Tachykinin acts upstream of autocrine Hedgehog signaling during nociceptive sensitization in Drosophila
Figures
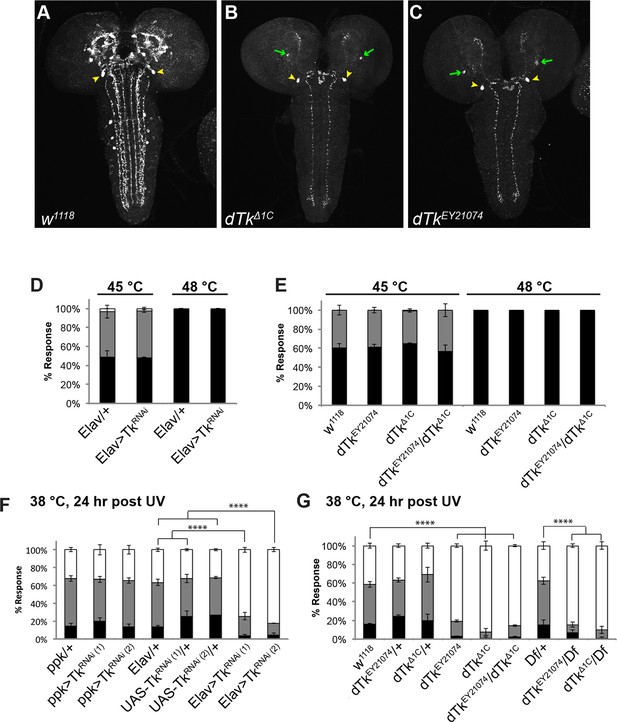
Tachykinin is expressed in the larval brain and required for thermal allodynia.
(A–C) Dissected larval brain wholemounts of the indicated genotypes immunostained with a guinea pig antiserum to DTK6. Arrowheads, large immunoreactive descending neurons. Arrows, remaining neurons immunoreactive to anti-DTK6. (A) w1118 (B) dTkΔ1C (C) dTkEY21074 (D) Baseline responses to thermal stimulation in the absence of injury at 45°C and 48°C when Tachykinin is targeted by RNAi in all neurons. Larvae of indicated genotypes were stimulated for up to 20 s with a thermal probe set to the indicated temperatures. The resulting behavior was categorized as “no withdrawal” (white) if a 360 º aversive roll did not occur, “slow withdrawal” (gray), if the roll occurred between 6 and 20 s of probe contact, or “fast withdrawal” (black), if the roll occurred within 5 s of probe contact. Percent behavioral responses were plotted as mean ± SEM. This scheme was employed for all behavioral quantitation in this study. (E) Baseline responses to thermal stimulation at 45°C and 48°C of dTk mutant alleles and relevant controls. (F–G) UV-induced thermal allodynia. (F) RNAi targeting dTk and controls. (1) and (2) refer to non-overlapping UAS-RNAi transgenes targeting Tachykinin. (G) Mutant alleles of dTk and controls. All behavior experiments throughout were performed in triplicate sets of n = 30 unless noted otherwise. Statistical significance was determined by the chi-square test. Same statistical significance markers were used throughout all figures. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.

Tachykinin is not expressed in class IV md nociceptive sensory neurons.
Dissected larval epidermal whole mounts (genotype: ppk-eGFP) immunostained with anti-DTK and anti-GFP antibodies. GFP (green in merge); anti-DTK (magenta in merge).
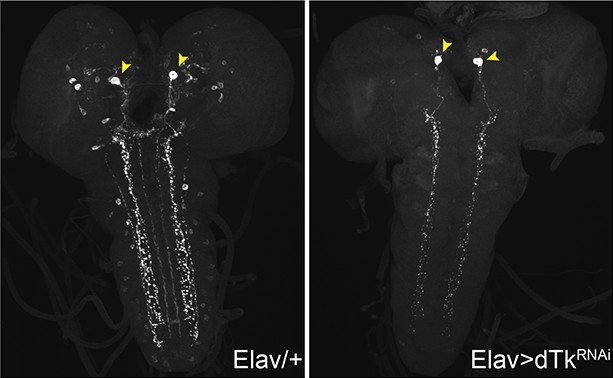
Dissected larval brain whole mounts of Elav/+ and Elav>TKRNAi immunostained with anti-LemTRP.
