Transcriptome profiling of tendon fibroblasts at the onset of embryonic muscle contraction reveals novel force-responsive genes
Figures
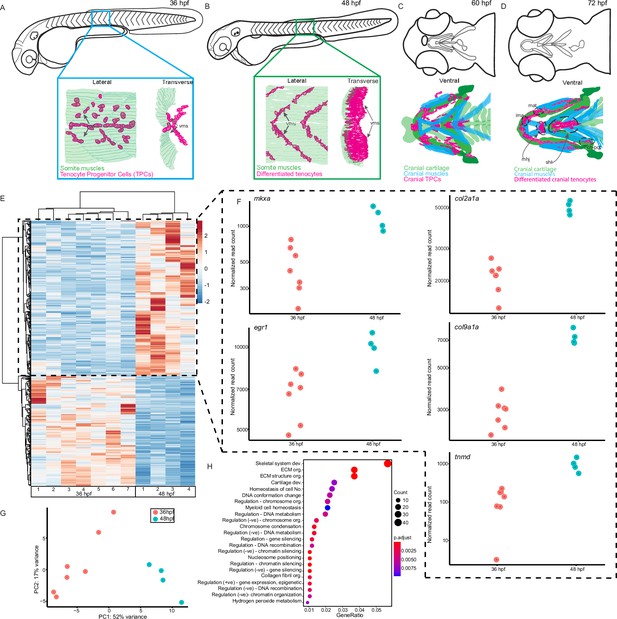
Onset of embryonic muscle contraction regulates transcription in tenocytes.
(A–D) Diagrams depicting changes in tenocyte distribution and morphology during onset of trunk and cranial muscle contractions, (A) 36 hpf when twitching movements are sporadic and (B) 48 hpf when embryos become free swimming. Lateral views of 36 (A) and 48 hpf embryos (B). Insets show lateral and transverse views of migrating tenocyte progenitors (A) and differentiated tenocytes at somite boundaries with polarized, branched projections (B). Ventral views of the embryonic head in 60 hpf (C) and 72 hpf (D) embryos just prior to and during the onset of jaw movements. Cartilage (green), tenocytes (magenta), and muscles (cyan) showing tenocyte elongation, particularly in the sternohyoid tendon (sht) and condensation, as well as the mandibulohyoid junction (mhj). (E) Heatmaps from bulk RNA-sequencing (RNA-seq) showing the top 1000 differentially expressed genes (DEGs) between 36 and 48 hpf. p < 0.05. (F) Elevated expression of tenocyte marker genes mkxa, tnmd, and egr1 and extracellular matrix (ECM) genes col2a1a, col9a1a in RNA-seq experiments at 48 hpf. Datapoints represent normalized read counts of single biological replicates at each color-coded timepoint (n = 7 for 36 hpf, n = 4 for 48 hpf). (G) Elevated expression of cartilage marker genes col2a1a and col9a1a in 48 hpf samples. (H) PCA of individual replicates showing separation of experimental conditions by timepoint. (I) GO analysis using Biological Process (BP) terms of top 2788 DEGs by adjusted p-value.
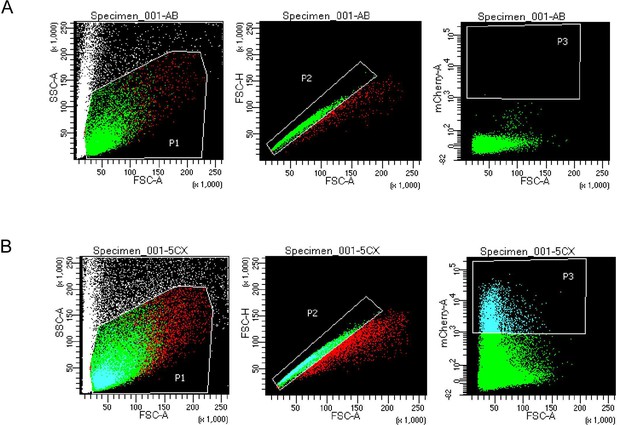
FACS gating thresholds for mCherry+ cells.
(Left to right for each) Forward Scatter A (FSC-A) versus Side Scatter A (SSC-A) shows P1 threshold. FSC-A versus Forward Scatter H (FSC-H) shows P2 threshold to select for single cells. FSC-A versus mCherry-A shows P3 fluorescence gating for mCherry+ cells. (A) P3 selection gating allowed selection of cells with strong mCherry signal based on Negative control AB (WT) sample versus mCherry expressing tenocytes (48 hpf) FACS gating. Established P3 gating selected for mCherry-positive cells in all 36 and 48 hpf Tg(scxa:mCherry) samples. (B) Thresholds used in 36 and 48 hpf Tg(scxa:mCherry) samples for FACS prior to bulk RNA-sequencing (RNA-seq).
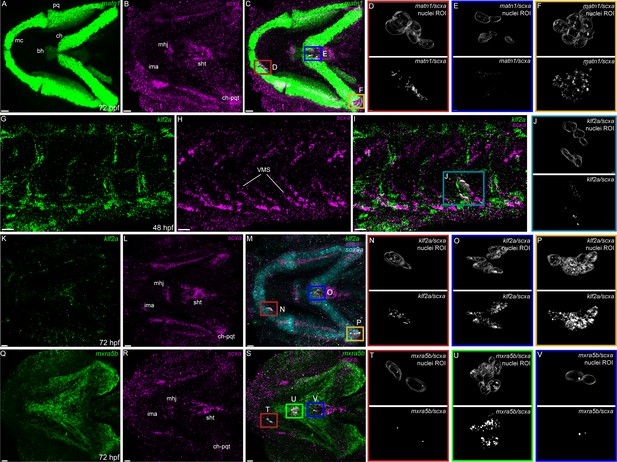
Expression of matn1, klf2a, and mxra5b with scxa in cranial and trunk tenocytes.
