PRDM16 functions as a co-repressor in the BMP pathway to suppress neural stem cell proliferation
Figures
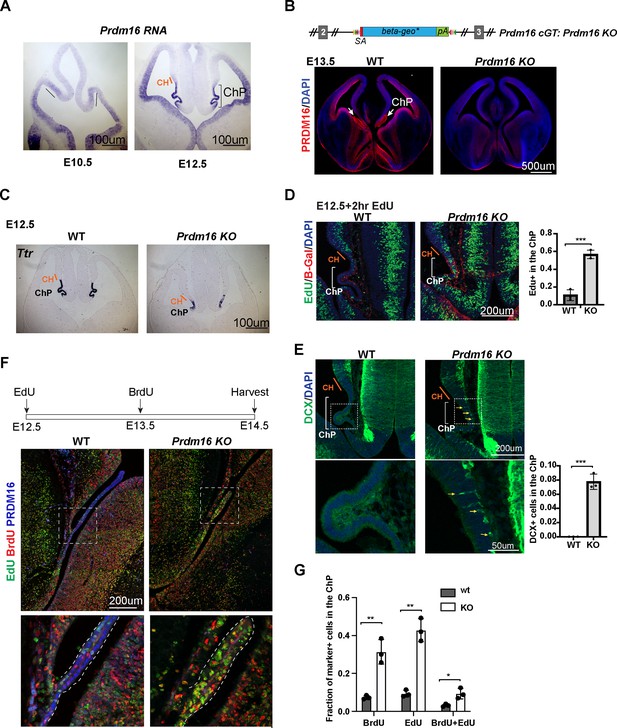
Prdm16 mutant choroid plexus (ChP) cells fail to exit the cell cycle and retain neural progenitor identity.
(A) RNA in situ hybridization using a Prdm16 probe shows Prdm16 expression in E10.5 and E12.5 brains. The line and bracket mark the developing CH and ChP. (B) Top: Schematic of the Prdm16 GT allele, as reported in Strassman et al., 2017. Bottom: immunostaining of PRDM16 in E13.5 control and null mutant brains. Red: PRDM16; blue: DAPI. White arrows indicate the ChP in the control brain. (C) RNA in situ hybridization for Ttr on E12.5 control and Prdm16 null mutant forebrains. Cortical hem (CH) regions are outlined based on morphology. (D) Two-hour EdU labeling in E12.5 control and Prdm16 homozygous brains. Sections are co-stained with anti-β-Gal antibody to identify mutant ChP cells in the KO brain. Note: the β-Geo trap is absent in wild-type animals. Brackets mark ChP regions. Red: β-Gal; blue: DAPI; green: EdU. (E) Immunostaining for Doublecortin (DCX) on E12.5 control and Prdm16 homozygous brain slices. Bottom panels show magnified views of the ChP (dashed rectangles). Yellow arrows indicate some of the DCX-positive cells. Right panels show quantification of EdU+ (D) and DCX+ cells (E). Biological replicates: N=3. Error bars represent standard deviation (SD). Statistical significance is calculated using unpaired t-test. ***p<0.001. (F) Schematic of the double S-phase labeling experiment (top). Brain sections from E14.5 control and prdm16 KO embryos were immunostained with PRDM16 and BrdU antibodies and processed for EdU detection. Bottom panels show magnified views of the ChP region. (G) Quantification of EdU+ and BrdU+ cells from the double labeling experiment. Biological replicates: N=3. Error bars represent standard deviation (SD). Statistical significance is calculated using unpaired t-test. **p<0.01. *p<0.05.
-
Figure 1—source data 1
Counting of EdU+ and DAPI stained cells, related to Figure 1D.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Counting of Dcx+ and DAPI stained cells, related to Figure 1E.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig1-data2-v1.xlsx
-
Figure 1—source data 3
Counting of EdU+, BrdU+, double positive and DAPI stained cells, related to Figure 1G.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig1-data3-v1.xlsx
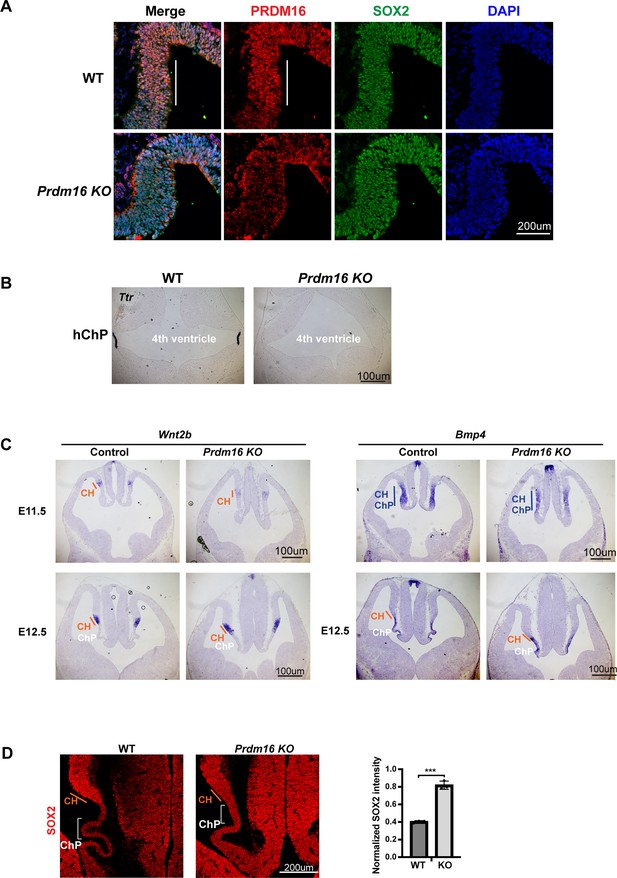
Prdm16 depletion affects choroid plexus (ChP) epithelial specification without altering dorsal midline patterning.
(A) Immunostaining of PRDM16 and SOX2 on E10.5 control and homozygous brain sections. ChP regions are outlined in white. (B) RNA in situ hybridization for Ttr in E12.5 control and Prdm16 null mutant hindbrains. (C) RNA in situ hybridization using Wnt2b and Bmp4 probes on control and mutant forebrains at E11.5 and E12.5. (D) SOX2 immunostaining on E12.5 brain sections. ChP areas are marked with brackets. Relative SOX2 signal intensity between ChP and neurogenic regions is quantified. Error bars represent standard deviation (SD), n=3. Statistical analysis by unpaired t-test (***p<0.001; **p<0.01; *p<0.05; n.s., not significant).
-
Figure 1—figure supplement 1—source data 1
Measurement of SOX2 signal intensity, related to Figure 1—figure supplement 1D.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig1-figsupp1-data1-v1.xlsx
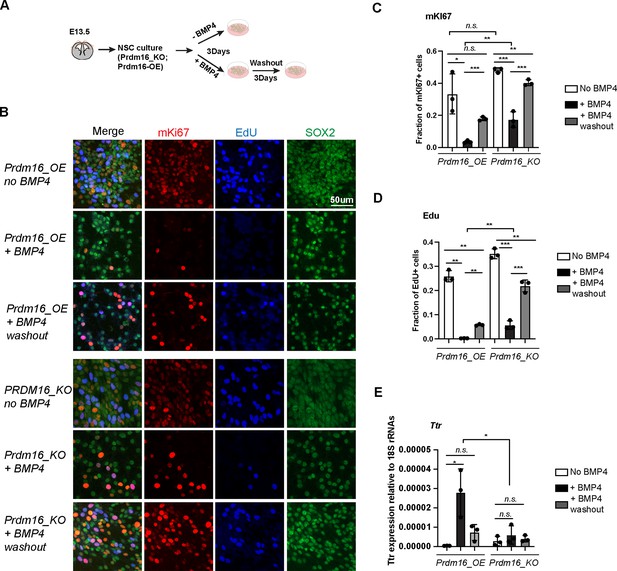
Prdm16 is required for BMP4-induced neural stem cell (NSC) quiescence.
(A) Schematic of the NSC culture assay. (B) Immunostaining of mKI67 in red and SOX2 in green and EdU labeling in blue on indicated NSC genotypes and treatment. (C–D) Quantification of the fraction of mKI67+ (C) and EdU+ (D) cells among the total cell number marked by SOX2. (E) RT-qPCR measurement of Ttr levels relative to 18 S RNAs. Biological replicates: N=3. Error bars represent standard deviation (SD). Statistical significance is calculated using unpaired t-test. ***p<0.001; **p<001; *p<0.05; n.s., non-significant.
-
Figure 2—source data 1
Counting of mKI67 + and DAPI-stained cells, related to Figure 2C.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Counting of EdU+ and DAPI-stained cells, related to Figure 2D.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig2-data2-v1.xlsx
-
Figure 2—source data 3
RTqPCR measurement of Ttr and 18srRNA expression, related to Figure 2E.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig2-data3-v1.xlsx
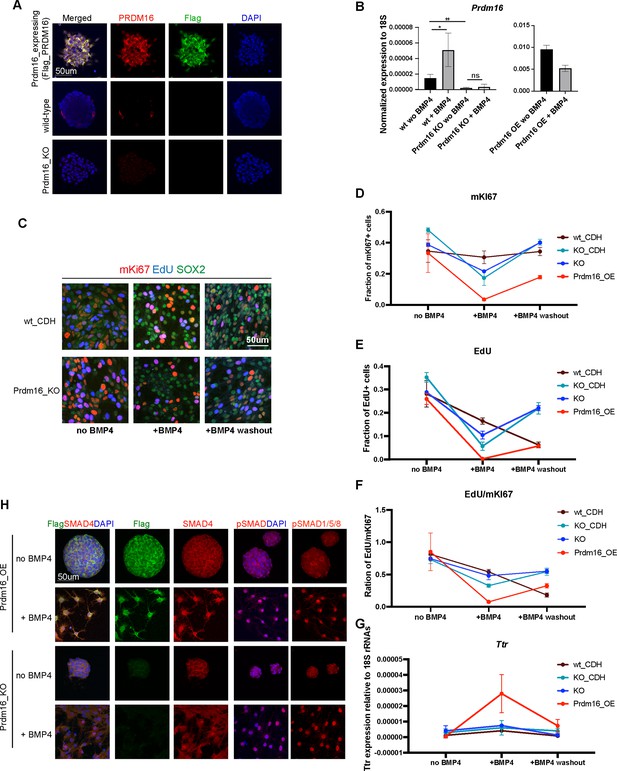
Neural stem cell (NSC) culture assay reveals PRDM16 function in NSCs.
(A) Immunostaining for PRDM16 and FLAG with DAPI in NSCs expressing Flag_Prdm16, wild-type (WT) NSCs, and Prdm16_KO NSCs. PRDM16 is sparsely expressed and cytoplasmic in WT NSCs, whereas FLAG_Prdm16 is robustly expressed in both cytoplasm and nucleus. (B) RT-qPCR measurement of Prdm16 mRNA levels normalized to the 18 S rRNAs in the indicated genotypes and condition, from three biological replicates, each with two technical replicates. Statistical significance was calculated using unpaired t-test (**p<0.01; *p<0.05; n.s., non-significant). (C) Immunostaining of dissociated NSCs on Matrigel for mKI67, EdU, and SOX2. WT_CDH denotes WT NSCs infected with the pCDH vector. (D–F). Quantification of marker-positive cell fractions under three culture conditions. KO_CDH indicates Prdm16_KO NSCs infected with the pCDH vector. (G) RT-qPCR measurement of Ttr mRNA levels normalized to 18 S rRNAs, from three biological replicates, each with two technical replicates. (H) Immunostaining for SMAD4, pSMAD1/5/8 and FLAG together with DAPI in NSCs. Nuclear localization of SMAD4 and pSMAD1/5/8 are enhanced following BMP4 treatment.