https://doi.org/10.7554/eLife.10735.005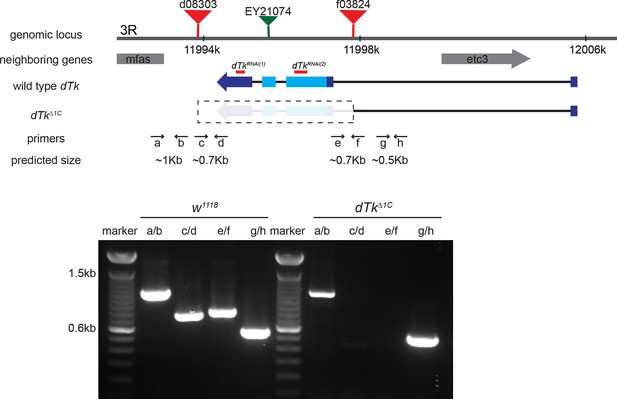
Schematic of the dTk locus.
Various genomic features and characterization tools are indicated to the left. FRT-containing transposon insertion alleles d08303 and f03824 were used to make a deletion allele of dTk, dTkΔ1C. Primers a through h were used to molecularly map the deleted and neighboring regions. Diagnostic PCR using genomic DNA of w1118 control and dTkΔ1C. In homozygous dTkΔ1C, PCR products were produced only with primers pairs of a/b and g/h that flank the deleted region.
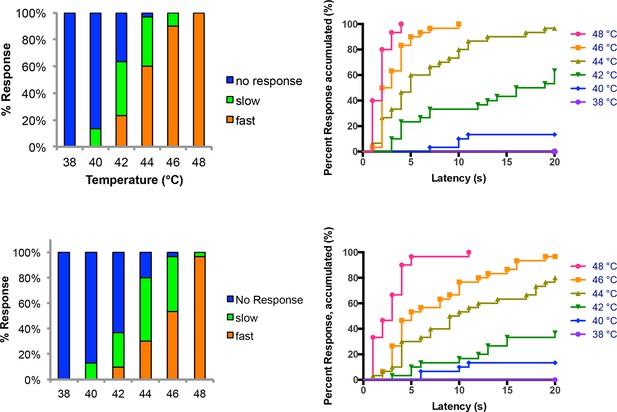
Temperature versus behavior dose response curves.
Control larvae were tested with thermal stimuli of temperature ranging from 38 to 48°C and their aversive rolling behaviors were monitored. Two sets of data were generated by two experimenters and plotted in parallel. The same set of data was plotted in two different displays: categorical bar graphs (left) and in non-categorical line graphs of the accumulated percent response on the Y-axis versus latency on the X-axis (right).
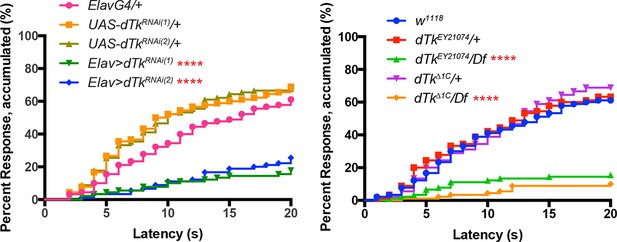
Alternative data presentation of thermal allodynia (a subset of Figure 1F and a subset of Figure 1G) in non-categorical line graphs of accumulated percent response as a function of measured latency.
Statistical tests were performed using Log-rank (Mantel-Cox) test.
