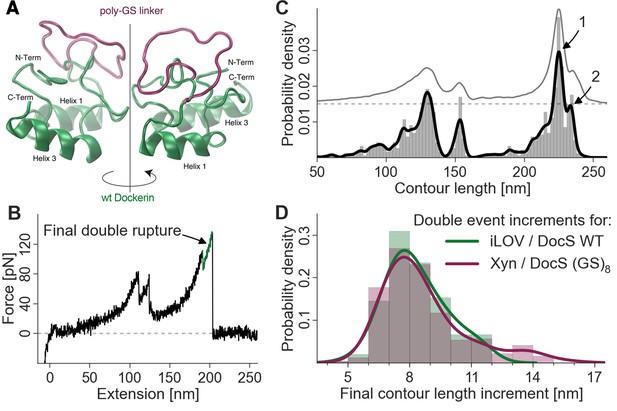
Probing the final contour length increment with Poly (GS) inserts.
(A) Structural homology model overlay of wild-type and mutant Doc containing a (GS)8-linker between helix 1 and helix 3. The wild-type Doc is shown in green. The 16 amino acid long GS-insert is shown in purple (Kelley and Sternberg, 2009) (remaining Doc domain not shown). (B) Typical force extension trace with final double rupture event depicted in green (arrow). (C) Histogram and kernel density estimate of the transformation of the single force extension trace in panel B into contour length space (black) and kernel density estimate of the whole dataset of single molecule Xyn-Doc:Coh-CBM traces bearing xylanase fingerprint and final double rupture (gray, offset in y-direction for readability) in contour length space. (D) Histograms (bars, bin width: 1 nm), kernel density estimates (drawn lines, bandwidth: 0.75 nm, gaussian kernel), and statistical test (Kolmogorov-Smirnov, ‘KS test’) are each calculated on the raw data of the final increments (peak-to-peak distances) in contour length space (x-distance between arrow 1 and 2 in panel (C). Maxima for final double event increments lie at 7.75 nm and 7.73 nm for iLOV-Coh:Doc (wild-type)-sfGFP (N = 255) and Xyn-Doc (GS)8:Coh-CBM (N = 320) final ruptures, respectively (a two-sample KS test on the raw data indicates no significant difference in the data distributions (p-value of 21.7%).
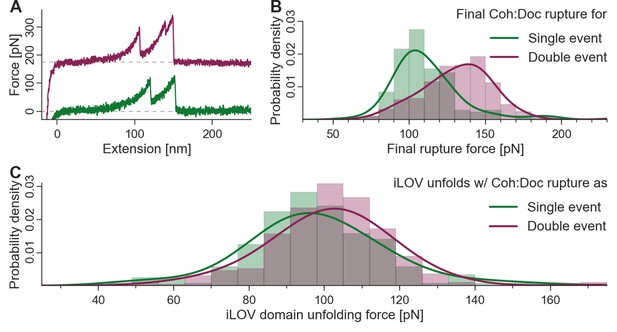
Biasing of unfolding force distributions by dual binding mode.
(A) Typical force traces showing iLOV unfolding with final single (green) and double (purple) complex ruptures. The curve terminating in a double peak is offset in the y-direction for clarity. (B) Final complex rupture force distribution for single and double events. Double events are more mechanically stable. (C) iLOV domain unfolding forces for final single (green) and double (red) events at a pulling velocity of 800 nm/s. Histograms (bars), kernel density estimates (lines), and statistical tests are each obtained from the raw data. Maxima for iLOV unfolding lie at 96.0 pN and 102.7 pN for single (N = 172) and double (N = 277) final ruptures, respectively. A two-sample Kolmogorov-Smirnov test showed significant differences in the data distributions (p-value of 0.09%). Since the data were all recorded with a single cantilever and both event types were distributed equally throughout the runtime of the measurement, no systematic biasing is expected. Because of the lower force distribution of final single peaks, the iLOV unfolding force distribution is truncated compared to final double peak force traces, supporting the notion that the binding mode is set prior to mechanical loading of the complex.