Ventral cranial (A–F, K–V) and lateral trunk (G–J) views of 72 hpf (A–F, K–V) and 48 hpf (G–J) embryos showing isHCR of matn1 (A, C–F), klf2a (G, I–K, M–P), and mxra5b (Q, S–V) in combination with scxa (B–F, H–J, L–P, R–V). (D–F, J, N–P, T–V) Higher magnification views of tenocyte nuclei in marked ROI. (C, D, M, N, S, T) ROI and panels outlined in magenta show magnified views of 3D volumes of tenocytes associated with imt. (I, J) ROI and panels outlined in cyan show magnified views of 3D volume of VMS tenocytes. (C, E, M, O, S, V) ROI and panels outlined in royal blue show magnified views of 3D volume of tenocytes associated with sht enthesis. (C, F, M, P) ROI and panels outlined in yellow show magnified views of 3D volumes of tenocytes associated with ch-pqt. (S, U) ROI and panels outlined in green show magnified views of 3D volumes of tenocytes associated with mhj. Each magnified view of ROI displays a translucent outline of the nuclear 3D volume with white puncta representing voxel colocalizations of isHCR as depicted by the colocalization function in Imaris (see Methods). mc – Meckel’s cartilage, pq – palatoquadrate, ch – ceratohyal, bh – basihyal cartilage, ima – intermadibularis anterior tendon, mhj – mandibulohyoid junction, sht – sternohyoideus tendon, ch-pqt – ceratohyal-palatoquadrate tendon, sb – somite boundary. Scale bars = 20 µm.
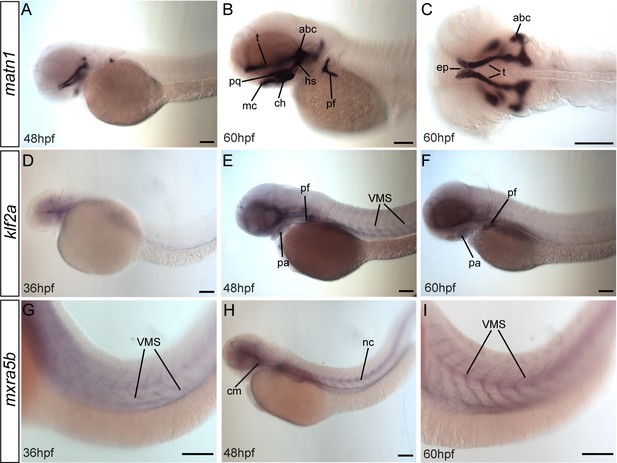
matn1, klf2a, and mxra5b are expressed in musculoskeletal tissues of developing embryos.
Lateral (A, B, D–I) and ventral (C) views of embryos showing expression of matn1 (A–C), klf2a (D–F), and mxra5b (G, H). (A–C) 48 hpf embryos show matn1 expression in cartilage progenitors at 48 hpf and in differentiated cartilages (and associated tenocytes) at 60 hpf (B, C). Lateral views of 36 hpf (D), 48 hpf (E), and 60 hpf (F) embryos show klf2a expression in pharyngeal mesenchyme (D), skeletal progenitors and in tenocytes along VMS (E, F). Lateral views of 36 hpf (G), 48 hpf (H), and 60 hpf (I) embryos show mxra5b expression in tenocytes along horizontal myosepta (HMS) along the notochord and VMS. Scale bars = 100 μm. Abbreviations: abc = anterior basicranial commissure, ch = ceratohyal cartilage, ep = ethmoid plate, hs = hyosymplectic cartilage, mc = meckel’s cartilage, nc = notochord, pf = pectoral fin, pq = palatoquadrate cartilage, sb = somite boundaries, t = trabeculae cartilage.
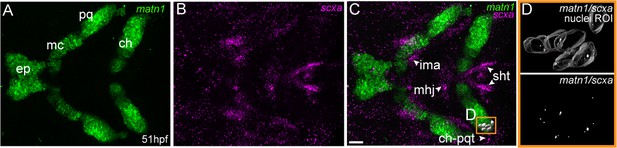
matn1 is expressed in differentiating cranial tenocytes.
Ventral view of the developing mandibular arch in a 51 hpf embryo showing in situ Hybridization Chain Reaction (isHCR) of matn1 (A, C, D) and scxa (B–D). (D) magnified view of yellow ROI (C) shows outline of tenocyte nuclear 3D volume with white puncta representing voxel colocalizations of matn1 and scxa as depicted by colocalization using Imaris (see methods). ep = ethmoid plate cartilage, ch = ceratohyal cartilage, mc = meckel’s cartilage, pq = palatoquadrate cartilage, ch-pqt = ceratohyal-palatoquadrate tendon, ima = intermandibularis anterior tendon, mhj = mandibulohyoid junction tendon, sht = sternohyoideus tendon. Scale bars = 20 µm.
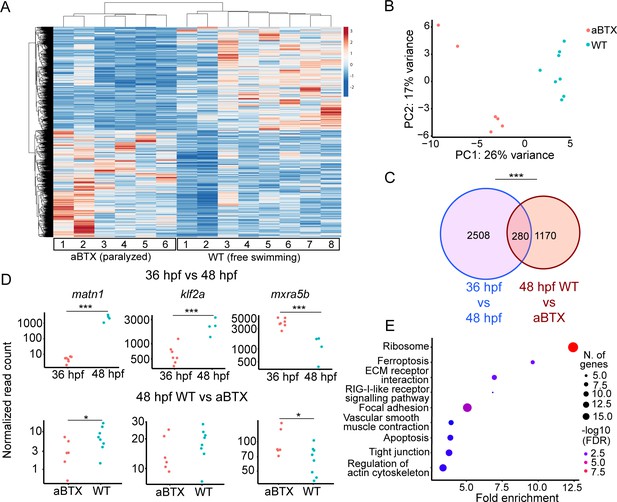
Paralysis regulates tenocyte gene expression in developing musculoskeletal system.
(A) Heatmap of differentially expressed genes (DEGs) from bulk RNA-sequencing (RNA-seq) between WT and aBTX-injected (aBTX-inj) paralyzed 48 hpf embryos (force perturbed). (B) PCA of individual replicates WT versus aBTX-inj embryos’ RNA-seq separate by experimental condition. (C) Venn diagram shows overlap of genes between developmental time-point and force perturbed RNA-seq experiments. (D) Comparison of normalized read counts between replicates of matn1, klf2a, and mxra5b in 36 versus 48 hpf and WT versus aBTX RNA-seq experiments. (E) KEGG pathway analysis plot shows enrichment of overlapping genes from (C). ns = not significant, *p < 0.05, ***p < 0.001.