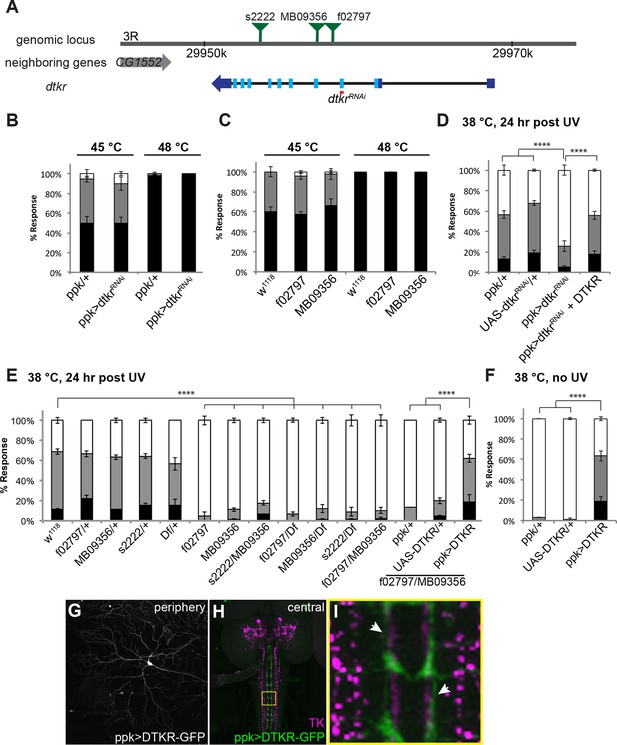
Tachykinin Receptor is required in class IV nociceptive sensory neurons for thermal allodynia.
(A) Schematic of the dtkr genomic locus. Location of transposon insertion alleles and targeted sequences of UAS-RNAi transgenes are shown. (B,C) Baseline thermal nociception at 45°C and 48°C. (B) dtkr RNAi in class IV neurons and controls. (C) dtkr mutant alleles and controls. (D,E) UV-induced thermal allodynia at 38°C. (D) dtkr RNAi and rescue in class IV neurons. (E) dtkr mutant alleles and controls. (F) “Genetic” thermal allodynia in the absence of injury upon overexpression of DTKR in class IV neurons. (G–I) Dissected larval epidermal wholemounts (genotype: ppk>DTKR-GFP) immunostained with anti-LemTRP-1 (reacts to DTKs) and anti-GFP. (G) DTKR-GFP expression in class IV neuron soma and dendrites. (H) Larval brain wholemount. GFP (green); anti-DTK (magenta). Yellow Box indicates close-up shown in I. (I) Axonal tracts expressing DTKR-GFP in class IV neurons juxtaposed with TK-expressing cells in the VNC. Arrows, regions where GFP-expressing axons are closely aligned with DTK-expressing axons.
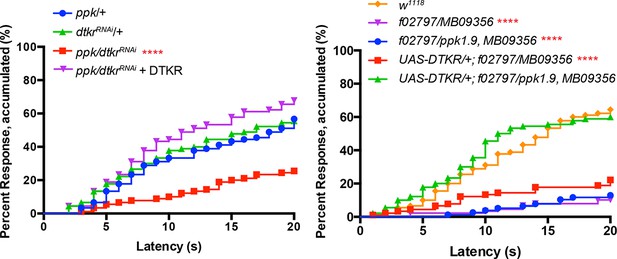
Alternative data presentation of thermal allodynia (Figure 2D and a subset of Figure 2E) in non-categorical line graphs of accumulated percent response as a function of measured latency.
Statistical tests were performed using Log-rank (Mantel-Cox) test.
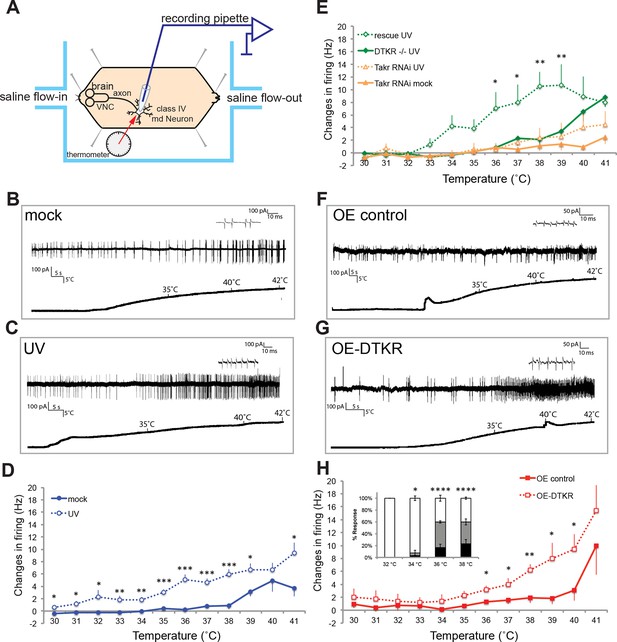
Class IV neurons display temperature-dependent changes in firing rates that are modulated by Tachykinin signaling.