-
Figure 2—figure supplement 1—source data 1
RTqPCR measurement of Prdm16 and 18srRNA expression, related to Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig2-figsupp1-data1-v1.xlsx
-
Figure 2—figure supplement 1—source data 2
Counting of mKI67+ and DAPI-stained cells, related to Figure 2—figure supplement 1D.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig2-figsupp1-data2-v1.xlsx
-
Figure 2—figure supplement 1—source data 3
Counting of EdU+ and DAPI-stained cells, related to Figure 2—figure supplement 1E.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig2-figsupp1-data3-v1.xlsx
-
Figure 2—figure supplement 1—source data 4
RTqPCR measurement of Ttr and 18srRNA expression, related to Figure 2—figure supplement 1G.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig2-figsupp1-data4-v1.xlsx
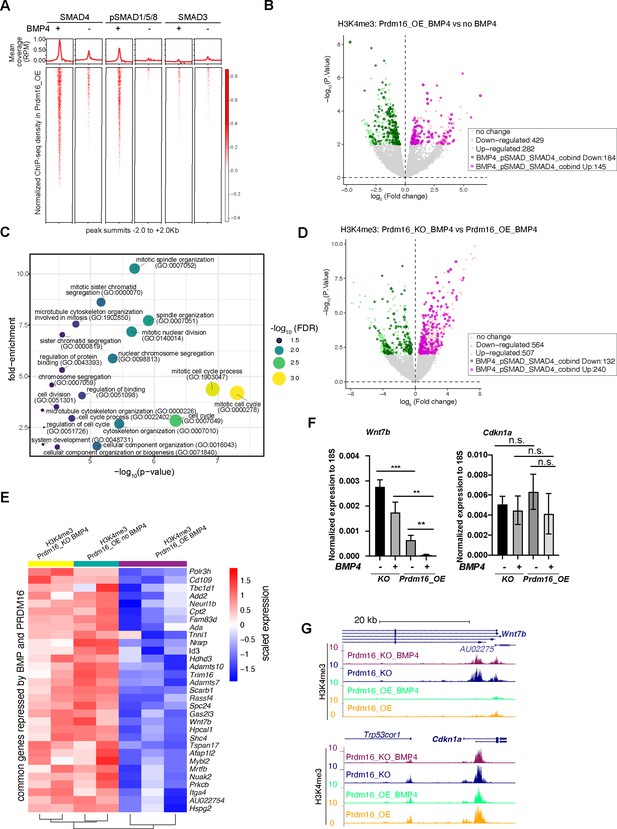
BMP signaling represses cell proliferation genes to induce cell quiescence.
(A) Heatmaps of ChIP-seq coverage (normalized by library depths and input) centered at peak summits for SMAD4, pSMAD1/5/8 and SMAD3 in Prdm16_OE cells with or without BMP4. (B) Volcano plot displaying differential H3K4me3 signal at TSS in Prdm16_OE cells with BMP4 versus untreated controls. Genes with significant changes (p<0.01) are colored. (C) Bubble plot of GO terms significantly enriched among SMAD-repressed genes (highlighted in dark green in panel B). (D) Volcano plot of differential H3K4me3 signal at TSS comparing Prdm16_KO and Prdm16_OE cells, both treated with BMP4. Significantly changed genes (p<0.01) are colored. (E) Heatmap of normalized H3K4me3 read counts at the transcription start sites (TSS) of 31 genes co-repressed by PRDM16 and SMADs (overlapping genes from dark green in B and dark pink in D). (F) RT-qPCR quantification of Wnt7b and Cdkn1a mRNA levels across indicated genotypes and treatment conditions. Data represent three biological replicates, each with two technical replicates. Statistical significance was calculated using unpaired t-test (***p<0.001; **p<0.01; n.s., non-significant). (G) Genome browser tracks of H3K4me3 CUT&TAG profiles at the Wnt7b and Cdkn1a loci.
-
Figure 3—source data 1
RTqPCR measurement of Wnt7b, Cdkn1a, and 18srRNA expression, related to Figure 3F.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig3-data1-v1.xlsx
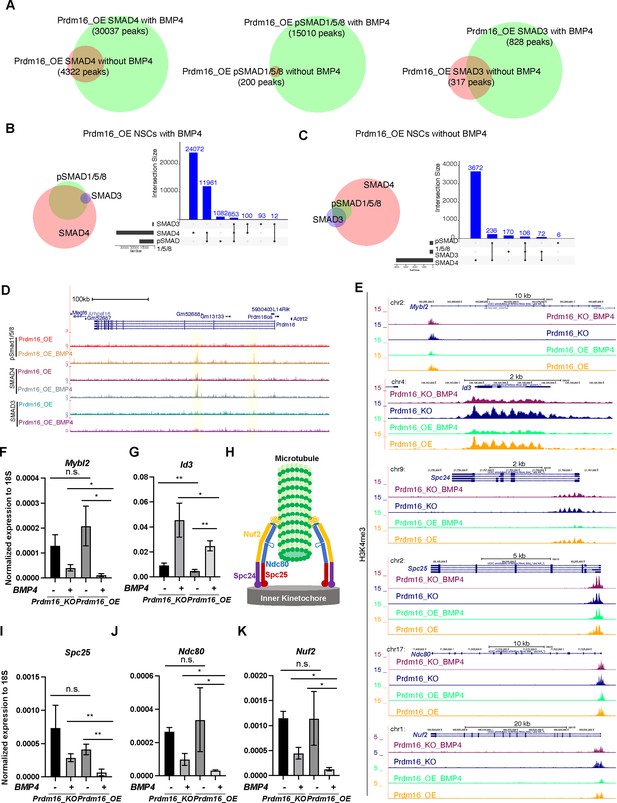
Overlapping analysis of SMAD4, pSMAD1/5/8, and SMAD3 CUT&TAG peaks.
(A) Venn diagrams showing overlapping SMAD4, pSMAD1/5/8, and SMAD3 ChIP-seq peaks in Prdm16_OE NSCs with or without BMP4. (B–C) Overlap Venn diagrams and bar plots for SMAD4, pSMAD1/5/8, and SMAD3 ChIP-seq peaks in Prdm16_OE NSCs with BMP4 (B) and without BMP4 (C). (D) Genome browser tracks of the Prdm16 locus showing increased SMAD4 and pSMAD1/5/8 binding at the two highlighted enhancer regions. (E) Genome browser tracks showing the H3K4me4 CUT&TAG signals at the indicated loci. (F–G, I–K). RT-qPCR measurement of the indicated genes normalized to 18 S rRNAs, from three biological replicates. Error bars represent standard deviation (SD). (H) Schematic of the NDC80 complex. Statistical significance is calculated using unpaired t-test. **p<001; * p<0.05; n.s., non-significant.
-
Figure 3—figure supplement 1—source data 1
RT-qPCR measurement of Mybl2 and 18srRNA expression, related to Figure 3—figure supplement 1F.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig3-figsupp1-data1-v1.xlsx
-
Figure 3—figure supplement 1—source data 2
RT-qPCR measurement of Id3 and 18srRNA expression, related to Figure 3—figure supplement 1G.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig3-figsupp1-data2-v1.xlsx
-
Figure 3—figure supplement 1—source data 3
RT-qPCR measurement of Spc25 and 18srRNA expression, related to Figure 3—figure supplement 1I.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig3-figsupp1-data3-v1.xlsx
-
Figure 3—figure supplement 1—source data 4
RT-qPCR measurement of Ndc80 and 18srRNA expression, related to Figure 3—figure supplement 1J.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig3-figsupp1-data4-v1.xlsx
-
Figure 3—figure supplement 1—source data 5
RT-qPCR measurement of Nuf2 and 18srRNA expression, related to Figure 3—figure supplement 1K.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig3-figsupp1-data5-v1.xlsx
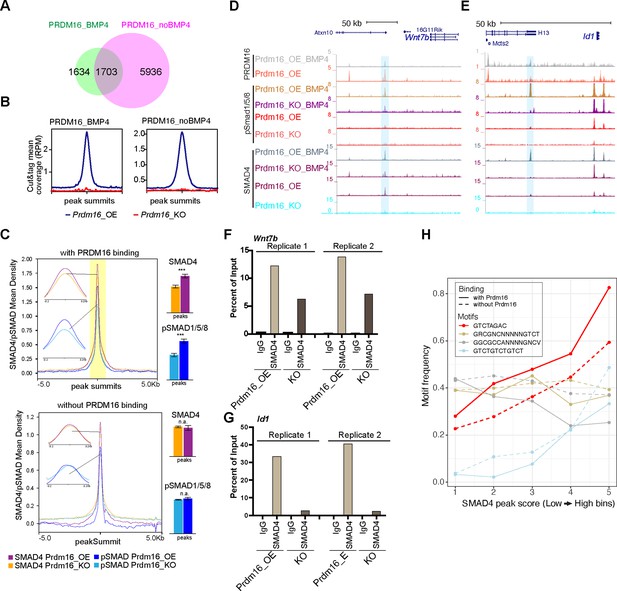
PRDM16 anchors SMAD proteins at specific genomic regions to mediate gene repression.
(A) Venn diagram showing overlap of PRDM16 CUT&TAG peak in BMP4-treated and untreated Prdm16_OE cells. (B) Average PRDM16 CUT&Tag signal (normalized to library size) at peak summits in BMP4-treated, untreated Prdm16_OE, and Prdm16_KO cells (4 replicates combined). (C) Metaplots of SMAD4 and pSMAD1/5/8 ChIP-seq coverage centered on peak summits with (top) and without (bottom) PRDM16 co-binding in BMP4-treated Prdm16_OE and Prdm16_KO cells. Bar plots (right) show average coverage; significant reduction in the KO cells is seen only at PRDM16 co-bound sites (***p<0.001; **p<0.01; *p<0.05; n.a. p>0.05). (D–E) Genome browser view of Wnt7b and Id1 loci showing PRDM16 CUT&TAG and SMAD ChIP-seq tracks. Co-bound regions are highlighted. (F–G) ChIP-qPCR validation of SMAD4 binding at highlighted regions in D-E from Prdm16_OE and Prdm16_KO cells (2 replicates). (H) Frequency of four SMAD motif types across SMAD ChIP-seq peaks binned by binding strength. The palindromic motif GTCTAGAC is most enriched at PRDM16 co-bound sites (solid red) and notably reduced in non-co-bound peaks (dashed red).
-
Figure 4—source data 1
ChIP-qPCR measurement of SMAD4 and PRDM16 co-bound site in Wnt7b, related to Figure 4F.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig4-data1-v1.xlsx
-
Figure 4—source data 2
ChIP-qPCR measurement of SMAD4 and PRDM16 co-bound site in Id1, related to Figure 4G.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig4-data2-v1.xlsx
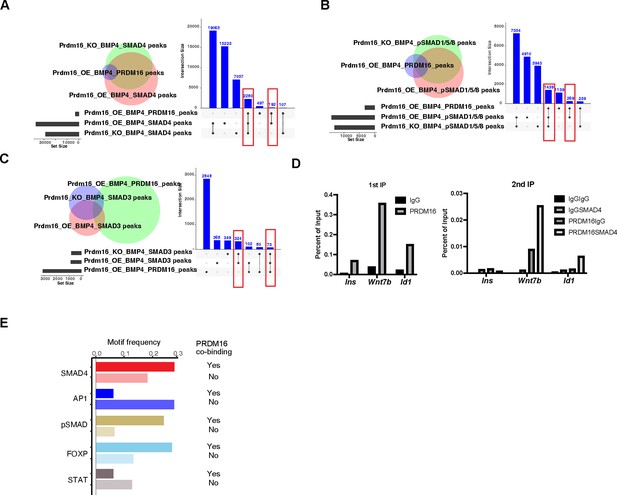
PRDM16 assists SMAD4/pSMAD1/5/8 genomic binding to their co-bound sites.
(A–C) Venn diagrams and interaction plots showing overlap between PRDM16 CUT&TAG peaks in Prdm16_OE cells with BMP4, SMAD4 (A), pSMAD1/5/8 (B), SMAD3 (C) ChIP-seq peaks in Prdm16_OE, and Prdm16_KO cells with BMP4. (D) Sequential ChIP-qPCR using IgG or PRDM16 antibodies followed by SMAD4 antibody. The Ins gene serves as a negative control. (E) Frequency of each DNA motif in SMAD4/pSMAD1/5/8 ChIP-seq peaks with or without PRDM16 co-binding in BMP4-treated Prdm16_OE cells.
