Aurkb/PP1-mediated resetting of Oct4 during the cell cycle determines the identity of embryonic stem cells
Figures
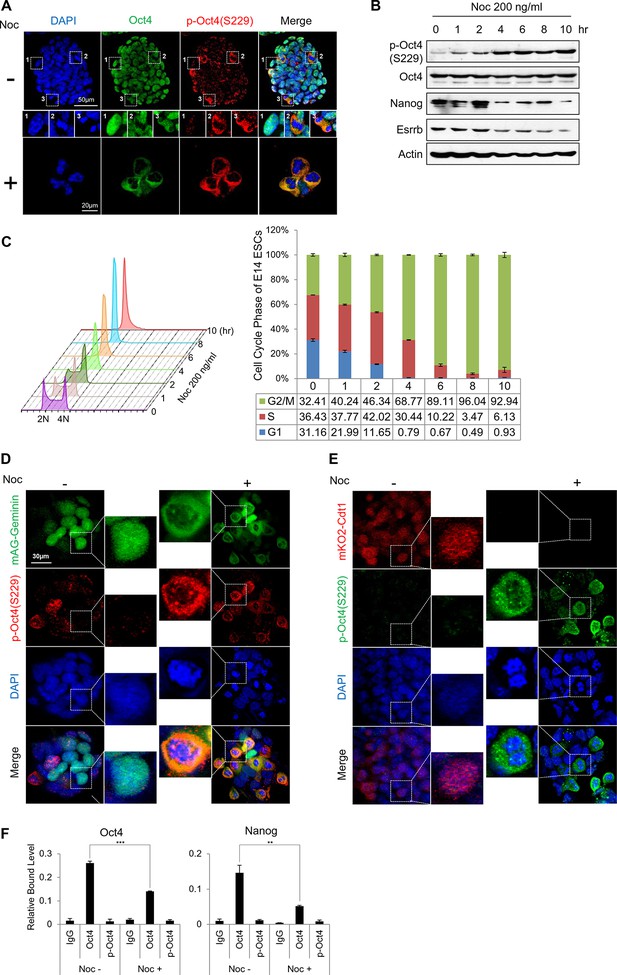
Phosphorylated Oct4 at serine 229 is enriched in G2/M phase and dissociated from chromatin.
(A) Immunostaining of E14 ESCs treated with or without nocodazole (NOC, 200 ng/ml) for 10 hr. Oct4 was stained with anti-Oct4 (green), p-Oct4(S229) was stained with anti-p-Oct4(S229) (red), and DNA was stained with DAPI (blue). White boxes represent cells at various stages. Shown are interphase (1), metaphase (2), and anaphase (3) cells. Scale bars were shown. (B) E14 ESCs were treated with nocodazole (200 ng/ml) for the indicated times and immunoblotted with the indicated antibodies. Phosphorylation levels of Oct4 at serine 229 were gradually induced during nocodazole treatment. (C) Histograms of the proportions of nocodazole-treated (200 ng/ml) E14 ESCs at various stages in the cell cycle. Cells were stained with PI and DNA contents were analyzed by FACS (1x104 cells/sample). (D and E) Fluorescence images of E14 ESCs expressing mKO2-Cdt1 and mAG-Geminin (FUCCI reporter). Shown are green (mAG-geminin) and red (mKO2-Cdt1) fluorescence. E14 ESCs expressing FUCCI reporter were left untreated or treated with nocodazole (NOC, 200 ng/ml) for 10 hr. p-Oct4(S229) was stained with anti-p-Oct4(S229) (red, Figure 1E; green, Figure 1F), and DNA was stained with DAPI (blue). Scale bars, 30 μm (F) ChIP-qPCR assay was performed with anti-IgG, anti-Oct4, and anti-p-Oct4(S229) in E14 ESCs with or without nocodazole (NOC, 200 ng/ml) for 10 hr. Values represent mean ± standard deviation (n≥3). (**p<0.01, ***p<0.001)
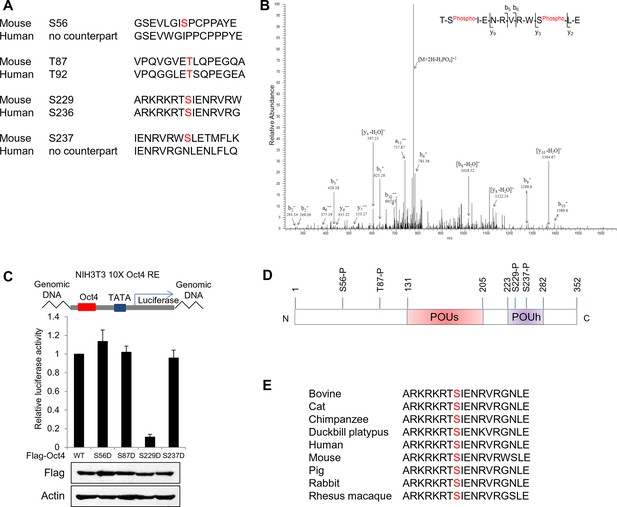
Identification of Oct4 phosphorylation at serine 229 residue.