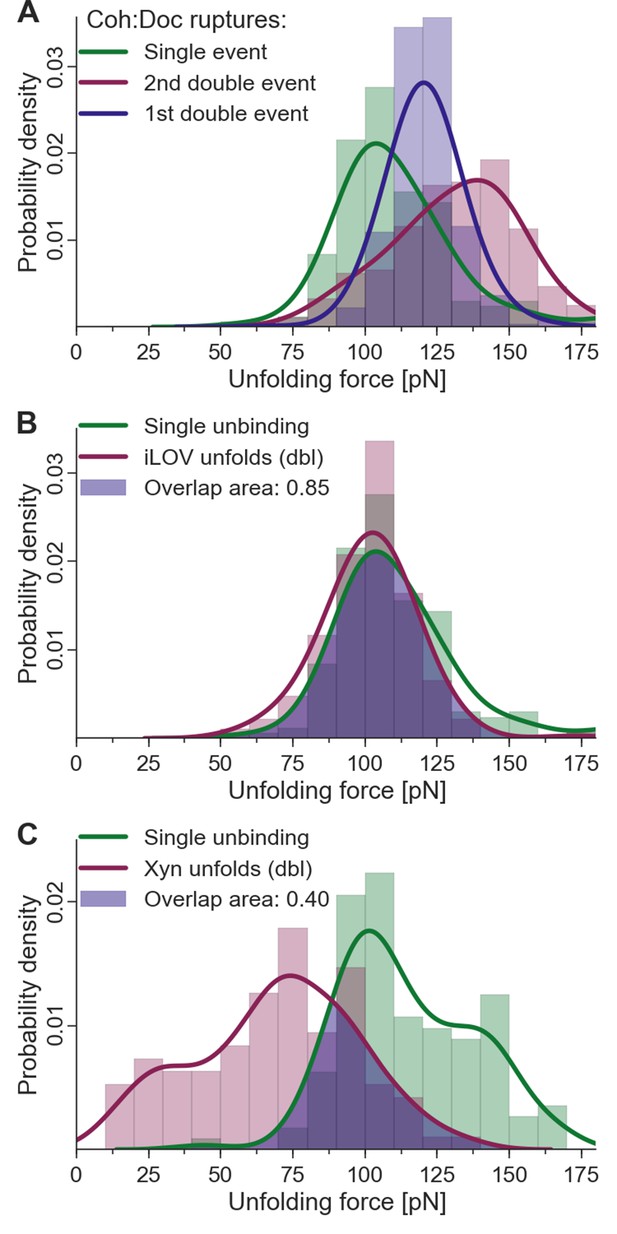
Fingerprint unfolding and complex unbinding forces.
(A) Rupture force distribution of final complex ruptures for single (green), first (purple) and second (red) double unbinding events. (B) Overlap area (purple) of iLOV domain unfolding force distribution (red) (iLOV-doubles curve class) with the rupture force distribution (green) for single-event complex ruptures. (C) Overlap area (purple) of Xyn domain unfolding force distribution (red) (Xyn-doubles curve class) with the rupture force distribution (green) for single-event complex ruptures. Overlaps in probability distributions allow normalizing single-event counts to double events to account for different biasing caused by the different unfolding forces of the fingerprint domain. Biasing occurs, because for overlapping force distributions of fingerprint unfolding and complex ruptures, unbinding events are more likely to take place without fingerprint unfolding if the two distributions are closer together. For the Coh:Doc unbinding, this effect is more pronounced for the weaker single ruptures. Because double events are also biased, this still does not give a true quantification, but only compensates for the differences of biasing. The non-bell-evans-like shape of the single rupture peaks, especially in the region of the 1st double event peak (A) suggests that this class of curves does not contain a single type of unbinding mechanism, but rather a superposition of different event types.