-
Figure 4—figure supplement 1—source data 1
ChIP-qPCR measurement of SMAD4 and PRDM16 co-bound sites in the sequential ChIP experiment, related to Figure 4—figure supplement 1D.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig4-figsupp1-data1-v1.xlsx
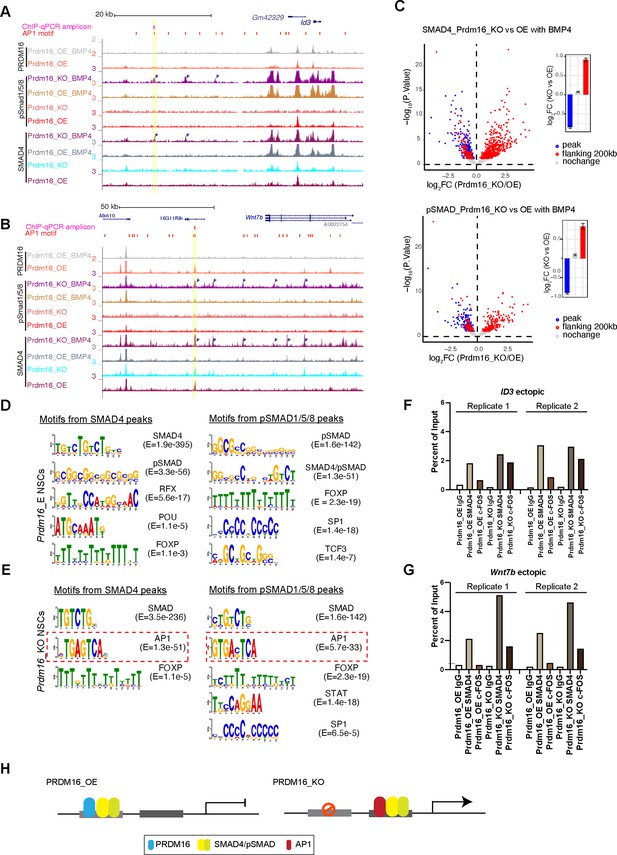
The SMAD complex shifts genome binding in the absence of Prdm16.
(A–B) Genome browser views of the Id3 (top) and Wnt7b (bottom) loci showing ectopic SMAD peaks (arrows) in Prdm16 KO cells. (C) Volcano plots showing differential ChIP-seq signal for SMAD4 and pSMAD1/5/8 in BMP4-treated Prdm16_KO cells versus BMP4-treated Prdm16_OE cells. Blue dots represent sites co-bound by PRDM16; red dots indicate sites not bound by PRDM16 but located within 200 kb of a PRDM16-cobound region. Mean log2 fold-change values are plotted to the right. (D–E) De novo motif discovery from SMAD4 and pSMAD1/5/8 ChIP-seq peaks in Prdm16_OE cells (D) and Prdm16_KO cells (E). (F–G) ChIP-qPCR validation of increased SMAD4 and c-FOS occupancy at the indicated ectopic binding sites in Prdm16_KO versus Prdm16_OE cells. Locations of PCR amplicons are shown in A and B. (H) Model illustrating enhancer switching: in the absence of PRDM16, SMAD complexes alter their co-factors and function as transcriptional activators.
-
Figure 5—source data 1
ChIP-qPCR measurement of ectopic SMAD4 and AP1 site in Id3, related to Figure 5F.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig5-data1-v1.xlsx
-
Figure 5—source data 2
ChIP-qPCR measurement of ectopic SMAD4 and AP1 site in Wnt7b, related to Figure 5G.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig5-data2-v1.xlsx
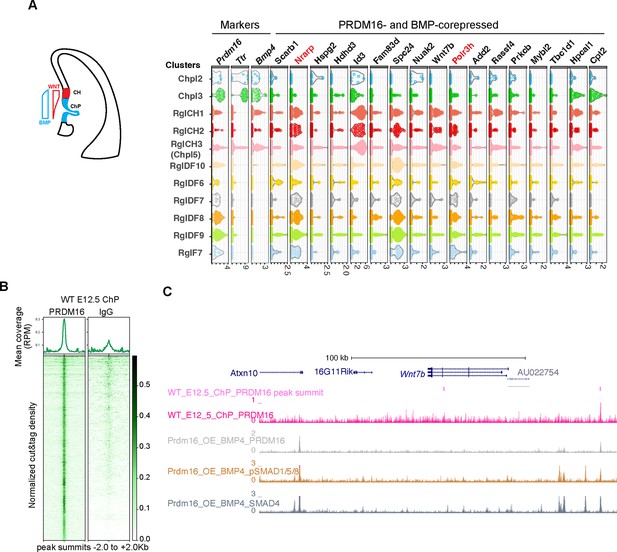
PRDM16 and SMADs co-regulated genes exhibit differential expression in choroid plexus (ChP) and neural cells.
(A) Schematic illustrating the cortical hem (CH) and ChP regions, which exhibit opposing Wnt and BMP signaling gradients. Shown alongside is a violin plot of scRNA-seq expression at E12.5 for the indicated genes across cell type clusters (Pyrowolakis et al., 2004). Cell clusters were defined based on marker gene expression from E9.5-E18.5 brain tissues, including choroid plexus clusters 1–5 (Chpl), with Chpl5 newly identified in this study based on Ttr expression as well as radial glia clusters: RglCH 1–3, RglDF 6–10, and RglF7. CH: cortical hem; DF: dorsal forebrain; F: forebrain. Gene expression per cell is represented as log2-normalized counts. (B) Metaplots and heatmaps showing CUT&TAG signal for PRDM16 and IgG, centered on peak summits and normalized for library size, from E12.5 dorsal midline tissues. (C) Genome browser view of the Wnt7b locus, displaying CUT&TAG tracks for PRDM16 and ChIP-seq tracks for SMAD proteins.
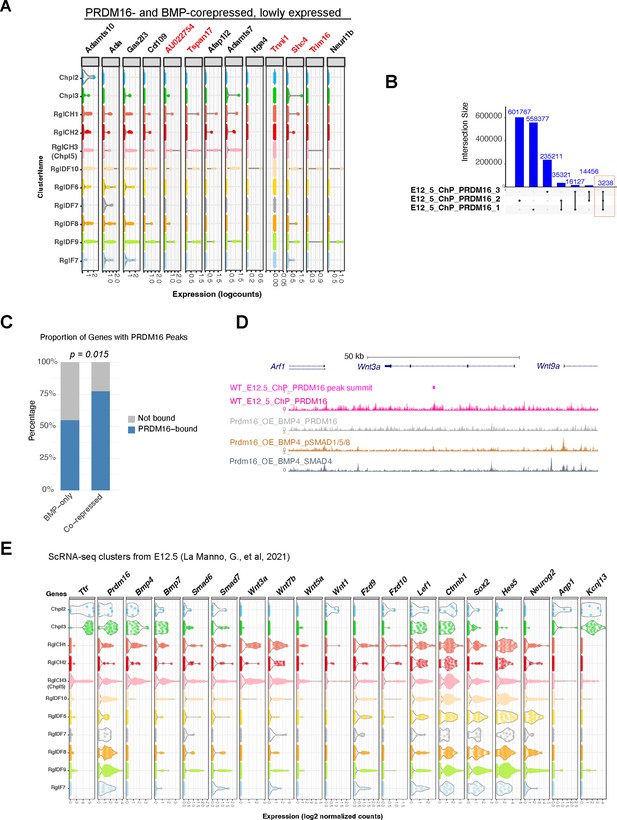
Wnt genes are potential targets regulated by PRDM16.
(A) Violin plots of scRNA-seq expression at E12.5 for the indicated genes in cell type clusters (reanalyzed from La Manno et al., 2021 as in Figure 6A). Genes shown here have lower expression than those in main Figure 6A. Red text indicates genes without PRDM16 binding in E12.5 ChP. (B) Intersection plot of PRDM16 CUT&Tag peaks across three biological replicates; commonly shared peaks are highlighted. (C) Bar graph showing proportion of genes with PRDM16 binding in E12.5 ChP. (D) Genome browser view of Wnt3a locus showing PRDM16 CUT&Tag and SMAD ChIP-seq tracks. (E) Violin plots of gene expression across reanalyzed cell type clusters.
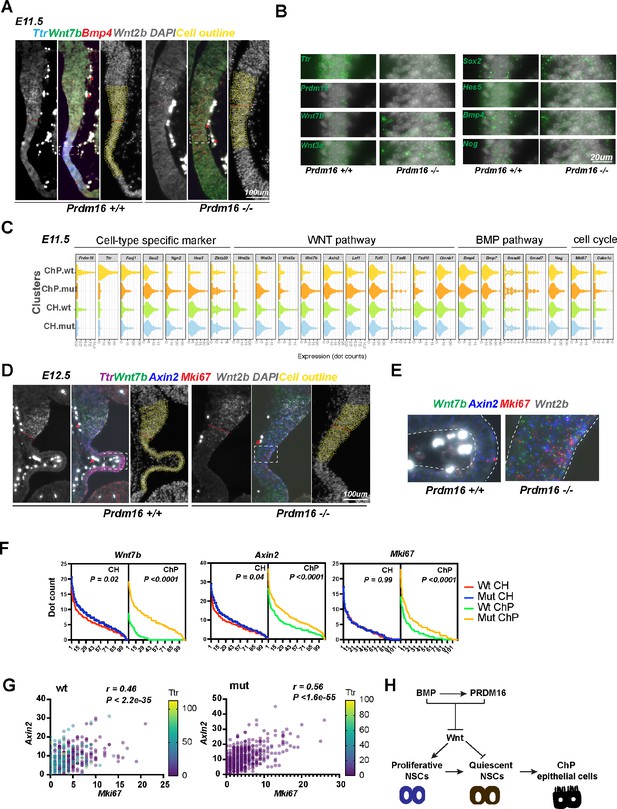
PRDM16 suppresses Wnt signaling and cell proliferation in choroid plexus (ChP) epithelial cells.
(A) SCRINSHOT images of E11.5 wild-type and Prdm16 mutant brains probed for Bmp4, Ttr, Wnt7b and Wnt2b. Red arrows indicate autofluorescent blood cells (non-specific signal in all channels). Cells quantified are circled in yellow, and the red dashed line marks the cortical hem (CH) and ChP boundary. (B) Enlarged views of the boxed region in (A), showing increased expression of Wnt and neural genes and decreased Ttr and Prdm16 in mutants. (C) Violin plots displaying dot counts for the 24 genes in 330 CH and 330 ChP cells from three wild-type and three Prdm16 mutant samples. (D) SCRINSHOT images of E12.5 wild-type and Prdm16 mutant brains with probes for Axin2, Ttr, Wnt7b, Wnt2b, and Mki67. (E) Enlarged views of the boxed areas in (D), with ChP regions outlined by dashed lines. (F) Distribution of dot counts for indicated genes in wild-type (wt) and Prdm16 mutant (mut) CH and ChP cells. X-axis: 110 cells per replicate; Y-axis: mean dot count across three biological replicates. P values calculated using area under the ROC curve (Prism Graphpad). (G) Multi-variant linear regression of Mki67, Axin2 and Ttr expression across 660 CH and ChP cells per genotype. Pearson correlation and significance were calculated using Prism Graphpad. (H) Model illustrating the regulatory circuit through which PRDM16 promotes the transition from proliferative to quiescent NSCs, a prerequisite step for ChP epithelial cell specification.
-
Figure 7—source data 1
Dot counts in three pairs of wt and Prdm16 mutant CH (1-330) and ChP (331-660) cells from E11.5 brains, related to Figure 7C.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig7-data1-v1.xlsx
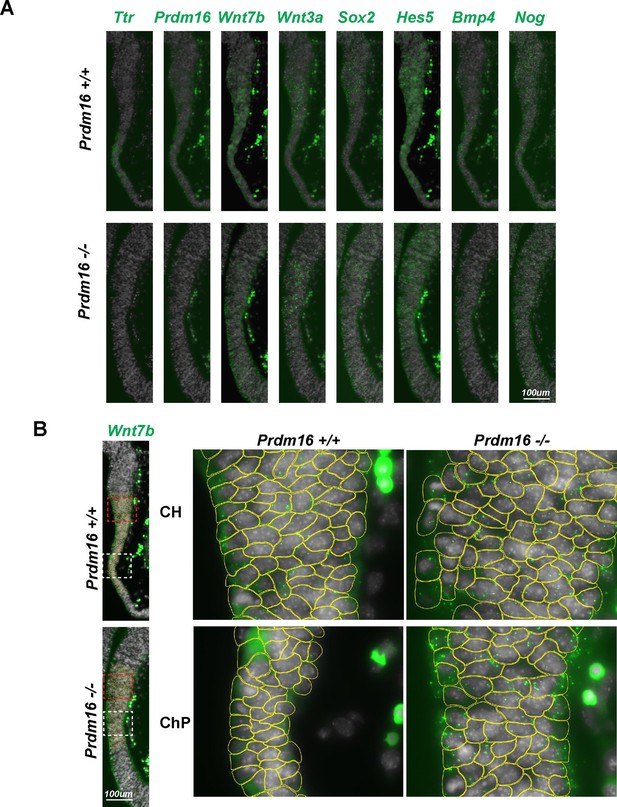
Full and exemplar segmentation images.