(A) Schematic diagram of assay setup. (B,C,F,G) Sample recording traces of the indicated genotypes in response to temperature ramping. (B) ppk1.9-Gal4, ppk-eGFP/+ mock (C) ppk1.9-Gal4, ppk-eGFP/+ 24 hr following UV (F) ppk-Gal4/+ (G) ppk-Gal4>DTKR-GFP. (D) Changes in firing rates from larvae in (B) and (C) in response to temperature ramping. n = 11 (mock), and 21 (UV). (E) Changes in firing rates between ppk-Gal4>dtkrRNAi (mock and UV), dtkrMB09356/f02797 (UV), and class IV neuron-specific rescue of dtkrMB09356/f02797 (UV) in response to temperature ramping. n = 12 (RNAi mock), 11 (RNAi UV), 17 (dtkr mutant), and 12 (rescue). (H) Changes in firing rates between Gal4 only control and class IV specific overexpression of DTKR in response to temperature ramping without tissue damage. n = 9 (control), 12 (Overexpression). Inset, Behavioral response to innocuous temperatures when DTKR is overexpressed in class IV neurons without tissue damage. *= P<0.05, **= P<0.01, ***= P<0.001. Statistical significance was determined by either Two-way ANOVA test with Bonferroni correction or two-tailed unequal variance Student’s t-Test for electrophysiology, or by Chi-square analysis for behavior analysis.
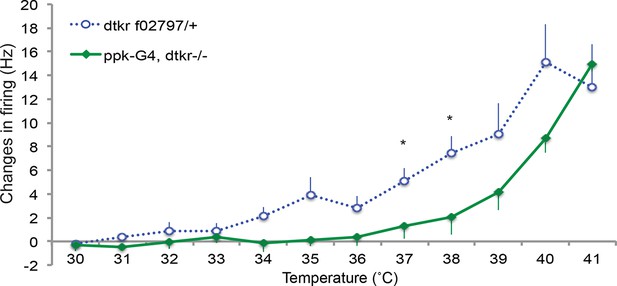
Control genotypes for electrophysiology recordings of class IV neurons.
Changes in firing rates of heterozygote dtkr (dtkrf02797/+) and Gal4 alone control in dtkr mutant background (ppk1.9Gal4, dtkrf02797/dtkrm09356) in response to temperature ramping upon UV irradiation. n = 8 (heterozygote), 7 (Gal4 control).
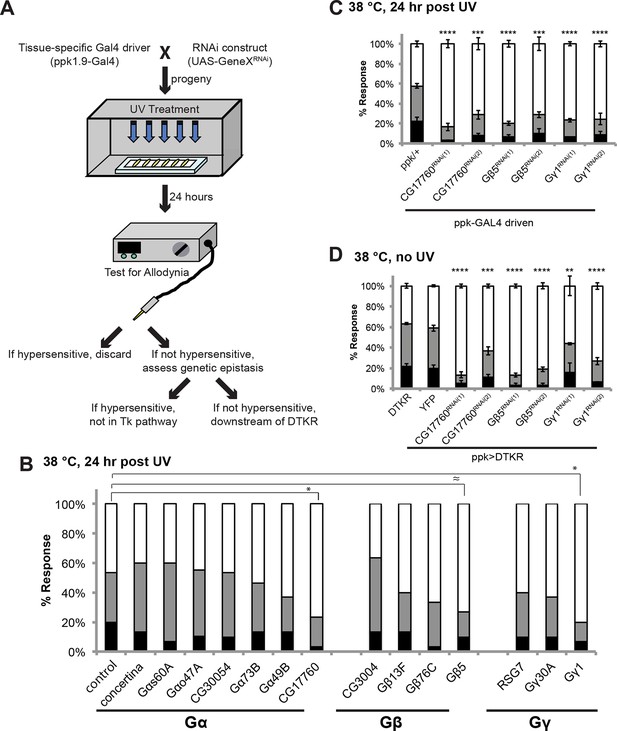
Specific Trimeric G proteins act downstream of DTKR in class IV neurons in thermal allodynia.