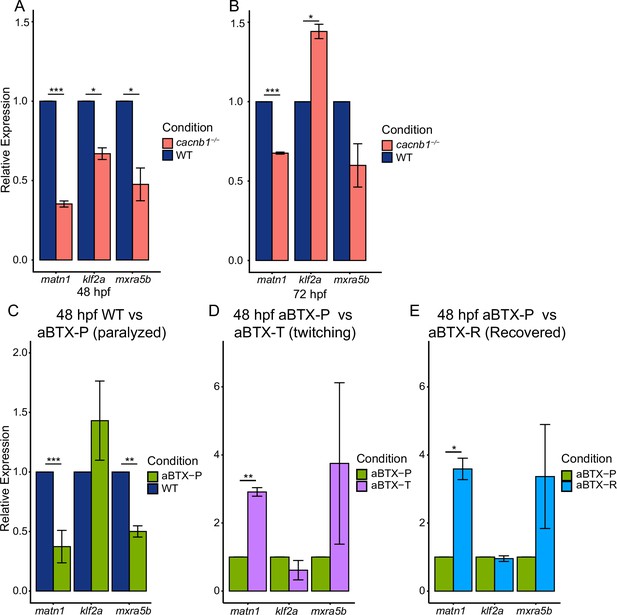
Paralysis regulates gene expression of matn1, klf2a, and mxra5b in developing embryos.
Bar plots showing global changes in relative expression from RT-qPCR of matn1, klf2a, and mxra5b genes between WT and cacnb1−/− mutant embryos at 48 hpf (A) and 72 hpf (B). Bar plots show global changes in relative expression of matn1, klf2a, and mxra5b between 48 hpf uninjected WT controls (blue) and aBTX-injected paralyzed (green) embryos (C), aBTX-injected paralyzed (green) and aBTX-injected ‘Twitching’ (partially recovered, magenta) embryos (D), and between aBTX-injected paralyzed (green) and aBTX-injected, ‘Recovered’ (cyan) embryos (E) at 48 hpf (right). ns = not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
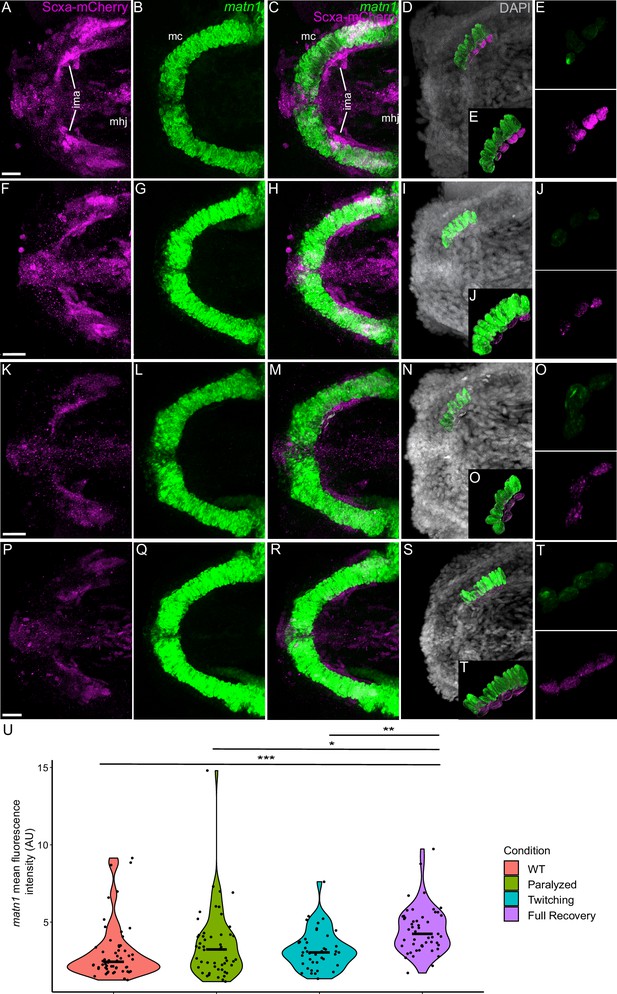
Mechanical force differentially regulates expression of matn1 in ima enthesis tenocytes.
Ventral views of Meckel’s cartilage and associated tenocytes showing in situ Hybridization Chain Reaction (isHCR) of matn1 (green) and anti-mCherry immunofluorescence (magenta) marking the tenocytes in Tg(scxa:mCherry) embryos at 72 hpf in WT uninjected (WT) (A–E), aBTX-inj (Paralyzed) (F–J), partially recovered aBTX-inj (Twitching) (K–O), and completely recovered aBTX-inj (Full Recovery) (P–T) conditions at ima enthesis. (D, I, N, S) Grayscale images showing nuclei stained with DAPI with ROIs showing isolated 3D volumes of chondrocytes (green) and enthesis tenocytes (magenta) based on DAPI signal. (E, J, O, T) Insets showing magnified views of the 3D volumes of tenocytes associated with ima enthesis depicting expression of matn1 and stained for mCherry. (U) Violin plot showing changes in mean fluorescence intensity of matn1 in ima enthesis tenocyte nuclei between WT (n = 8), Paralyzed (n = 8), Twitching (n = 6), and Full Recovery (n = 7) with ~8 nuclei measured per embryo. p-value calculated with linear mixed effects model with Tukey post hoc test. *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars = 20 µm.
-
Figure 4—source data 1
Measurements of matn1 isHCR signal intensity in ima enthesis tenocytes.
- https://cdn.elifesciences.org/articles/105802/elife-105802-fig4-data1-v2.xlsx
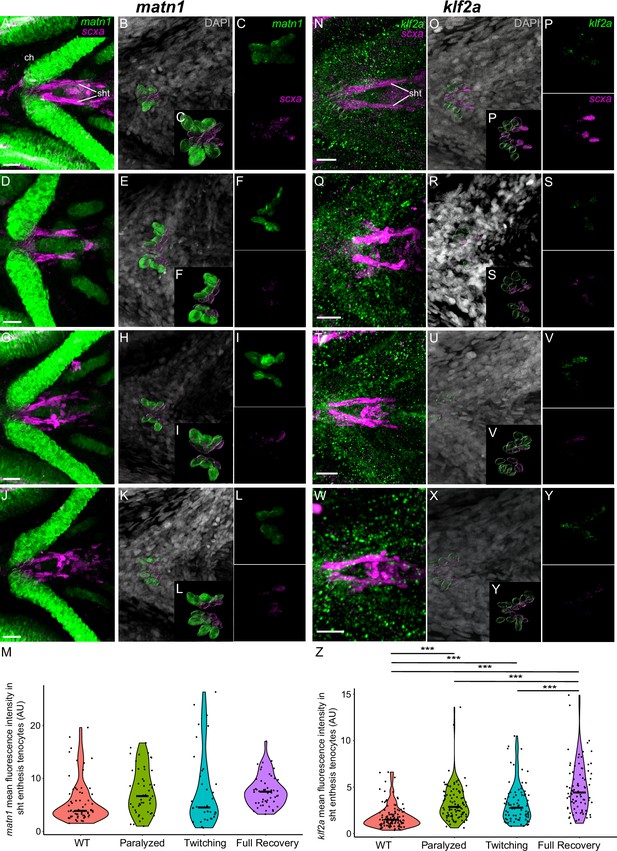
Mechanical force regulates expression of matn1 and klf2a in sht enthesis tenocytes.