(A) Full images of SCRINSHOT signals for genes shown in Figure 7B. (B) Example of image segmentation for SCRINSHOT dot quantification.
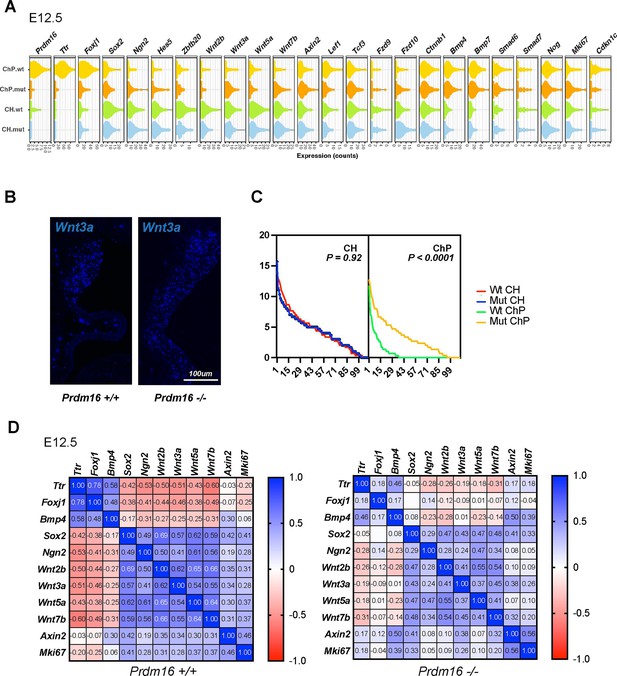
Altered expression of Wnt pathway and proliferation genes in Prdm16 mutant choroid plexus (ChP) cells.
(A) Violin plots showing expression changes of 24 examined genes (including ChP, neural, BMP pathway, Wnt pathway genes, and Mki67) across 110 cortical hem (CH) and 110 ChP cells per sample (330 cells per genotype, three wild-type (WT), and mutant pairs). (B) SCRINSHOT images showing Wnt3a mRNA in WT and Prdm16 mutant brains. (C) Dot quantification of Wnt3a expression in CH and ChP regions across three replicates. X-axis: cell ID; Y-axis: average dot count. P value was calculated using area under the ROC curve in Prism Graphpad. (D) Heatmap of gene expression correlations. In WT cells, Bmp genes cluster with Ttr and Foxj1, while Wnt genes cluster with Mki67. In mutants, BMP genes correlation with ChP markers is reduced, while Wnt- Mki67 correlations is maintained.
-
Figure 7—figure supplement 2—source data 1
Dot counts in three pairs of wt and Prdm16 mutant CH (1-330) and ChP (331-660) cells from E12.5 brains, related to Figure 7—figure supplement 2A.
- https://cdn.elifesciences.org/articles/104076/elife-104076-fig7-figsupp2-data1-v1.xlsx
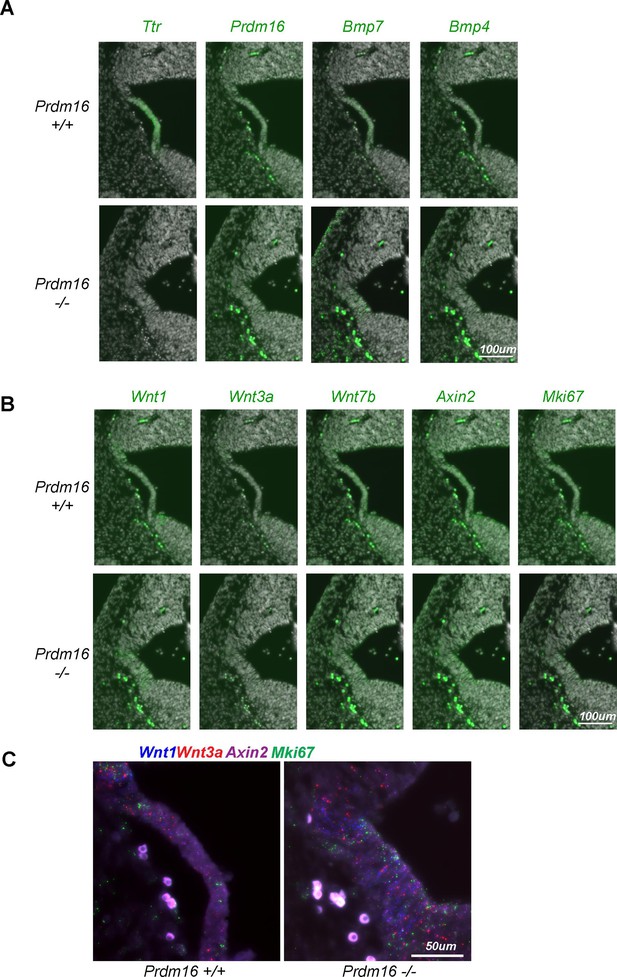
Expression of Bmp and Wnt genes in the developing fourth ventricle choroid plexus (ChP).
(A–B) SCRINSHOT images showing mRNA levels of ChP marker and BMP genes (A), and Wnt genes and Mki67 (B), in wild-type and Prdm16 mutant brains. Green: mRNA signals; gray: DAPI. (C) Enlarged view of SCRINSHOT images. Note the increased dot count for Wnt genes and Mki67 in the mutant.
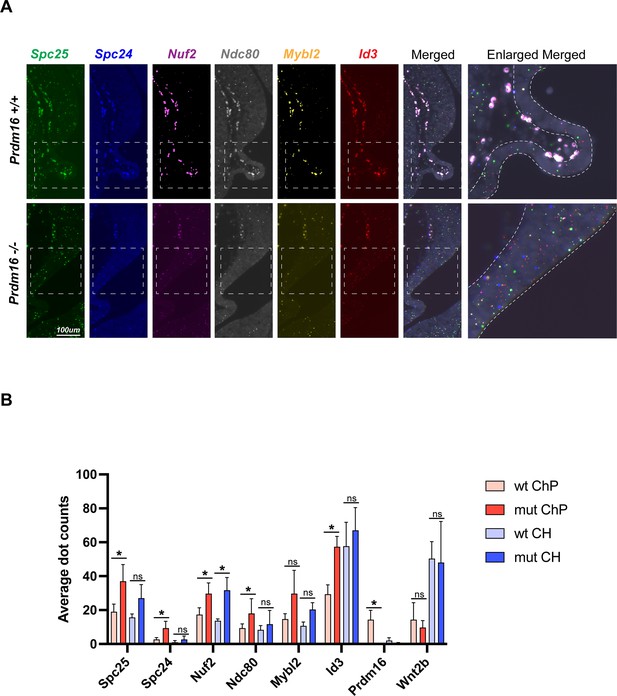
Increased expression of additional PRDM16 target genes in Prdm16 mutant choroid plexus (ChP).
(A) SCRINSHOT images showing mRNA levels of indicated genes in wild-type and Prdm16 mutant brains; enlarged views at right. Gene expression is elevated in mutants. (B) Quantification of dot counts in cortical hem (CH) and ChP regions (110 cells each) across three replicates. Prdm16 and Wnt2b are included as controls. Error bars denote SD. Statistical analysis via paired t-test (*p<0.05; n.s., not significant).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | FVB/NJ | Strassman, A., et al., 2017 | A gift from Bryan C. Bjork’s laboratory | |
| Genetic reagent (Mus musculus) | Prdm16 GT | Strassman, A., et al., 2017 | A gift from Bryan C. Bjork’s laboratory | |
| Antibody | Rabbit polyclonal anti-PRDM16 | Gift from Bryan Bjork’s lab | IF (1:500–1000), WB (1:2000) Cut&Tag (1:150) | |
| Antibody | Goat polyclonal anti-SOX2 | R&D systems | AF2018 | IF (1:1000) |
| Antibody | Rabbit polyclonal anti-Ki67 | Abcam | ab15580 | IF (1:1000) |
| Antibody | Goat polyclonal anti-Doublecortin | SANTA CRUZ Biotechnology | sc-8066 | IF (1:500) |
| Antibody | beta Galactosidase Polyclonal Antibody | Thermo Fisher | A-11132 | IF (1:500) |
| Antibody | Rabbit polyclonal anti-SMAD4 | Proteintech | 10231–1-AP | IF (1:500) WB (1:2000) ChIP (1:150) |
| Antibody | Rabbit polyclonal anti-pSMAD1/5/8 | Millipore | AB3848-1 | IF (1:500) WB (1:2000) ChIP (1:150) |
| Antibody | Secondary antibodies conjugated to Cy3 or Cy5 or Alexa Fluor-488 | Jackson ImmunoResearch | IF (1:1000) | |
| Antibody | mouse anti-Rabbit IgG light chain specific | Jackson ImmunoResearch | Cat# 211-032-171 | WB (1:40000) |
| Antibody | Rabbit polyclonal anti-SMAD3 | abcam | ab227223 | IF (1:500) WB (1:1000) ChIP (1:150) |
| Antibody | Rabbit monoclonal anti-C-Fos | Invitrogen | MA5-15055 | ChIP (1:150) |
| Antibody | Rabbit polyclonal to Histone H3 (tri methyl K4) | abcam | ab8580 | CUT&TAG (1:150) |
| Antibody | IgG from rabbit serum | SIGMA-ALDRICH | I5006 | ChIP (1:150) CUT&TAG (1:150) |
| Antibody | Guinea Pig anti-Rabbit IgG (Heavy & Light Chain) Antibody - Preadsorbed | antibodies-online | ABIN101961 | ChIP (1:150) CUT&TAG (1:150) |
| Cell line | 293t | ATCC | CRL-3216 | |
| Sequence-based reagent | Ttr ISH F | This paper | PCR primers | GCTCTAGAGGATGGCTTCCCTTCGAC |
| Sequence-based reagent | Ttr ISH R | This paper | PCR primers | CCCAAGCTTAAAAATGCTTCCCGGCAT |
| Sequence-based reagent | Wnt2b ISH F | This paper | PCR primers | GCTCTAGAGAAGCTGCAGGGTGAGGAT |
| Sequence-based reagent | Wnt2b ISH R | This paper | PCR primers | GGAATTCTCATATCGCCTCCTCAGGTAGT |
| Sequence-based reagent | Bmp4 ISH F | This paper | PCR primers | CGGGATCCCCTGCAGCGATCCAGTCT |
| Sequence-based reagent | Bmp4 ISH R | This paper | PCR primers | GGAATTCGCCCAATCTCCACTCCCT |
| Sequence-based reagent | 18srRNA qRTF | This paper | RTqPCR primers | GTAACCCGTTGAACCCCATT |
| Sequence-based reagent | 18srRNA qRTR | This paper | RTqPCR primers | CCATCCAATCGGTAGTAGCG |
| Sequence-based reagent | Mki67 qRTF1F | This paper | RTqPCR primers | TGCCTCAGATGGCTCAAAGA |
| Sequence-based reagent | Mki67 qRTF1R | This paper | RTqPCR primers | TCTGCCAGTGTGCTGTTCTA |
| Sequence-based reagent | Wnt7b qRTF | This paper | RTqPCR primers | TGCCTTCACCTATGCCATCA |
| Sequence-based reagent | Wnt7b qRTR | This paper | RTqPCR primers | GTCACAGCCACAATTGCTCA |
| Sequence-based reagent | Prdm16 qRTF | This paper | RTqPCR primers | CACCGGGTCAGAGGAGAAAT |
| Sequence-based reagent | Prdm16 qRTR | This paper | RTqPCR primers | CGACATGTCAGGGCTCCTAT |
| Sequence-based reagent | Cdkn1a qRTF | This paper | RTqPCR primers | CAAAGTGTGCCGTTGTCTCT |
| Sequence-based reagent | Cdkn1a qRTR | This paper | RTqPCR primers | CGAAGTCAAAGTTCCACCGT |
| Sequence-based reagent | Mybl2 qRTF | This paper | RTqPCR primers | CCTGGAGCAGAGAGACAACA |
| Sequence-based reagent | Mybl2 qRTR | This paper | RTqPCR primers | GTACTGGCATTGCTGGTCTG |
| Sequence-based reagent | Ndc80 qRTF | This paper | RTqPCR primers | CGCCTCTCTATGCAGGAGTT |
| Sequence-based reagent | Ndc80 qRTR | This paper | RTqPCR primers | TTCCGATGTCGGTTTGTGTG |
| Sequence-based reagent | Nuf2 qRTF | This paper | RTqPCR primers | GAACAAACCGGACAAGTCGT |
| Sequence-based reagent | Nuf2 qRTR | This paper | RTqPCR primers | CTTCCTGGTGCACATTGCTT |
| Sequence-based reagent | Spc24 qRTF | This paper | RTqPCR primers | TGCTTTGAGACAGCATGGAG |
| Sequence-based reagent | Spc24 qRTR | This paper | RTqPCR primers | AGCCCTCTCTAACCTCTCCT |
| Sequence-based reagent | Spc25 qRTF | This paper | RTqPCR primers | AGAGTCGGAAGAGCTGACTG |
| Sequence-based reagent | Spc25 qRTR | This paper | RTqPCR primers | CGCTGATTTCTGCAGTCCTT |
| Sequence-based reagent | Wnt7b Co-bound enhancer ChIP qPCRF | This paper | ChIPqPCR primers | GTCATGTGCTTTCAGGGCAT |
| Sequence-based reagent | Wnt7b Co-bound enhancer ChIP qPCRR | This paper | ChIPqPCR primers | TCCTAGGAGTCACCATGGAA |
| Sequence-based reagent | Id1 Co-bound enhancer ChIP qPCRF | This paper | ChIPqPCR primers | GAGGCCACTCCAGTTCCAAT |
| Sequence-based reagent | Id1 Co-bound enhancer ChIP qPCRR | This paper | ChIPqPCR primers | GCGGTTCACATCTGGAGTCT |
| Sequence-based reagent | Wnt7b ectopic enhancer ChIP qPCRF | This paper | ChIPqPCR primers | CAGCGGTCAAGTTTAAGCGA |
| Sequence-based reagent | Wnt7b ectopic enhancer ChIP qPCRR | This paper | ChIPqPCR primers | CCACAGATGTCCCAGAGTGA |
| Sequence-based reagent | Id3 ectopic enhancer ChIP qPCRF | This paper | ChIPqPCR primers | CTTGGGCAATGTGAAAGGCT |
| Sequence-based reagent | Id3 ectopic enhancer ChIP qPCRR | This paper | ChIPqPCR primers | CAGTTGGTCCCTGTCAGTGT |
| Sequence-based reagent | Prdm16_PL_4 | This paper | SCRINSHOT padlock probe | /5Phos/AAGTCTTCAGAGATCTGCTTTTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCAGAACTTCTCGCTACCC |
| Sequence-based reagent | Ttr_Pl-1 | This paper | SCRINSHOT padlock probe | /5Phos/CTTCTACAAACTTCTCATCTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCTACTATCTTCTTTCAGTTCTACTCTGTACACTC |
| Sequence-based reagent | Foxj1_PL_1 | This paper | SCRINSHOT padlock probe | CGTGATCCACTTGTAGATGGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCGGAAGTAGCAGAAGTTGTC |
| Sequence-based reagent | Foxj1_PL_2 | This paper | SCRINSHOT padlock probe | /5Phos/GCCAAGAAGGTCTCATCAAAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGATGCTGTAGGAAGGATGTG |
| Sequence-based reagent | Foxj1_PL_3 | This paper | SCRINSHOT padlock probe | /5Phos/CACTTTGATGAAGCACTTGTTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTCATCCTTCTCCCGAGG |
| Sequence-based reagent | Sox2_Pl-2 | This paper | SCRINSHOT padlock probe | /5Phos/TCGGCAGCCTGATTCCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCACGTCTATCTTCTTTTAGTTCAAATATATACATGGATTC |
| Sequence-based reagent | Sox2_Pl-3 | This paper | SCRINSHOT padlock probe | /5Phos/CTCTAAATATTACTGGTAAAGACTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCACGTCTATCTTCTTTCATCGCCCGGAGTCTAG |
| Sequence-based reagent | Ngn2_Pl-1 | This paper | SCRINSHOT padlock probe | /5Phos/TTGGTTTGACAGGTGAAATTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCATCGCTATCTTCTTTCACATCAGAGAGGGAAAGT |
| Sequence-based reagent | Ngn2_Pl-2 | This paper | SCRINSHOT padlock probe | /5Phos/ACGCTTGCATTCAACCCTTTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCATCGCTATCTTCTTTGAGAATTCAGCCTAAATTTCC |
| Sequence-based reagent | Ngn2_Pl-3 | This paper | SCRINSHOT padlock probe | /5Phos/ATAAACCAGAGCTGGTCTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCATCGCTATCTTCTTTCCATAAATCCTCACATCTTCA |
| Sequence-based reagent | Hes5_Pl-1 | This paper | SCRINSHOT padlock probe | /5Phos/CAAGAGTTCAAATTCTAAGTACTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCAGCTCTATCTTCTTTTGTGTTCTCCCACATGAC |
| Sequence-based reagent | Hes5_Pl-2 | This paper | SCRINSHOT padlock probe | /5Phos/CACACATTCTCTAAGAATGACTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCAGCTCTATCTTCTTTCCCAAATGACAACTCTGCA |
| Sequence-based reagent | Hes5_Pl-3 | This paper | SCRINSHOT padlock probe | /5Phos/TTAAGGATCATCGTGGAGACTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCAGCTCTATCTTCTTTCACCCATACAAAGGAATCCT |
| Sequence-based reagent | Zbtb20_PL-1 | This paper | SCRINSHOT padlock probe | /5Phos/CCATTTCCTGCTTGATGTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCATAGTAGTCATAGTCATCTT |
| Sequence-based reagent | Zbtb20_PL-2 | This paper | SCRINSHOT padlock probe | /5Phos/TACTTATCCGTCAGACACATTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGTAAGAAAGAGAGAAAGGTAC |
| Sequence-based reagent | Zbtb20_PL-3 | This paper | SCRINSHOT padlock probe | /5Phos/GAATGTGTCGAGTGGATGAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGAGCGTGAGAGTTTGTCAGT |
| Sequence-based reagent | Wnt2b_Pl-1 | This paper | SCRINSHOT padlock probe | /5Phos/TGAAGACAGACCTCTCCTATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCCGTACTATCTTCTTTTTAGTGCAACTCAAATCACTC |
| Sequence-based reagent | Wnt2b_Pl-2 | This paper | SCRINSHOT padlock probe | /5Phos/GTAATGGATGTTGTCACTACATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCCGTACTATCTTCTTTCTTGGCAAAGCGAACACC |
| Sequence-based reagent | Wnt2b_Pl-3 | This paper | SCRINSHOT padlock probe | /5Phos/CGTGCAGTAGTTAACAGTTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCCGTACTATCTTCTTTCTGTTTAAGTCGATCGTGGAA |
| Sequence-based reagent | Wnt3a_Pl-1 | This paper | SCRINSHOT padlock probe | /5Phos/CCTCTCTACTCAGGATAGAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCCATAGAGATTCCTAGAGAAC |
| Sequence-based reagent | Wnt3a_Pl-2 | This paper | SCRINSHOT padlock probe | /5Phos/ATGAGACCCTCACAGGTAGGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCAGAAACCGGGTCCTTAGGT |
| Sequence-based reagent | Wnt3a_Pl-3 | This paper | SCRINSHOT padlock probe | /5Phos/CATGTATCTAGATAGAGCAGTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCACCCTGAAGCACCCTCT |
| Sequence-based reagent | Wnt5a_PL-1 | This paper | SCRINSHOT padlock probe | /5Phos/TTAAGAGCCACAGGACTGATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTAATGAATATTGTTGGGCAATAAG |
| Sequence-based reagent | Wnt5a_PL-2 | This paper | SCRINSHOT padlock probe | /5Phos/TAGAGACCACCAAGAATTAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTGAACAGGGTTATTCATACC |
| Sequence-based reagent | Wnt5a_PL-3 | This paper | SCRINSHOT padlock probe | /5Phos/CTTCATTGTTGTGTAAGTTCATGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTATACTGTCCTACGGCCTG |
| Sequence-based reagent | Wnt7b_PL-1 | This paper | SCRINSHOT padlock probe | /5Phos/AGGGATTTGTCTCCATTGGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCACTTTCCCAAAGAGAAGTAA |
| Sequence-based reagent | Wnt7b_PL-2 | This paper | SCRINSHOT padlock probe | /5Phos/CATTGTTGTGAAGGTTCATGAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGAACCTTTCTGCCCGCCT |
| Sequence-based reagent | Wnt7b_PL-3 | This paper | SCRINSHOT padlock probe | /5Phos/CCGGTCACAGCCACAATTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGTTGTAGTAGCCTTGCTTCTC |
| Sequence-based reagent | Axin2_PL-1 | This paper | SCRINSHOT padlock probe | /5Phos/AAGTCAAGAACACCTGGTAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCATATTCCAGGTAAATGTCAG |
| Sequence-based reagent | Axin2_PL-2 | This paper | SCRINSHOT padlock probe | /5Phos/TACGTGATAAGGATTGACTGGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCAAAGACATAGCCGGAACC |
| Sequence-based reagent | Axin2_PL-3 | This paper | SCRINSHOT padlock probe | /5Phos/TCCTGTATGGAATTTCTTCTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTTTGAGCCTTCAGCATCC |
| Sequence-based reagent | Lef1_Pl-1 | This paper | SCRINSHOT padlock probe | /5Phos/CATATTGGGCATCATTATGTAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCACTGCTATCTTCTTTCATGTACGGGTCGCTGTT |
| Sequence-based reagent | Lef1_Pl-2 | This paper | SCRINSHOT padlock probe | /5Phos/CGTGCTAGTTCATAGTATTTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCACTGCTATCTTCTTTGTGTAGCTGTCTCTCTTTC |
| Sequence-based reagent | Lef1_Pl-3 | This paper | SCRINSHOT padlock probe | /5Phos/CTTCATCAGACACTTCACAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCACTGCTATCTTCTTTGGGTTTGGCTATCTCTTTAG |
| Sequence-based reagent | Tcf3_PL_1 | This paper | SCRINSHOT padlock probe | /5Phos/AGGTCACTCAGTTCCTTGTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTACATCATGCTGAAGTCCAGG |
| Sequence-based reagent | Tcf3_PL_2 | This paper | SCRINSHOT padlock probe | /5Phos/GAGACCTGTCTCATCCAGAATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTCTCTGCTCATGCTTCGATG |
| Sequence-based reagent | Tcf3_PL_3 | This paper | SCRINSHOT padlock probe | /5Phos/TCTTTCTCCTCCCGCTTGATTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTGATGCGATTTCCTCATCC |
| Sequence-based reagent | Fzd9-PL-1 | This paper | SCRINSHOT padlock probe | /5Phos/CACATAGAAAGGAAGATAATCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCAAGGAGTAGACATTGTAG |
| Sequence-based reagent | Fzd9-PL-2 | This paper | SCRINSHOT padlock probe | /5Phos/TTCATAAACGTAGCAGACAATGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGAAGTCCATGTTGAGGCG |
| Sequence-based reagent | Fzd9-PL-3 | This paper | SCRINSHOT padlock probe | /5Phos/CACTGGACCCTGCCCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTAAATTACATCATTAAATAACTCTG |
| Sequence-based reagent | Fzd10-PL-1 | This paper | SCRINSHOT padlock probe | /5Phos/CTCAAGGTAGACTTCATGTATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTACATTCAGCTGTTCTGAAC |
| Sequence-based reagent | Fzd10-PL-2 | This paper | SCRINSHOT padlock probe | /5Phos/TGGAGAGGAAGATGATAGGATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCGAATAAACGCAGTAGCACA |
| Sequence-based reagent | Fzd10-PL-3 | This paper | SCRINSHOT padlock probe | /5Phos/GATTGTTCATCTTACACTTGTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCAGTCAGGTGTCTTGGTCT |
| Sequence-based reagent | Ctnnb1_Pl-1 | This paper | SCRINSHOT padlock probe | /5Phos/GTGTCAACATCTTCTTCCTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCATGCCTATCTTCTTTCATTCATAAAGGACTTGGGAG |
| Sequence-based reagent | Ctnnb1_Pl-2 | This paper | SCRINSHOT padlock probe | /5Phos/GTCGCTGCATCTGAAAGGTTTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCATGCCTATCTTCTTTCTTCCATCCCTTCCTGCTTA |
| Sequence-based reagent | Ctnnb1_Pl-3 | This paper | SCRINSHOT padlock probe | /5Phos/CAGGCCAGCTGATTGCTATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCATGCCTATCTTCTTTGATTTACAGGTCAGTATCAAAC |
| Sequence-based reagent | Bmp4_Pl-1 | This paper | SCRINSHOT padlock probe | /5Phos/AGAATTAACAGAGTTGACTAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCATCTATCTTCTTTAACAGGCCTTAGGGATACT |
| Sequence-based reagent | Bmp4_Pl-2 | This paper | SCRINSHOT padlock probe | /5Phos/GTGATGGAAACTCCTCACAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCATCTATCTTCTTTGATGTTCTCCAGATGTTCTTC |
| Sequence-based reagent | Bmp4_Pl-3 | This paper | SCRINSHOT padlock probe | /5Phos/GTGGTTGAATGGGAACGTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCATCTATCTTCTTTCAGTTTGTGTGGTATGTGTAG |
| Sequence-based reagent | Bmp7_Pl-1 | This paper | SCRINSHOT padlock probe | /5Phos/TTCTTCAGGATGACATTAGAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCTACGCTATCTTCTTTGACCACCATGTTTCTGTAC |