(A) Schematic of genetic screening strategy for testing G-protein subunit function by in vivo tissue-specific RNAi in class IV neurons. (B) UV-induced thermal allodynia on targeting the indicated G protein subunits by RNAi. n = 30 larvae per genotype. ≈ P = 0.082, * P<0.05. Statistical significance was determined by Fisher’s exact test. (C) UV-induced thermal allodynia for the three putative hits from the mini-screen in A. (1) and (2) indicate non-overlapping RNAi transgenes. (D) Suppression of UAS-DTKR-induced “genetic” allodynia by co-expression of UAS-RNAi transgenes targeting the indicated G protein subunits. Seven sets of n=30 for ppk>DTKR-GFP controls, triplicate sets of n=30 for the rest.
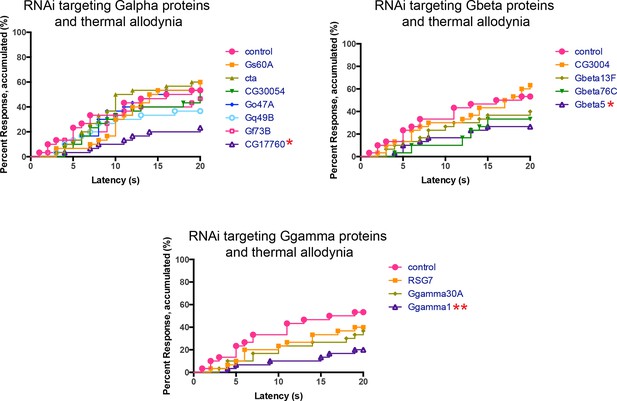
Alternative data presentation of UV-induced thermal allodynia on targeting G protein subunits by RNAi (Figure 4B) in non-categorical line graphs of accumulated percent response as a function of measured latency.
Statistical tests were performed using Log-rank (Mantel-Cox) test.
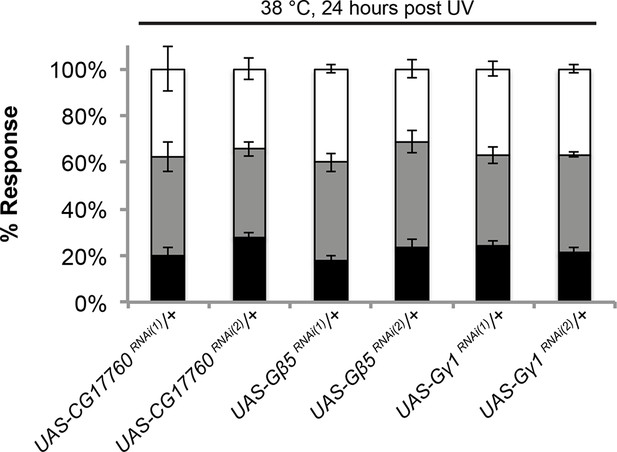
UAS alone controls of RNAi targeting G protein subunits do not exhibit defects in UV-induced thermal allodynia.
Flies bearing the indicated UAS-RNAi transgenes were crossed to w1118 and their progeny were tested. Triplicate sets of n = 30.
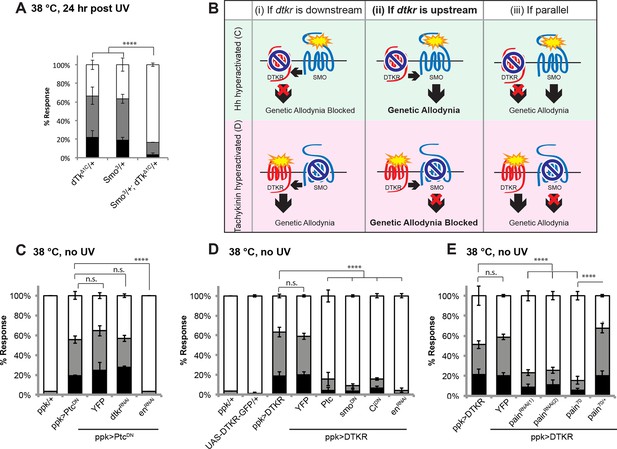
Tachykinin signaling is upstream of Smoothened and Painless in thermal allodynia.