Ventral views of ceratohyal (ch) cartilage and associated tenocytes showing in situ Hybridization Chain Reaction (isHCR) of matn1 (green) (A–L) and klf2a (green) (N–Y) and anti-mCherry immunofluorescence (magenta) marking the tenocytes in Tg(scxa:mCherry) embryos at 72 hpf in WT uninjected (WT) (A–C, N–P), aBTX-inj (Paralyzed) (D–F, Q–S), partially recovered aBTX-inj (Twitching) (G–I, T–V), and completely recovered aBTX-inj (Full Recovery) (J–L, W–Y) conditions at sht enthesis. (B, E, H, K, O, R, U, X) Grayscale images showing nuclei stained with DAPI with ROIs showing isolated 3D volumes of chondrocytes (green) and sternohyoideus-ceratohyal (sht) enthesis tenocytes (magenta) based on DAPI signal. (C, F, I, L, P, S, V, Y) Insets showing magnified views of the 3D volumes of tenocytes associated with sht enthesis depicting expression of matn1 and stained for mCherry. (M) Violin plot showing changes in mean fluorescence intensity of matn1 in sht enthesis tenocyte nuclei between WT (n = 8), Paralyzed (n = 8), Twitching (n = 6), and Full Recovery (n = 7) with ~8 nuclei measured per embryo. (Z) Violin plot showing changes in mean fluorescence intensity of klf2a in sht enthesis tenocyte nuclei between WT (n = 15), Paralyzed (n = 16), Twitching (n = 14), and Full Recovery (n = 11) with ~8 nuclei measured per embryo. p-values calculated with linear mixed effects model with Tukey post hoc test. ***p < 0.001. Scale bars = 20 µm.
-
Figure 4—figure supplement 1—source data 1
Measurements of matn1 and klf2a isHCR signal intensity in sht enthesis tenocytes.
- https://cdn.elifesciences.org/articles/105802/elife-105802-fig4-figsupp1-data1-v2.xlsx
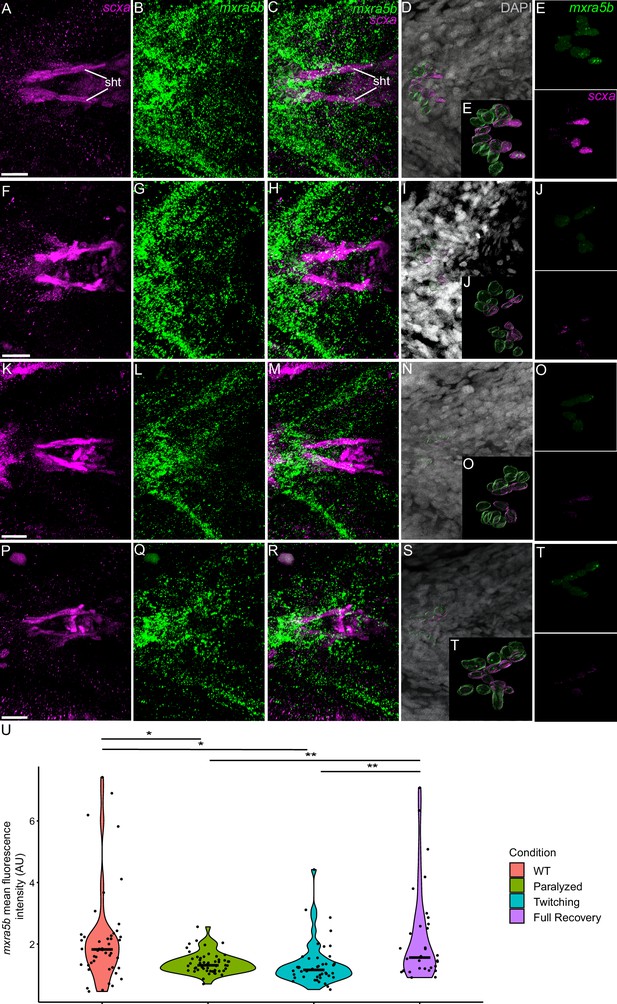
Mechanical force differentially regulates expression of mxra5b in sht enthesis tenocytes.
Ventral views of ceratohyal (ch) cartilage and associated tenocytes showing in situ Hybridization Chain Reaction (isHCR) of mxra5b (green) and anti-mCherry immunofluorescence (magenta) marking the tenocytes in Tg(scxa:mCherry) embryos at 72 hpf in WT uninjected (WT) (A–E), aBTX-inj paralyzed (F–J), partially recovered aBTX-inj (Twitching) (K–O), and completely recovered aBTX-inj (Full Recovery) (P–T) conditions at sht enthesis. (D, I, N, S) Grayscale images showing nuclei stained with DAPI with ROIs showing isolated 3D volumes of chondrocytes (green) and sht enthesis tenocytes (magenta) based on DAPI signal. (E, J, O, T) Insets showing magnified views of the 3D volumes of tenocytes associated with sht enthesis depicting expression of mxra5b and stained for mCherry. (U) Violin plot showing changes in mean fluorescence intensity of mxra5b in sht enthesis tenocyte nuclei between WT (n = 7), Paralyzed (n = 8), Twitching (n = 8), and Full Recovery (n = 4) with ~8 nuclei measured per embryo. p-value calculated with linear mixed effects model with Tukey post hoc test. *p < 0.05, **p < 0.01. Scale bars = 20 µm.
-
Figure 5—source data 1
Measurements of mxra5b isHCR signal intensity in sht enthesis tenocytes.