| Sequence-based reagent | Bmp7_Pl-2 | This paper | SCRINSHOT padlock probe | /5Phos/AGTCCTTATAGATCCTGAATTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCTACGCTATCTTCTTTCAAATCGCTCCCGGATGT |
| Sequence-based reagent | Bmp7_Pl-3 | This paper | SCRINSHOT padlock probe | /5Phos/TGTCGCAGAACCTTCTGTTATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCTACGCTATCTTCTTTCTCAGATCCAGAAACCAATC |
| Sequence-based reagent | Smad6_PL-1 | This paper | SCRINSHOT padlock probe | /5Phos/ACTAAGGCTTGTGGATAACTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTTTCCCTGCAGGATCCGAA |
| Sequence-based reagent | Smad6_PL-2 | This paper | SCRINSHOT padlock probe | /5Phos/CAGTGTAAGACAATGTAGAATCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTAGTTGGTGGCCTCGGTTT |
| Sequence-based reagent | Smad6_PL-3 | This paper | SCRINSHOT padlock probe | /5Phos/AGTAGGATCTCCAGCCAACTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCATTGCTATCTGTGGTTGTTG |
| Sequence-based reagent | Smad7_PL-1 | This paper | SCRINSHOT padlock probe | /5Phos/GCCTGCAGTTGGTTTGAGAATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGAAGGTACAGCATCTGGACA |
| Sequence-based reagent | Smad7_PL-2 | This paper | SCRINSHOT padlock probe | /5Phos/GTACCAGCTGACTCTTGTTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGATCTTGCTCCGCACTTTCT |
| Sequence-based reagent | Smad7_PL-3 | This paper | SCRINSHOT padlock probe | /5Phos/CCTCTTGACTTCCGAGGAATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGTAAGATTCACAGCAACACAG |
| Sequence-based reagent | Nog_PL-1 | This paper | SCRINSHOT padlock probe | /5Phos/AGGGTCTGGATGTTCGATGATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTTCTCCTTAGGGTCAAAGAT |
| Sequence-based reagent | Nog_PL-2 | This paper | SCRINSHOT padlock probe | /5Phos/GATATAAATAGCAGGTTAACATTATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCATGCGAAGGGTACTGG |
| Sequence-based reagent | Nog_PL-3 | This paper | SCRINSHOT padlock probe | /5Phos/TGCATTACAGGAACCAGAAATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCATTCCTACACAGTTAAACAG |
| Sequence-based reagent | mKi67_PL_1 | This paper | SCRINSHOT padlock probe | /5Phos/GGACTTTCCTGGAAATTCTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTTGATGGTTCCTTTCCAAG |
| Sequence-based reagent | mKi67_PL_2 | This paper | SCRINSHOT padlock probe | /5Phos/ACTTTCCTTGCATCTTTGAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGAACCTTCACTTAATTCACC |
| Sequence-based reagent | mKi67_PL_3 | This paper | SCRINSHOT padlock probe | /5Phos/CTGAGGCTTTATCAAGATTTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCAGAGACTCCTTTCTCTTC |
| Sequence-based reagent | Cdkn1c_PL_1 | This paper | SCRINSHOT padlock probe | /5Phos/CAAGTTTAAGAGCAATCTAATGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTTTCGACTGTCTGGTCAC |
| Sequence-based reagent | Cdkn1c_PL_2 | This paper | SCRINSHOT padlock probe | /5Phos/CGCATGGATAAATACAAATATTAATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGTGGCAGAGGGATCCA |
| Sequence-based reagent | Cdkn1c_PL_3 | This paper | SCRINSHOT padlock probe | /5Phos/GCCTCTAAACTAACTCATCTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCAAGTTCTCTCTGGCCGTTA |
| Sequence-based reagent | Id3-1 | This paper | SCRINSHOT padlock probe | /5Phos/AGAGTAGAGATAGAGAGGGAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTCAGCGCCTTCATGTTGG |
| Sequence-based reagent | Id3-2 | This paper | SCRINSHOT padlock probe | /5Phos/GGGCTGGGTTAAGATCGAATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTGAGTTCAGGGTAAGTGAAGA |
| Sequence-based reagent | Id3-3 | This paper | SCRINSHOT padlock probe | /5Phos/TGCTGCCTTGGCAGAGTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGAAACCAGAAGAACAGCTCTTA |
| Sequence-based reagent | Spc24-1 | This paper | SCRINSHOT padlock probe | /5Phos/GCTGCTCTTCTATAGAATACTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTCAACAATTAAGAGCACTG |
| Sequence-based reagent | Spc24-2 | This paper | SCRINSHOT padlock probe | /5Phos/AAATCAATGAACGCACTCACTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTGTCTCAAAGCAAGTAGAT |
| Sequence-based reagent | Spc24-3 | This paper | SCRINSHOT padlock probe | /5Phos/CACAGCTAACAGACACACATTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTCTAACCTCTCCTGGAATA |
| Sequence-based reagent | Nrarp-1 | This paper | SCRINSHOT padlock probe | /5Phos/GGTACCATGCTCTCAGAATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTATAATAGAAGTCACTAGGAAG |
| Sequence-based reagent | Nrarp-2 | This paper | SCRINSHOT padlock probe | /5Phos/ACTTCATACAGATAGAAGAGACTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTTAGGAGTGTGCCACAG |
| Sequence-based reagent | Nrarp-3 | This paper | SCRINSHOT padlock probe | /5Phos/TAGTATGAGAGATTCACAGTCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCCTTCCACTCATTCAGCAA |
| Sequence-based reagent | Rassf4-1 | This paper | SCRINSHOT padlock probe | /5Phos/ACTGAGGTCTTGTGGTTATAAATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCCATAGGCAGGAGTGAAC |
| Sequence-based reagent | Rassf4-2 | This paper | SCRINSHOT padlock probe | /5Phos/CATCAAGAAGATCTTGACAATTTTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTCGCTCAAGTCAGCTTC |
| Sequence-based reagent | Rassf4-3 | This paper | SCRINSHOT padlock probe | CTCGGTGTCTTAGCTGAAAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGATCAGAGTCCCTTCTTCCT |
| Sequence-based reagent | Nuak2-1 | This paper | SCRINSHOT padlock probe | /5Phos/TTGCTGCTATTCTCAAACACTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTACTCCATGACAATCACAATC |
| Sequence-based reagent | Nuak2-2 | This paper | SCRINSHOT padlock probe | /5Phos/GGACTTCTTAAGAGAATGTTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCCATGTCATTCTCCTTTCG |
| Sequence-based reagent | Nuak2-3 | This paper | SCRINSHOT padlock probe | /5Phos/GTCGAGACTTCTTAAGGATGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGTAGTAACCAGATTCACGCT |
| Sequence-based reagent | Fam83d-1 | This paper | SCRINSHOT padlock probe | /5Phos/AATTGTCCGAACTGTCATCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTGCATAGTAGATATTTCCTGT |
| Sequence-based reagent | Fam83d-2 | This paper | SCRINSHOT padlock probe | /5Phos/GTAGAAGCCTCAGAGTCAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGAAATAGTCTTCATCGCTGATG |
| Sequence-based reagent | Fam83d-3 | This paper | SCRINSHOT padlock probe | /5Phos/GTTGGTAGAGGCTCTGGTAATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTATGTCCCTCACAGAAACCAG |
| Sequence-based reagent | Mybl2-1 | This paper | SCRINSHOT padlock probe | /5Phos/AAGTAGCAGCTATGGCAATCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTATGTCCGAGTTCTTTAGCAG |
| Sequence-based reagent | Mybl2-2 | This paper | SCRINSHOT padlock probe | /5Phos/TTCTGATGATGGATACTTCTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTACTTCTGATCTGGGAGTAC |
| Sequence-based reagent | Mybl2-3 | This paper | SCRINSHOT padlock probe | /5Phos/TTGACCAACTCAATGACCTTTTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTGTTTGGTGCCATACTTC |
| Sequence-based reagent | Gas2-1 | This paper | SCRINSHOT padlock probe | /5Phos/TTCCTGAAGACTTTGATGAATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTAAGTTTCCAGTACTCTTCT |
| Sequence-based reagent | Gas2-2 | This paper | SCRINSHOT padlock probe | /5Phos/CTGTTATCTCTTTCCCTAATATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTTCTCCATGAAGGTTTCTG |
| Sequence-based reagent | Gas2-3 | This paper | SCRINSHOT padlock probe | /5Phos/TCTTTGAATTTCTCCTGCACATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTTGTTGGCATCCATACTC |
| Sequence-based reagent | Ada-1 | This paper | SCRINSHOT padlock probe | /5Phos/GAGAGCTTCATCCTCGATTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTTTCTTTCAGTAGTCTGTTGTA |
| Sequence-based reagent | Ada-2 | This paper | SCRINSHOT padlock probe | /5Phos/AAACTTGGCCAGGAAGCCTTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTAATCACAGGCATGTAGTAGTC |
| Sequence-based reagent | Ada-3 | This paper | SCRINSHOT padlock probe | /5Phos/CAGCTCCAACACCTCAAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTCTTCTGATTGTACTTCTTACA |
| Sequence-based reagent | Hspg2-1 | This paper | SCRINSHOT padlock probe | /5Phos/CTTCCTGTAGGCATCTTTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGAAGGTGATCTTGATCTCAAA |
| Sequence-based reagent | Hspg2-2 | This paper | SCRINSHOT padlock probe | /5Phos/TGGATATCAGGGTCCAAGATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTATGATGTTATTGCCCGTAATC |
| Sequence-based reagent | Hspg2-3 | This paper | SCRINSHOT padlock probe | /5Phos/CTTGAGCCATGTACCTGATGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTTCACCTGGAGAAGTCTCAGT |
| Sequence-based reagent | shc4-1 | This paper | SCRINSHOT padlock probe | /5Phos/TCATGGTCTCTGTCTTCAGGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCCGGGATGGAGTTGTAATAG |
| Sequence-based reagent | shc4-2 | This paper | SCRINSHOT padlock probe | /5Phos/AGTCCTGTTTAGATTGGATTCTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGGTGCAGGTGCAGGTCC |
| Sequence-based reagent | shc4-3 | This paper | SCRINSHOT padlock probe | /5Phos/GGTTGTCAAGGTTCATCAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTGGTGATTTGCAATAATCTGTT |
| Sequence-based reagent | Nuf2-1 | This paper | SCRINSHOT padlock probe | /5Phos/AATCACTTTCAATCTGGTCCTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCAGTTTCTTTAGTTCCGATG |
| Sequence-based reagent | Nuf2-2 | This paper | SCRINSHOT padlock probe | /5Phos/GGCTTTCATGTAAATCATGTTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTACTCCATACACTAACTGTAA |
| Sequence-based reagent | Nuf2-3 | This paper | SCRINSHOT padlock probe | /5Phos/GTGGTTACTTGCTCACAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTTGTGGATTTCTTGATTAACA |
| Sequence-based reagent | Ndc80-1 | This paper | SCRINSHOT padlock probe | /5Phos/ATACTCTCTGTCGGCTTTATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTATCTTCAGACATGAATTCTTC |
| Sequence-based reagent | Ndc80-2 | This paper | SCRINSHOT padlock probe | /5Phos/GGATCCATGTCCGCTAGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTAATATACCAAGTTGACTATTCCT |
| Sequence-based reagent | Ndc80-3 | This paper | SCRINSHOT padlock probe | /5Phos/CTGTTCATTTAGTGCTTTGTTTTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTCCTCCAGTCTTGCAAT |
| Sequence-based reagent | Spc25-1 | This paper | SCRINSHOT padlock probe | /5Phos/CCCATGTTTAGGTAGACTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTAGATACCTCTATCATCTAGGT |
| Sequence-based reagent | Spc25-2 | This paper | SCRINSHOT padlock probe | /5Phos/ACAGATCCGCTGATTTCTGTCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTCTAGTCCAAGGTAATCTCTAT |
| Sequence-based reagent | Spc25-3 | This paper | SCRINSHOT padlock probe | /5Phos/AAATGGTTTCCCTCTTCCTATCCTCTATGATTACTGACTGCGTCTATTTAGTGGAGCCGCCCCTATCTTCTTTATTAGCTTTGTTAGCAGTGG |
| Sequence-based reagent | Prdm16_Det_4 | This paper | SCRINSHOT detection probe | TCGCUACCCAAGUCTTCAGA/3Cy3Sp/ |
| Sequence-based reagent | Ttr_Det-1 | This paper | SCRINSHOT detection probe | TACTCUGTACACUCCTTCUACAAACTTC-FITC |
| Sequence-based reagent | Foxj1_Det_1 | This paper | SCRINSHOT detection probe | AGAAGTUGTCCGTGAUCCAC/3Cy5Sp/ |
| Sequence-based reagent | Foxj1_Det_2 | This paper | SCRINSHOT detection probe | AGGAAGGAUGUGGCCAAG/3Cy5Sp/ |
| Sequence-based reagent | Foxj1_Det_3 | This paper | SCRINSHOT detection probe | CGAGGCACUTTGAUGAAGCA/3Cy5Sp/ |
| Sequence-based reagent | Sox2_Det-2 | This paper | SCRINSHOT detection probe | ATATAUACATGGATUCTCGGCAGC-cy3 |
| Sequence-based reagent | Sox2_Det-3 | This paper | SCRINSHOT detection probe | CGGAGTCUAGCTCTAAAUATTACTGG-cy3 |
| Sequence-based reagent | Ngn2_Det-1 | This paper | SCRINSHOT detection probe | GGGAAAGUTTGGTTUGACAGGT-Cy3 |
| Sequence-based reagent | Ngn2_Det-2 | This paper | SCRINSHOT detection probe | GCCTAAAUTTCCACGCUTGC-Cy3 |
| Sequence-based reagent | Ngn2_Det-3 | This paper | SCRINSHOT detection probe | ATCCTCACAUCTTCAAUAAACCAGAG-Cy3 |
| Sequence-based reagent | Hes5_Det-1 | This paper | SCRINSHOT detection probe | CCCACAUGACCAAGAGUTCAAAT-Cy3 |
| Sequence-based reagent | Hes5_Det-2 | This paper | SCRINSHOT detection probe | CAACTCUGCACACACAUTCTCT-Cy3 |
| Sequence-based reagent | Hes5_Det-3 | This paper | SCRINSHOT detection probe | CAAAGGAAUCCTTUAAGGAUCATCGT-Cy3 |
| Sequence-based reagent | Zbtb20_Det-1 | This paper | SCRINSHOT detection probe | GTCATAGUCATCTTCCAUTTCCTGC/3Cy5Sp/ |
| Sequence-based reagent | Zbtb20_Det-2 | This paper | SCRINSHOT detection probe | GAGAAAGGUACTACTTAUCCGTCAG/3Cy5Sp/ |
| Sequence-based reagent | Zbtb20_Det-3 | This paper | SCRINSHOT detection probe | AGTTTGUCAGTGAAUGTGTCGA/3Cy5Sp/ |
| Sequence-based reagent | Wnt2b_Det-1 | This paper | SCRINSHOT detection probe | CAACUCAAAUCACTCUGAAGACAGA-Cy5 |
| Sequence-based reagent | Wnt2b_Det-2 | This paper | SCRINSHOT detection probe | CGAACACCGUAATGGAUGTTGT-Cy5 |
| Sequence-based reagent | Wnt2b_Det-3 | This paper | SCRINSHOT detection probe | ATCGUGGAACGUGCAGUAGT-Cy5 |
| Sequence-based reagent | Wnt3a_Det-1 | This paper | SCRINSHOT detection probe | TTCCUAGAGAACCCUCTCTACTC-Cy5 |
| Sequence-based reagent | Wnt3a_Det-2 | This paper | SCRINSHOT detection probe | GTCCTTAGGUATGAGACCCUCA-Cy5 |
| Sequence-based reagent | Wnt3a_Det-3 | This paper | SCRINSHOT detection probe | AAGCACCCUCTCATGTAUCTAGATA-Cy5 |
| Sequence-based reagent | Foxj1_Det_2 | This paper | SCRINSHOT detection probe | TTGGGCAAUAAGTUAAGAGCCA/3Cy3Sp/ |
| Sequence-based reagent | Foxj1_Det_3 | This paper | SCRINSHOT detection probe | CAGGGTTAUTCATACCUAGAGACCA/3Cy3Sp/ |
| Sequence-based reagent | Sox2_Det-2 | This paper | SCRINSHOT detection probe | CGGCCUGCTTCATUGTTG/3Cy3Sp/ |
| Sequence-based reagent | Sox2_Det-3 | This paper | SCRINSHOT detection probe | AGAGAAGUAAAGGGATUTGTCTCC/36-FAM/ |
| Sequence-based reagent | Ngn2_Det-1 | This paper | SCRINSHOT detection probe | CCCGCCUCATTGUTGTGA/36-FAM/ |
| Sequence-based reagent | Ngn2_Det-2 | This paper | SCRINSHOT detection probe | CTTGCTUCTCCCGGUCAC/36-FAM/ |
| Sequence-based reagent | Ngn2_Det-3 | This paper | SCRINSHOT detection probe | AGGTAAATGUCAGAAGUCAAGAACA/36-FAM/ |
| Sequence-based reagent | Hes5_Det-1 | This paper | SCRINSHOT detection probe | CGGAACCUACGTGAUAAGGATT/36-FAM/ |
| Sequence-based reagent | Hes5_Det-2 | This paper | SCRINSHOT detection probe | TCAGCAUCCTCCTGUATGGA/36-FAM/ |
| Sequence-based reagent | Hes5_Det-3 | This paper | SCRINSHOT detection probe | TCGCUACCCAAGUCTTCAGA/3Cy3Sp/ |
| Sequence-based reagent | Zbtb20_Det-1 | This paper | SCRINSHOT detection probe | TACTCUGTACACUCCTTCUACAAACTTC-FITC |
| Sequence-based reagent | Zbtb20_Det-2 | This paper | SCRINSHOT detection probe | AGAAGTUGTCCGTGAUCCAC/3Cy5Sp/ |
| Sequence-based reagent | Zbtb20_Det-3 | This paper | SCRINSHOT detection probe | AGGAAGGAUGUGGCCAAG/3Cy5Sp/ |
| Sequence-based reagent | Wnt2b_Det-1 | This paper | SCRINSHOT detection probe | CGAGGCACUTTGAUGAAGCA/3Cy5Sp/ |
| Sequence-based reagent | Wnt2b_Det-2 | This paper | SCRINSHOT detection probe | ATATAUACATGGATUCTCGGCAGC-cy3 |
| Sequence-based reagent | Wnt2b_Det-3 | This paper | SCRINSHOT detection probe | CGGAGTCUAGCTCTAAAUATTACTGG-cy3 |
| Sequence-based reagent | Wnt3a_Det-1 | This paper | SCRINSHOT detection probe | GGGAAAGUTTGGTTUGACAGGT-Cy3 |
| Sequence-based reagent | Wnt3a_Det-2 | This paper | SCRINSHOT detection probe | GCCTAAAUTTCCACGCUTGC-Cy3 |
| Sequence-based reagent | Wnt3a_Det-3 | This paper | SCRINSHOT detection probe | ATCCTCACAUCTTCAAUAAACCAGAG-Cy3 |
| Sequence-based reagent | Wnt5a_Det-1 | This paper | SCRINSHOT detection probe | CCCACAUGACCAAGAGUTCAAAT-Cy3 |
| Sequence-based reagent | Wnt5a_Det-2 | This paper | SCRINSHOT detection probe | CAACTCUGCACACACAUTCTCT-Cy3 |
| Sequence-based reagent | Wnt5a_Det-3 | This paper | SCRINSHOT detection probe | CAAAGGAAUCCTTUAAGGAUCATCGT-Cy3 |
| Sequence-based reagent | Wnt7b_Det-1 | This paper | SCRINSHOT detection probe | GTCATAGUCATCTTCCAUTTCCTGC/3Cy5Sp/ |
| Sequence-based reagent | Wnt7b_Det-2 | This paper | SCRINSHOT detection probe | GAGAAAGGUACTACTTAUCCGTCAG/3Cy5Sp/ |
| Sequence-based reagent | Wnt7b_Det-3 | This paper | SCRINSHOT detection probe | AGTTTGUCAGTGAAUGTGTCGA/3Cy5Sp/ |
| Sequence-based reagent | Axin2_Det-1 | This paper | SCRINSHOT detection probe | CAACUCAAAUCACTCUGAAGACAGA-Cy5 |
| Sequence-based reagent | Axin2_Det-2 | This paper | SCRINSHOT detection probe | CGAACACCGUAATGGAUGTTGT-Cy5 |
| Sequence-based reagent | Axin2_Det-3 | This paper | SCRINSHOT detection probe | ATCGUGGAACGUGCAGUAGT-Cy5 |
| Sequence-based reagent | Lef1_Det-1 | This paper | SCRINSHOT detection probe | TCGCTGUTCATATUGGGCAT-FITC |
| Sequence-based reagent | Lef1_Det-2 | This paper | SCRINSHOT detection probe | TCTCTCTTUCCGTGCUAGTTCA-FITC |
| Sequence-based reagent | Lef1_Det-3 | This paper | SCRINSHOT detection probe | GCTATCTCTUTAGCTTCAUCAGACAC-FITC |
| Sequence-based reagent | Tcf3_Det_1 | This paper | SCRINSHOT detection probe | GAAGUCCAGGAGGUCACTC/36-FAM/ |