(A) Thermal allodynia in indicated dTk and smo heterozygotes and transheterozygotes. (B) Schematic of the expected results for genetic epistasis tests between the dTK and Hh pathways. (C) Suppression of Hh pathway-induced “genetic” allodynia by co-expression of UAS-dtkrRNAi. UAS-enRNAi serves as a positive control. (D–E) Suppression of DTKR-induced “genetic” allodynia. (D) Co-expression of indicated transgenes targeting the Hh signaling pathway and relevant controls. (E) Co-expression of indicated RNAi transgenes targeting TRP channel, painless.
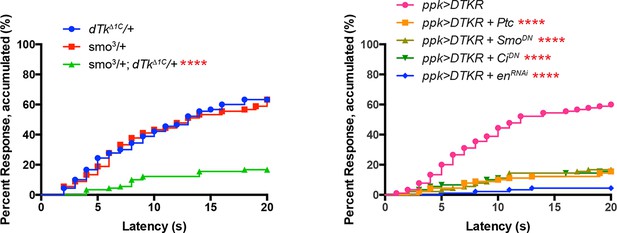
Alternative data presentation of thermal allodynia results (Figure 5A and Figure 5D) in non-categorical line graphs of accumulated percent response as a function of measured latency.
Statistical tests were performed using Log-rank (Mantel-Cox) test.
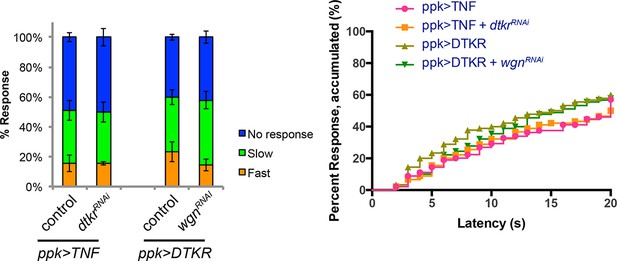
Genetic epistasis tests between DTKR and TNF pathway.
Co-expression of UAS-dtkrRNAi did not suppress TNF-induced genetic allodynia and co-expression of UAS-wengenRNAi did not suppress DTKR-induced genetic allodynia. Data presented in categorical bar graphs (left) and in non-categorical line graphs of accumulated percent response as a function of measured latency. Statistical tests were performed using Chi-Square analysis (bar graph) or Log-rank (Mantel-Cox) test (line graphs).
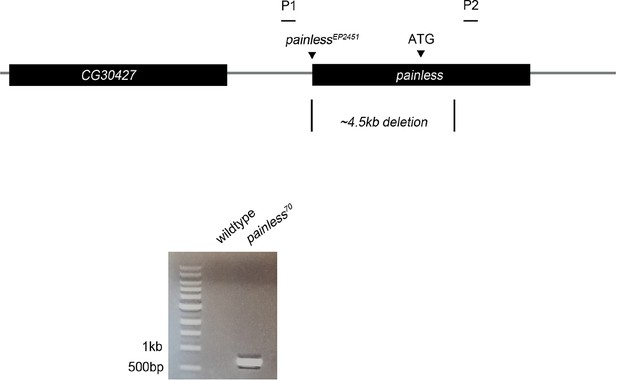
Schematic of painless genomic locus. painless70 was generated by imprecise excision of painlessEP2451, deleting 4.5 kb of surrounding sequence including the ATG of the A splice variant.
Primers P1/P2 were used to confirm the deletion in diagnostic PCR amplification of pain70 versus control w1118 genomic DNA.
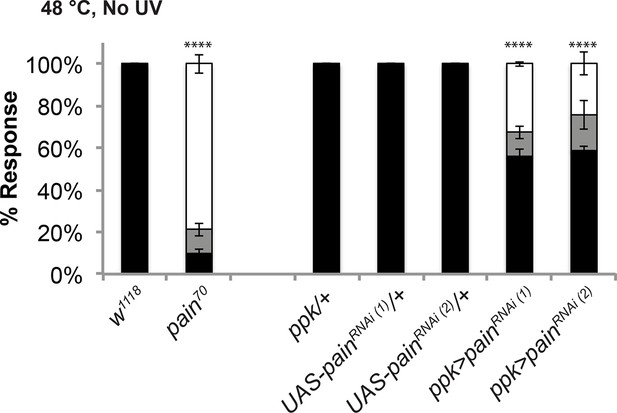
The pain70 deletion allele and UAS-painRNAi transgenes cause defects in baseline thermal nociception.