- https://cdn.elifesciences.org/articles/105802/elife-105802-fig5-data1-v2.xlsx

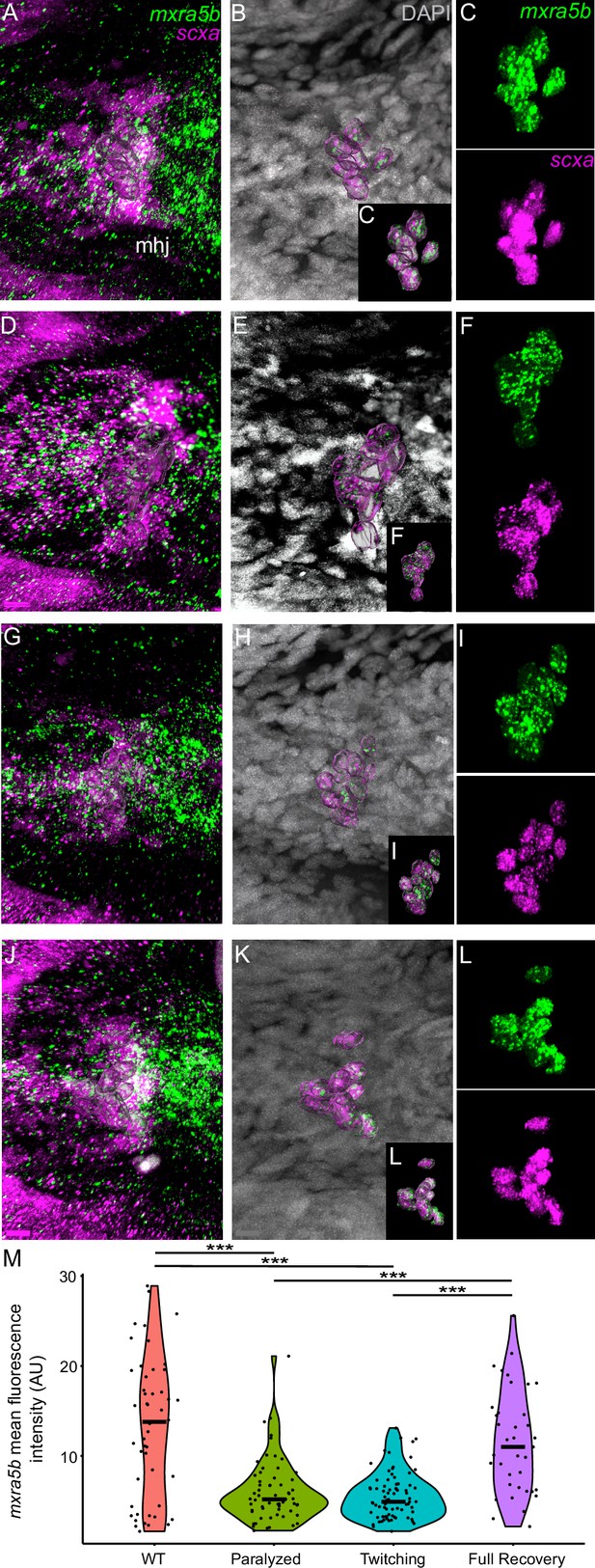
Mechanical force differentially regulates expression of mxra5b and klf2a in ima enthesis tenocytes.
Ventral views of Meckels cartilage and associated tenocytes showing in situ Hybridization Chain Reaction (isHCR) of klf2a (green) (A–L) and mxra5b (green) (N–Y) and anti-mCherry immunofluorescence (magenta) marking the tenocytes in Tg(scxa:mCherry) embryos at 72 hpf in WT uninjected (WT) (A–C, N–P), aBTX-inj (Paralyzed) (D–F, Q–S), partially recovered aBTX-inj (Twitching) (G–I, T–V), and completely recovered aBTX-inj (Full Recovery) (J–L, W–Y) conditions at ima enthesis. (B, E, H, K, O, R, U, X) Grayscale images showing nuclei stained with DAPI with ROIs showing isolated 3D volumes of chondrocytes (green) and ima enthesis tenocytes (magenta) based on DAPI signal. (C, F, I, L, P, S, V, Y) Insets showing magnified views of the 3D volumes of tenocytes associated with ima enthesis depicting expression of mxra5b and klf2a and stained for mCherry. (M) Violin plot showing changes in mean fluorescence intensity of klf2a in ima enthesis tenocyte nuclei between WT (n = 15), Paralyzed (n = 16), Twitching (n = 14), and Full Recovery (n = 11) with ~8 nuclei measured per embryo. (Z) Violin plot showing changes in mean fluorescence intensity of mxra5b in ima enthesis tenocyte nuclei between WT (n = 7), Paralyzed (n = 8), Twitching (n = 8), and Full Recovery (n = 4) with ~8 nuclei measured per embryo. p-values calculated with linear mixed effects model with Tukey post hoc test. ***p < 0.001. Scale bars = 20 µm.
-
Figure 5—figure supplement 1—source data 1
Measurements of klf2a and mxra5b isHCR signal intensity in ima enthesis tenocytes.
- https://cdn.elifesciences.org/articles/105802/elife-105802-fig5-figsupp1-data1-v2.xlsx
Mechanical force regulates expression of mxra5b in mhj myotendinous junction tenocytes.
Ventral views of mandibulohyoid (mhj) myotendinous junction (MTJ) associated tenocytes showing in situ Hybridization Chain Reaction (isHCR) of mxra5b (green) and anti-mCherry immunofluorescence (magenta) marking the tenocytes in Tg(scxa:mCherry) embryos at 72 hpf in WT uninjected (WT) (A–C), aBTX-inj (Paralyzed) (D–F), partially recovered aBTX-inj (Twitching) (G–I), and completely recovered aBTX-inj (Full Recovery) (J–L) conditions. (B, E, H, K) Grayscale images showing nuclei stained with DAPI with ROIs showing isolated 3D volumes of mhj tenocytes (magenta) based on DAPI signal. (C, F, I, L) Insets showing magnified views of the 3D volumes of tenocytes associated with mhj MTJ depicting expression of mxra5b and stained for mCherry. (M) Violin plot showing changes in mean fluorescence intensity of mxra5b in mhj MTJ tenocyte nuclei between WT (n = 7), Paralyzed (n = 8), Twitching (n = 8), and Full Recovery (n = 4) with ~10 nuclei measured per embryo. p-value calculated with linear mixed effects model with Tukey post hoc test. ***p < 0.001. Scale bars = 20 µm.
-
Figure 5—figure supplement 2—source data 1
Measurements of mxra5b isHCR signal intensity in mhj MTJ tenocytes.
- https://cdn.elifesciences.org/articles/105802/elife-105802-fig5-figsupp2-data1-v2.xlsx

Mechanical force differentially regulates expression of mxra5b and klf2a in sht myotendinous junction tenocytes.