| Sequence-based reagent | Tcf3_Det_2 | This paper | SCRINSHOT detection probe | GCTTCGAUGGAGACCUGTC/36-FAM/ |
| Sequence-based reagent | Tcf3_Det_3 | This paper | SCRINSHOT detection probe | GATTTCCUCATCCTCTUTCTCCTC/36-FAM/ |
| Sequence-based reagent | Fzd9-Det-1 | This paper | SCRINSHOT detection probe | GTAGACATUGTAGCACAUAGAAAGGA/36-FAM/ |
| Sequence-based reagent | Fzd9-Det-2 | This paper | SCRINSHOT detection probe | TGAGGCGTUCATAAACGUAGC/36-FAM/ |
| Sequence-based reagent | Fzd9-Det-3 | This paper | SCRINSHOT detection probe | TAAATAACUCTGCACUGGACCC/36-FAM/ |
| Sequence-based reagent | Fzd10-Det-1 | This paper | SCRINSHOT detection probe | GCTGTUCTGAACCUCAAGGTA/3Cy5Sp/ |
| Sequence-based reagent | Fzd10-Det-2 | CGCAGUAGCACAUGGAGAG/3Cy5Sp/ | ||
| Sequence-based reagent | Fzd10-Det-3 | This paper | SCRINSHOT detection probe | TGTCTTGGUCTGATTGTUCATCTT/3Cy5Sp/ |
| Sequence-based reagent | Ctnnb1_Det-1 | This paper | SCRINSHOT detection probe | GACUTGGGAGGUGTCAACAT-Cy5 |
| Sequence-based reagent | Ctnnb1_Det-2 | This paper | SCRINSHOT detection probe | CCTGCUTAGTCGCUGCA-Cy5 |
| Sequence-based reagent | Ctnnb1_Det-3 | This paper | SCRINSHOT detection probe | TTACAGGUCAGTAUCAAACCAGG-Cy5 |
| Sequence-based reagent | Bmp4_Det-1 | This paper | SCRINSHOT detection probe | GCCTTUGGGATACUAGAATUAACAGAG-FITC |
| Sequence-based reagent | Bmp4_Det-2 | This paper | SCRINSHOT detection probe | CAGAUGTTCTUCGTGAUGGAAACT-FITC |
| Sequence-based reagent | Bmp4_Det-3 | This paper | SCRINSHOT detection probe | GTGGUATGTGUAGGTGGUTGAATG-FITC |
| Sequence-based reagent | Bmp7_Det-1 | This paper | SCRINSHOT detection probe | CATGTTTCUGTACTTCTUCAGGATGA-Cy5 |
| Sequence-based reagent | Bmp7_Det-2 | This paper | SCRINSHOT detection probe | TCCCGGAUGTAGTCCUTATAGATC-Cy5 |
| Sequence-based reagent | Bmp7_Det-3 | This paper | SCRINSHOT detection probe | GAAACCAAUCTGUCGCAGAAC-Cy5 |
| Sequence-based reagent | Smad6_Det-1 | This paper | SCRINSHOT detection probe | CAGGAUCCGAAACUAAGGCTT/36-FAM/ |
| Sequence-based reagent | Smad6_Det-2 | This paper | SCRINSHOT detection probe | CCTCGGUTTCAGTGUAAGACAA/36-FAM/ |
| Sequence-based reagent | Smad6_Det-3 | This paper | SCRINSHOT detection probe | CTGTGGUTGTTGAGUAGGATCT/36-FAM/ |
| Sequence-based reagent | Smad7_Det-1 | This paper | SCRINSHOT detection probe | CUGGACAGCCUGCAGT/36-FAM/ |
| Sequence-based reagent | Smad7_Det-2 | This paper | SCRINSHOT detection probe | CGCACUTTCTGUACCAGCT/36-FAM/ |
| Sequence-based reagent | Smad7_Det-3 | This paper | SCRINSHOT detection probe | ACACAGCCUCTTGACUTCC/36-FAM/ |
| Sequence-based reagent | Nog_Det-1 | This paper | SCRINSHOT detection probe | GTCAAAGAUAGGGTCUGGATGTTC/3Cy5Sp/ |
| Sequence-based reagent | Nog_Det-2 | This paper | SCRINSHOT detection probe | GCGAAGGGUACTGGGATAUAAATAG/3Cy5Sp/ |
| Sequence-based reagent | Nog_Det-3 | This paper | SCRINSHOT detection probe | CTACACAGUTAAACAGUGCATTACAG/3Cy5Sp/ |
| Sequence-based reagent | mKi67_Det_1 | This paper | SCRINSHOT detection probe | CCTTUCCAAGGGACUTTCCT/3Cy3Sp/ |
| Sequence-based reagent | mKi67_Det_2 | This paper | SCRINSHOT detection probe | ACTTAATUCACCACTTUCCTTGCA/3Cy3Sp/ |
| Sequence-based reagent | mKi67_Det_3 | This paper | SCRINSHOT detection probe | CTTTCTCTUCCTGAGGCUTTATCA/3Cy3Sp/ |
| Sequence-based reagent | Cdkn1c_Det-1 | This paper | SCRINSHOT detection probe | TCTGGUCACCAAGTUTAAGAGC/36-FAM/ |
| Sequence-based reagent | Cdkn1c_Det-2 | This paper | SCRINSHOT detection probe | GGGAUCCACGCAUGGATAA/36-FAM/ |
| Sequence-based reagent | Cdkn1c_Det-3 | This paper | SCRINSHOT detection probe | TGGCCGUTAGCCTCUAAAC/36-FAM/ |
| Sequence-based reagent | Id3-Det-1 | This paper | SCRINSHOT detection probe | CCTTCATGUTGGAGAGUAGAGATAG/3Cy3Sp/ |
| Sequence-based reagent | Id3-Det-2 | This paper | SCRINSHOT detection probe | TAAGUGAAGAGGGCUGGGT/3Cy3Sp/ |
| Sequence-based reagent | Id3-Det-3 | This paper | SCRINSHOT detection probe | GAACAGCUCTTATGCUGCCT/3Cy3Sp/ |
| Sequence-based reagent | Spc24-Det-1 | This paper | SCRINSHOT detection probe | GAGCACUGGCTGCUCTT/36-FAM/ |
| Sequence-based reagent | Spc24-Det-2 | This paper | SCRINSHOT detection probe | AAAGCAAGUAGATAAAUCAATGAACGC/36-FAM/ |
| Sequence-based reagent | Spc24-Det-3 | This paper | SCRINSHOT detection probe | CCTGGAAUACACAGCUAACAGAC/36-FAM/ |
| Sequence-based reagent | Nrarp-Det-1 | This paper | SCRINSHOT detection probe | AGTCACUAGGAAGGGUACCATG/3Cy5Sp/ |
| Sequence-based reagent | Nrarp-Det-2 | This paper | SCRINSHOT detection probe | GUGCCACAGACUTCAUACAGATAG/3Cy5Sp/ |
| Sequence-based reagent | Nrarp-Det-3 | This paper | SCRINSHOT detection probe | ACTCATUCAGCAATAGUATGAGAGAUTC/3Cy5Sp/ |
| Sequence-based reagent | Rassf4-Det-1 | This paper | SCRINSHOT detection probe | AGGAGUGAACACUGAGGTCT/3Cy3Sp/ |
| Sequence-based reagent | Rassf4-Det-2 | This paper | SCRINSHOT detection probe | TCAGCUTCCAUCAAGAAGAUCTTG/3Cy3Sp/ |
| Sequence-based reagent | Rassf4-Det-3 | This paper | SCRINSHOT detection probe | CCTTCUTCCTCUCGGTGTC/3Cy3Sp/ |
| Sequence-based reagent | Nuak2-Det-1 | This paper | SCRINSHOT detection probe | GACAAUCACAAUCTTGCTGCUATTC/36-FAM/ |
| Sequence-based reagent | Nuak2-Det-2 | This paper | SCRINSHOT detection probe | TCTCCTTUCGGGACTUCTTAAGA/36-FAM/ |
| Sequence-based reagent | Nuak2-Det-3 | This paper | SCRINSHOT detection probe | TCACGCTGUCGAGACUTCT/36-FAM/ |
| Sequence-based reagent | Fam83d-Det-1 | This paper | SCRINSHOT detection probe | GTAGATATTUCCTGTAATUGTCCGAACT/36-FAM/ |
| Sequence-based reagent | Fam83d-Det-2 | This paper | SCRINSHOT detection probe | TTCATCGCUGATGGUAGAAGC/36-FAM/ |
| Sequence-based reagent | Fam83d-Det-3 | This paper | SCRINSHOT detection probe | ACCAGGUTGGUAGAGGCT/36-FAM/ |
| Sequence-based reagent | Mybl2-Det-1 | This paper | SCRINSHOT detection probe | GTTCTTUAGCAGAAGUAGCAGCT/3Cy5Sp/ |
| Sequence-based reagent | Mybl2-Det-2 | This paper | SCRINSHOT detection probe | CTGGGAGUACTTCTGAUGATGG/3Cy5Sp/ |
| Sequence-based reagent | Mybl2-Det-3 | This paper | SCRINSHOT detection probe | GCCAUACTTCTUGACCAACUCAAT/3Cy5Sp/ |
| Sequence-based reagent | Gas2-Det-1 | This paper | SCRINSHOT detection probe | CCAGTACUCTTCTTTCCUGAAGAC/36-FAM/ |
| Sequence-based reagent | Gas2-Det-2 | This paper | SCRINSHOT detection probe | TGAAGGTTUCTGCTGTTAUCTCTTTC/36-FAM/ |
| Sequence-based reagent | Gas2-Det-3 | This paper | SCRINSHOT detection probe | CATCCATACUCTCTTTGAAUTTCTCCT/36-FAM/ |
| Sequence-based reagent | Ada-Det-1 | This paper | SCRINSHOT detection probe | AGTAGUCTGTTGUAGAGAGCUTCAT/36-FAM/ |
| Sequence-based reagent | Ada-Det-2 | This paper | SCRINSHOT detection probe | CATGTAGUAGTCAAACUTGGCCA/36-FAM/ |
| Sequence-based reagent | Ada-Det-3 | This paper | SCRINSHOT detection probe | GTACTTCUTACACAGCUCCAACA/36-FAM/ |
| Sequence-based reagent | Hspg2-Det-1 | This paper | SCRINSHOT detection probe | CTTGATCUCAAACTTCCUGTAGGC/3Cy3Sp/ |
| Sequence-based reagent | Hspg2-Det-2 | This paper | SCRINSHOT detection probe | GCCCGTAAUCTGGAUATCAGG/3Cy3Sp/ |
| Sequence-based reagent | Hspg2-Det-3 | This paper | SCRINSHOT detection probe | AGAAGTCUCAGTCTUGAGCCA/3Cy3Sp/ |
| Sequence-based reagent | Shc4-Det-1 | This paper | SCRINSHOT detection probe | GGAGTTGUAATAGTCAUGGTCTCTG/3Cy3Sp/ |
| Sequence-based reagent | Shc4-Det-2 | This paper | SCRINSHOT detection probe | GCAGGUCCAGTCCUGTTTAG/3Cy3Sp/ |
| Sequence-based reagent | Shc4-Det-3 | This paper | SCRINSHOT detection probe | GCAATAAUCTGTTGGTUGTCAAGG/3Cy3Sp/ |
| Sequence-based reagent | Nuf2-Det-1 | This paper | SCRINSHOT detection probe | AGTTCCGAUGAATCACTUTCAATCT/3Cy5Sp/ |
| Sequence-based reagent | Nuf2-Det-2 | This paper | SCRINSHOT detection probe | TACACTAACUGTAAGGCTUTCATGTAA/3Cy5Sp/ |
| Sequence-based reagent | Nuf2-Det-3 | This paper | SCRINSHOT detection probe | ATTTCTTGAUTAACAGTGGUTACTTGC/3Cy5Sp/ |
| Sequence-based reagent | Ndc80-1 | This paper | SCRINSHOT detection probe | GACATGAATUCTTCATACUCTCTGTCG/3Cy3Sp/ |
| Sequence-based reagent | Ndc80-2 | This paper | SCRINSHOT detection probe | CAAGTUGACTATTCCUGGATCCA/3Cy3Sp/ |
| Sequence-based reagent | Ndc80-3 | This paper | SCRINSHOT detection probe | GTCTTGCAAUCTGTTCATUTAGTGC/3Cy3Sp/ |
| Sequence-based reagent | Spc25-Det-1 | This paper | SCRINSHOT detection probe | TCTATCATCUAGGTCCCAUGTTTAGG/3Cy5Sp/ |
| Sequence-based reagent | Spc25-Det-2 | This paper | SCRINSHOT detection probe | AGGTAATCUCTATACAGAUCCGCTG/3Cy5Sp/ |
| Sequence-based reagent | Spc25-Det-3 | This paper | SCRINSHOT detection probe | GTTAGCAGUGGAAATGGUTTCC/3Cy5Sp/ |