Quantitation of baseline responses to thermal stimulation at 48°C. painless70 was compared to w1118 controls, while two non-overlapping UAS-painlessRNAi transgenes (v39477 and 31510) expressed in class IV neurons were compared to Gal4 or UAS alone controls.
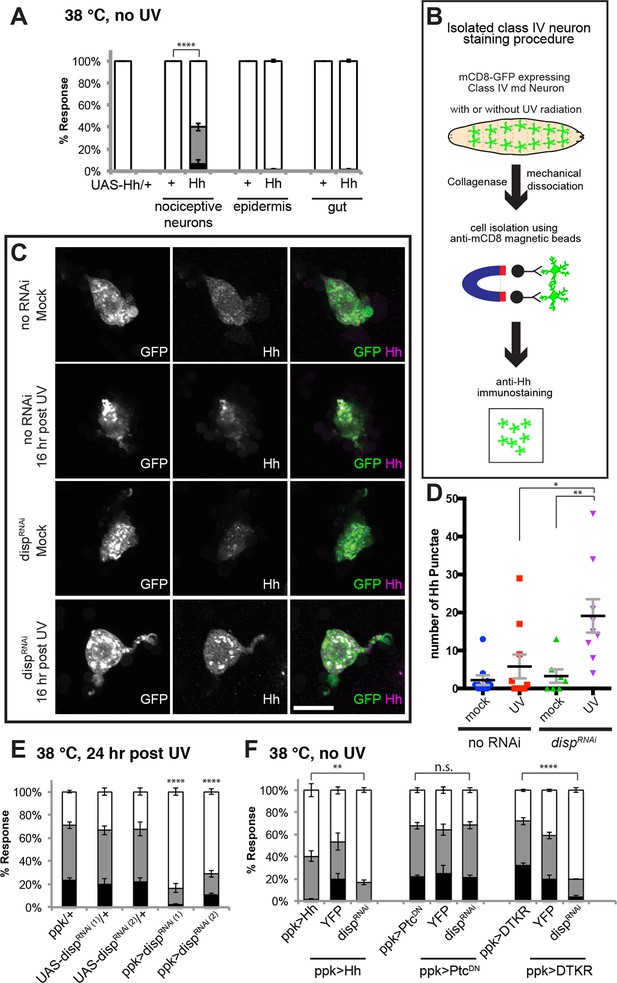
Tachykinin-induced Hedgehog is autocrine from class IV nociceptive sensory neurons.
(A) “Genetic” allodynia induced by ectopic Hh overexpression in various tissues. Tissue-specific Gal4 drivers, UAS controls and combinations are indicated. The Gal4 drivers used are ppk-Gal4 (class IV sensory neuron), A58-Gal4 (epidermis), and Myosin1A-Gal4 (gut). (B) Schematic of class IV neuron isolation and immunostaining. (C) Isolated class IV neurons stained with anti-Hh. mCD8-GFP (green in merge); anti-Hh (magenta in merge). (D) Number of Hh punctae in isolated class IV neurons from genotypes/conditions in (C). Punctae per image are plotted as individual points. Black bar; mean gray bracket; SEM. Statistical significance was determined by One-way ANOVA test followed by multiple comparisons with Tukey correction. (E) UV-induced thermal allodynia upon UAS-dispRNAi expression with relevant controls. (F) Suppression of “genetic” allodynia by co-expression of UAS-dispRNAi in class IV neurons. Genetic allodynia conditions were induced by Hh overexpression, PtcDN expression, or DTKR-GFP overexpression.