Ventral views of sht and associated myotendinous junction (MTJ) Tenocytes showing in situ Hybridization Chain Reaction (isHCR) of mxra5b (green) (A–L) and klf2a (green) (N–Y) and anti-mCherry immunofluorescence (magenta) marking the tenocytes in Tg(scxa:mCherry) embryos at 72 hpf in WT uninjected (WT) (A–C, N–P), aBTX-inj (Paralyzed) (D–F, Q–S), partially recovered aBTX-inj (Twitching) (G–I, T–V), and completely recovered aBTX-inj (Full Recovery) (J–L, W–Y) conditions at sht MTJ. (B, E, H, K, O, R, U, X) Grayscale images showing nuclei stained with DAPI with ROIs showing isolated 3D volumes of ima enthesis tenocytes (magenta) based on DAPI signal. (C, F, I, L, P, S, V, Y) Insets showing magnified views of the 3D volumes of tenocytes associated with sht enthesis depicting expression of mxra5b and klf2a and stained for mCherry. (M) Violin plot showing changes in mean fluorescence intensity of mxra5b in sht MTJ tenocyte nuclei between WT (n = 7), Paralyzed (n = 8), Twitching (n = 8), and Full Recovery (n = 4) with ~4 nuclei measured per embryo. (Z) Violin plot showing changes in mean fluorescence intensity of klf2a in ima enthesis tenocyte nuclei between WT (n = 15), Paralyzed (n = 16), Twitching (n = 14), and Full Recovery (n = 11) with ~4 nuclei measured per embryo. p-values calculated with linear mixed effects model with Tukey post hoc test. *p < 0.05, **p < 0.01. Scale bars = 20 µm.
-
Figure 5—figure supplement 3—source data 1
Measurements of mxra5b and klf2a isHCR signal intensity in sht MTJ tenocytes.
- https://cdn.elifesciences.org/articles/105802/elife-105802-fig5-figsupp3-data1-v2.xlsx
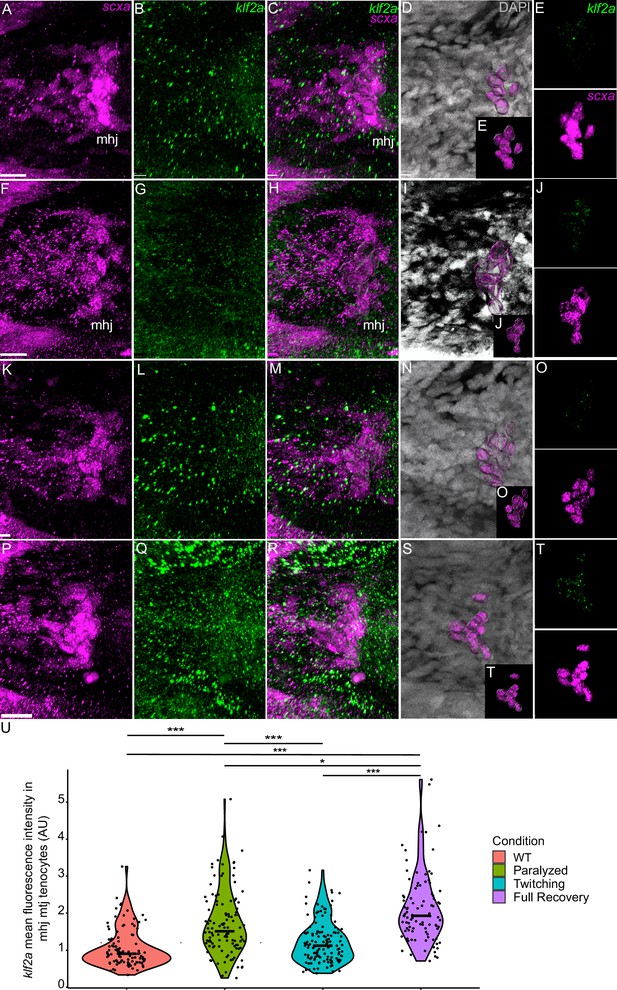
Mechanical force regulates expression of klf2a in mhj myotendinous junction tenocytes.
Ventral views of mandibulohyoid junction (mhj), myotendinous junction (MTJ) associated tenocytes showing in situ Hybridization Chain Reaction (isHCR) of klf2a (green) and anti-mCherry immunofluorescence (magenta) marking the tenocytes in Tg(scxa:mCherry) embryos at 72 hpf in WT uninjected (WT) (A–E), aBTX-inj (Paralyzed) (F–J), partially recovered aBTX-inj (Twitching) (K–O), and completely recovered aBTX-inj (Full Recovery) (P–T) conditions. (D, I, N, S) Grayscale images showing nuclei stained with DAPI with ROIs showing isolated 3D volumes of mhj tenocytes (magenta) based on DAPI signal. (E, J, O, T) Insets showing magnified views of the 3D volumes of tenocytes associated with mhj MTJ depicting expression of klf2a and stained for mCherry. (U) Violin plot showing changes in mean fluorescence intensity of klf2a in mhj MTJ tenocyte nuclei between WT (n = 17), Paralyzed (n = 15), Twitching (n = 14), and Full Recovery (n = 11) with ~10 nuclei measured per embryo. p-value calculated with linear mixed effects model with Tukey post hoc test. *p < 0.05, ***p < 0.001. Scale bars = 20 µm.
-
Figure 6—source data 1
Measurements of klf2a isHCR signal intensity in mhj MTJ tenocytes.
- https://cdn.elifesciences.org/articles/105802/elife-105802-fig6-data1-v2.xlsx
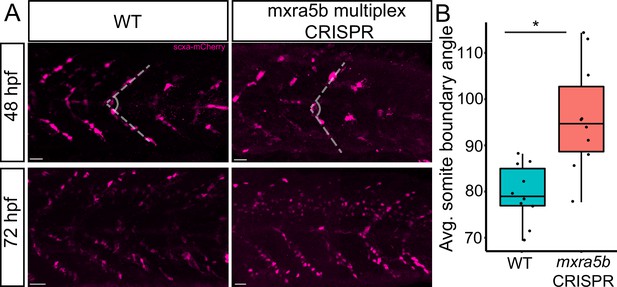
Loss of mxra5b function affects somite boundary structure.
(A) Lateral views of WT and mxra5b multiplex CRISPants at 48 and 72 hpf Tg(scx:mCherry) embryos stained with anti-mCherry to show tenocytes at the somite boundary (SB). (B) Quantification of somite boundary angle measurements of 48 hpf WT or mxra5b multiplex CRISPant embryos. p-value calculated with Watson’s U2 test. *p < 0.05.
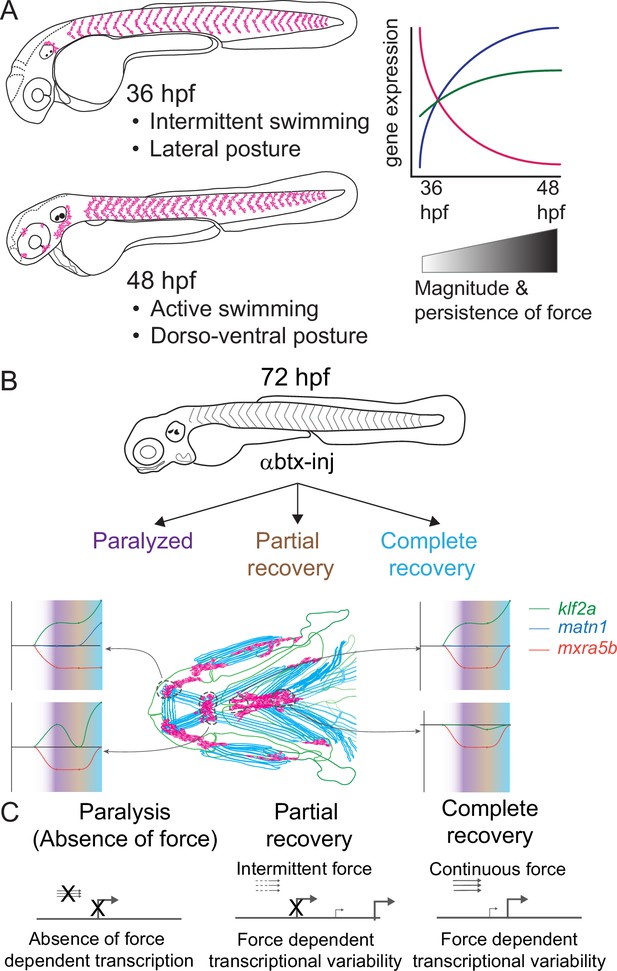
Model depicting role of mechanical force in regulating expression of genes in tenocytes during onset of active muscle contraction.
(A) Cartoon showing role of force in regulating tenocyte morphogenesis and gene expression in tenocytes between 36 and 48 hpf stages correlating with onset of active swimming The variability in gene expression is related to increase in both magnitude and persistence of muscle contraction force. (B) Representative model summarizing the multifaceted role of muscle contractile force on expression dynamics of matn1, klf2a, and mxra5b genes in cranial tendon attachments. (C) Force-responsive gene expression is more nuanced than a binary on/off control.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (Danio rerio) | AB | Schilling lab | RRID:NCBITaxon_7955 | |
| Genetic reagent (Danio rerio) | Tg(scxa:mCherry) | Galloway lab | fb301Tg; RRID:ZFIN_ZDB-GENO-180925-6 | scx BAC transgenic in AB background |
| Genetic reagent (Danio rerio) | cacnb1−/−; Tg(scxa:mCherry) | Schilling lab | Ir1092;fb301; RRID:ZFIN_ZDB-ALT-191023-1 | cacnb1 mutant in Tg(scx:mCherry) background |
| Sequence-based reagent | T7 sequence-tagged primers | This paper | Supplementary file 7 | 2 mM final concentration |
| Commercial assay or kit | Protoscript II first strand cDNA synthesis kit | New England Biolabs | Cat # E6560 | |
| Commercial assay or kit | T7 RNA polymerase | Millipore Sigma (Roche) | Cat # 10881767001 | |
| Commercial assay or kit | Monarch Total RNA Miniprep kit | New England Biolabs | Cat # T2010S | |
| Commercial assay or kit | DIG RNA labeling mix | Millipore Sigma (Roche) | Cat # 11277073910 | |
| Commercial assay or kit | MEGAshortscript T7 transcription kit | Thermo Fisher Scientific (Invitrogen) | Cat # AM1354 | |
| Commercial assay or kit | Luna Universal qPCR master mix | New England Biolabs | Cat # M3003S | |
| Commercial assay or kit | Zirconium beads | Benchmark Scientific | Cat # D1032-10 | |
| Commercial assay or kit | RNEasy Micro Kit | QIAGEN | Cat # 74004 | |
| Commercial assay or kit | 40 µm filter | Pluriselect-USA | Cat # 43-10040-50 | |
| Commercial assay or kit | HCR Buffers (v3.0) | Molecular Instruments | Hybridization buffer, Wash buffer, Amplifier buffer | |
| Antibody | Anti-Digoxigenin-AP, Fab fragments | Millipore Sigma (Roche) | Cat # 11093274910 RRID:AB_514497 | 1:2000 |
| Antibody | Rat monoclonal anti-mCherry antibody | Invitrogen (Thermo Fisher Scientific) | Cat # M11217 RRID:AB_2536611 | 1:500 |
| Antibody | Chicken polyclonal anti-GFP antibody | abcam | Cat # ab13970 RRID:AB_300798 | 1:1000 |
| Antibody | Mouse monoclonal anti-myosin heavy chain antibody | Developmental Studies Hybridoma Bank (DHSB) | Cat # A4.1025 RRID:AB_528356 | 1:200 |
| Antibody | Alexa Fluor 594 AffiniPure F(ab’)2 Fragment Donkey polyclonal anti-Rat IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat # 712-586-153 RRID:AB_2340691 | 1:1000 |
| Antibody | Alexa Fluor 488 AffiniPure F(ab’)2 Fragment Donkey polyclonal anti-Chicken IgY IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat # 703-546-155 RRID:AB_2340376 | 1:1000 |
| Antibody | Alexa Fluor 647 AffiniPure F(ab’)2 Fragment Donkey polyclonal anti-Mouse IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat # 715-606-151 RRID:AB_2340866 | 1:1000 |
| Chemical compound, drug | Nitro Blue Tetrazolium chloride solution (NBT) | Millipore Sigma (Roche) | Cat # 11383213001 PubChem CID: 9281 | |
| Chemical compound, drug | 5-Bromo-4-chloro-3-indolyl phosphate solution | Millipore Sigma (Roche) | Cat # 11383221001 PubChem CID: 81059 | |
| Chemical compound, drug | Ethylenediaminetetraacetic acid disodium salt | Millipore Sigma (Roche) | Cat # E5134 PubChem CID: 8759 | |
| Chemical compound, drug | Calcium chloride hexahydrate | Millipore Sigma (Roche) | Cat # 21108 PubChem CID: 6093252 | |
| Chemical compound, drug | Dulbecco’s phosphate-buffered saline (DPBS) 1× | Thermo Fisher Scientific (Gibco) | Cat # 14190144 | |
| Chemical compound, drug | Agarose low gelling temperature | Millipore Sigma (Sigma-Aldrich) | Cat # A9414 | |
| Chemical compound, drug | SSC buffer 20× | Millipore Sigma (Sigma-Aldrich) | Cat # S6639-1L | |
| Chemical compound, drug | DAPI | Millipore Sigma (Sigma-Aldrich) | Cat # D9542 PubChem CID: 2954 | |
| Sequence-based reagent | matn1-B1 | Molecular instruments | NM_001099740.2 | 20 probe set |
| Sequence-based reagent | mxra5b-B1 | Molecular instruments | XM_017357865.2 | 20 probe set |
| Sequence-based reagent | mxra5b-B1 | Molecular instruments | XM_017357865.2 | 20 probe set |
| Sequence-based reagent | klf2a-B3 | Molecular instruments | NM_131856.3 | 20 probe set |
| Sequence-based reagent | scxa-B2 | Molecular instruments | NM_001083069 | 18 probe set |