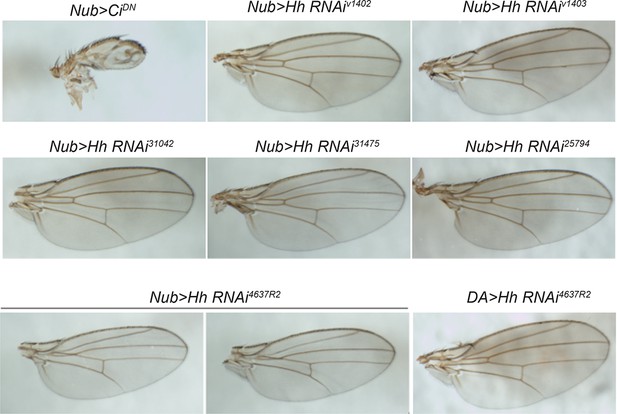
RNAi-mediated knockdown of hh was not effective.
Wing pattern phenotypes of various UAS-RNAi transgenes targeting hh using nubbin-GAL4 and daughterless-GAL4. UAS-CiDN was used as a positive control. None of the UAS-HhRNAi transgenes tested effectively interfered with wing patterning. This suggests that the lack of phenotype in thermal allodynia assays on expression of these transgenes is largely due to inefficient knockdown of hh function.
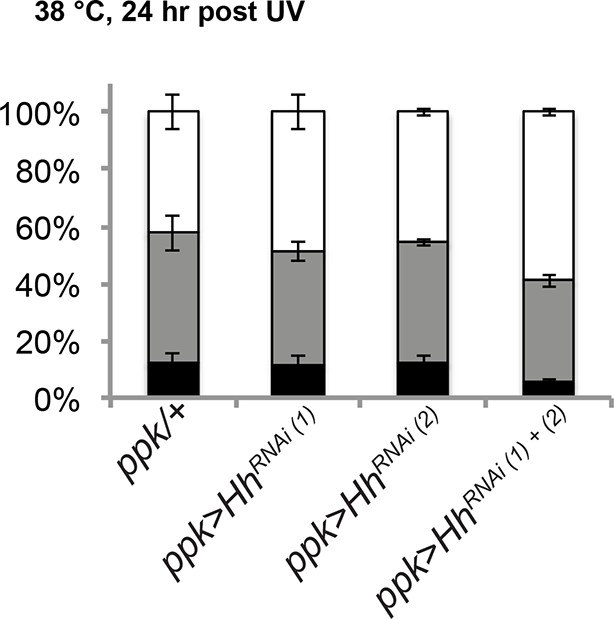
RNAi-mediated knockdown of hh was not effective in blocking thermal allodynia.
UAS-hhRNAi(1) refers to 4637R2 whereas UAS-hhRNAi(2) refers to v1403.
A few more examples of isolated class IV neurons stained with anti-Hh.
mCD8-GFP (green in merge); anti-Hh (magenta in merge).
Genetic allodynia in the absence of tissue injury upon overexpression of TNF in class IV neurons.
Knockdown of disp did not interfere with TNF-induced genetic allodynia.
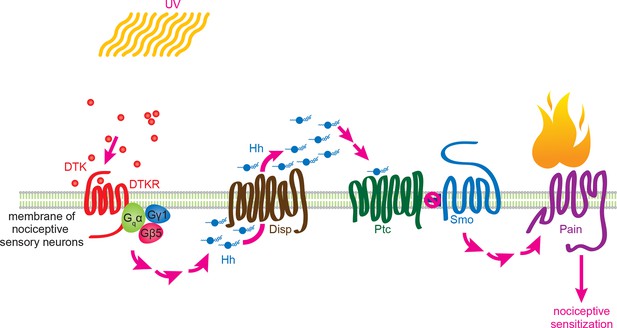
Working model for Tachykinin/Tachykinin Receptor function upstream of Hh signaling in UV-induced thermal allodynia.
Tachykinin ligands are released from the brain neurons targeting class IV nociceptive sensory neurons upon UV-induced tissue damage. DTKR is coupled to trimeric G proteins and the signaling cascade then induces Disp-dependent Hh release. Hh binds to Ptc in an autocrine fashion and activates the Smo downstream signaling cascade, followed by modification/activation of Painless. These series of signaling cascades result in thermal allodynia, where stimulation at a sub-threshold temperature induces pain behaviors (thermal nociceptive sensitization).