| Sequence-based reagent | B1-h1&h2- Alexa Fluor 488 amplifier hairpins | Molecular instruments | HCR RNA-FISH (v3.0) | |
| Sequence-based reagent | B2-h1&h2- Alexa Fluor 546 amplifier hairpins | Molecular instruments | HCR RNA-FISH (v3.0) | |
| Sequence-based reagent | B3-h1&h2- Alexa Fluor 647 amplifier hairpins | Molecular instruments | HCR RNA-FISH (v3.0) | |
| Other | BD FACSAria II Cell Sorter | Becton, Dickinson and Company | RRID:SCR_018934 | |
| Other | Bioanalyzer 2100 instrument | Agilent | RRID:SCR_018043 | |
| Other | HiSeq 4000 sequencing system | Illumina | RRID:SCR_016386 | |
| Other | NextSeq 550 system | Illumina | RRID:SCR_016381 | |
| Other | LightCycler 480 Real Time PCR System | Roche | RRID:SCR_018626 | |
| Other | SP8 Lightning Confocal microscope | Leica | RRID:SCR_018169 | |
| Other | Zeiss Axioplan 2 imaging system | Zeiss | RRID:SCR_020918 | |
| Other | MicroPublisher color RTV-5.0 CCD camera | QImaging | ||
| Other | BeadBug 3 microtube homogenizer | Benchmark Scientific | Cat # D1030 | |
| Peptide, recombinant protein | Protease (Subtilisin Carlsberg) from Bacillus licheniformis | Millipore Sigma (Sigma-Aldrich) | Cat # P5380 UniProtKB: P00780.SUBC_BACLI | |
| Peptide, recombinant protein | Collagenase Type IV from Hathewaya histolytica (Clostridium histolyticum) | Thermo Fisher Scientific (Gibco Life technologies) | Cat # 17104019 | |
| Peptide, recombinant protein | Deoxyribonuclease I (DNase I) from bovine pancreas | Millipore Sigma (Roche) | Cat # 10104159001 UniProtKB: P00639.DNAS1_BOVIN | |
| Peptide, recombinant protein | Bovine serum albumin stock solution (10%) | Miltenyi Biotec | Cat # 130-091-376 | |
| Recombinant DNA reagent | pmtb-t7-alpha-bungarotoxin | Addgene (Megason lab) | Cat # 69542 RRID:Addgene_69542 | |
| Software, algorithm | Spliced Transcripts Alignment to a Reference (STAR) v2.5.2a | Dobin lab | RRID:SCR_004463 | |
| Software, algorithm | Smart-seq2 single sample pipeline | Broad Institute | RRID:SCR_021228 | |
| Software, algorithm | RSEM v1.2.31 | Dewey lab | RRID:SCR_000262 | |
| Software, algorithm | DESeq2 v1.30.1 | Anders lab | RRID:SCR_015687 | |
| Software, algorithm | ClustVis | Vilo lab | RRID:SCR_017133 | |
| Software, algorithm | ClusterProfiler R package | Qing-Yu lab | RRID:SCR_016884 | |
| Software, algorithm | ShinyGO | Ge lab | RRID:SCR_019213 | |
| Software, algorithm | VennDiagram v1.7.3 | Boutros lab | RRID:SCR_002414 | |
| Software, algorithm | GeneOverlap v1.26.0 | Shen lab | RRID:SCR_018419 | |
| Software, algorithm | LightCycler Software | Roche | RRID:SCR_012155 | |
| Software, algorithm | Zeiss Zen Microscopy software | Zeiss | RRID:SCR_013672 | |
| Software, algorithm | Leica Application Suite X | Leica | RRID:SCR_013673 | |
| Software, algorithm | Imaris | Bitplane | RRID:SCR_007370 | |
| Other | Optical Biology Core at UCI | Department of Developmental Biology, UCI | RRID:SCR_026614 | Core facility |
| Other | Genomics Research and Technology Hub Core at UCI | Department of Biological Chemistry, UCI | RRID:SCR_026615 | Core facility |
| Other | Flow Cytometry Core at UCI | Stem Cell Research Center, UCI | RRID:SCR_026616 | Core facility |
Additional files
-
Supplementary file 1
Differentially expressed gene list of bulk RNA-seq of sorted mCherry+ tenocytes from 36 hpf vs 48 hpf embryos.
- https://cdn.elifesciences.org/articles/105802/elife-105802-supp1-v2.csv
-
Supplementary file 2
ShinyGO analysis of 36 vs. 48 hpf bulk RNA-seq differentially expressed genes.
- https://cdn.elifesciences.org/articles/105802/elife-105802-supp2-v2.csv
-
Supplementary file 3
DAVID analysis of 36 vs. 48 hpf bulk RNA-seq differentially expressed genes.
- https://cdn.elifesciences.org/articles/105802/elife-105802-supp3-v2.xlsx
-
Supplementary file 4
Differentially expressed gene list from bulk RNA-seq of sorted mCherry+ tenocytes from 48 hpf WT vs aBTX-injected embryos.
- https://cdn.elifesciences.org/articles/105802/elife-105802-supp4-v2.csv
-
Supplementary file 5
List of differentially expressed genes overlapping between 36 hpf vs 48 hpf bulk RNA-seq and 48 hpf WT vs a-BTX injected paralysis bulk RNA-seq.
- https://cdn.elifesciences.org/articles/105802/elife-105802-supp5-v2.csv
-
Supplementary file 6
ShinyGO analysis of differentially expressed genes overlapping between 36 hpf vs 48 hpf bulk RNA-seq and 48 hpf WT vs a-BTX injected paralysis bulk RNA-seq.
- https://cdn.elifesciences.org/articles/105802/elife-105802-supp6-v2.csv
-
Supplementary file 7
List of primers used for chromogenic in situ hybridizations and RT-qPCRs.
- https://cdn.elifesciences.org/articles/105802/elife-105802-supp7-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/105802/elife-105802-mdarchecklist1-v2.pdf