(A) Sequence alignment of detected phosphorylation regions of Oct4 between mouse and human. (B) Oct4 is phosphorylated at S229 in ESCs. Oct4 was purified using stably expressed Flag-Oct4 in ZHBTc4 ESCs by immunoprecipitation with anti-Flag. Phosphorylation sites on Oct4 were analyzed by nano-LC-ESI-MS/MS. (C) Oct4-driven transcriptional activity was measured with Oct4 mutants. Ten copies of Oct4-responsive element (10X Oct4 RE)-driven luciferase reporter gene was incorporated into the genome of NIH 3T3 cells. These stable cells were infected with retroviruses expressing Oct4 wild-type (WT) and phosphor mimic mutants. Luciferase activity was measured 4 days after infection. Values represent mean ± standard deviation (n≥3). (D) A schematic shows identified Oct4 phosphorylation sites. The S229 residue is located at POUh domain on Oct4. (E) Sequence alignment of Oct4 phosphorylated region (S229) between species.
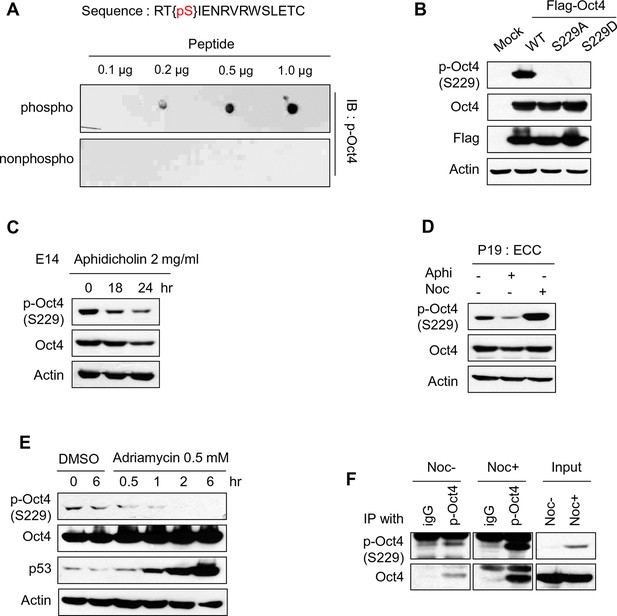
Characterization of Oct4 phosphorylation site at serine 229 and anti-p-Oct4(S229) antibody.
(A and B) Characterization of the antibody to p-Oct4(S229). By dot blot analysis, the antibody (p-Oct4(S229)) specifically recognized phosphorylated peptide (A). Lysates of HEK293T cells transfected with Flag-Oct4 wild type (WT), S229A, and S229D mutant were immunoblotted with indicated antibodies. The p-Oct4(S229) antibody only recognized wild type Oct4 (B). (C) By treatment of E14 ESCs with aphidicholin, which induced G1 phase arrest, phosphorylation of Oct4 at S229 was decreased. p-Oct4(S229) was detected by Western blot and Actin was used as an internal control. (D) The level of p-Oct4(S229) was only increased after treatment of nocodazole (Noc) in P19 ECC cells. P19 ECC cells were treated with aphidicholin (Aphi) or nocodazole (Noc) for 10 hr. Lysates from these cells were immunoblotted with indicated antibodies and Actin was used as an endogenous control. (E) DNA damage in E14 ESCs was induced by Adriamycin treatment. Gradual decrease of p-Oct4(S229) during adriamycin treatment was detected by Western blot. p53 was used as an positive control for inducing DNA damage in E14 ESCs. (F) p-Oct4(S229) was pulled down with anti-p-Oct4(S229) antibody under the same condition we performed ChIP assay followed by western blot.
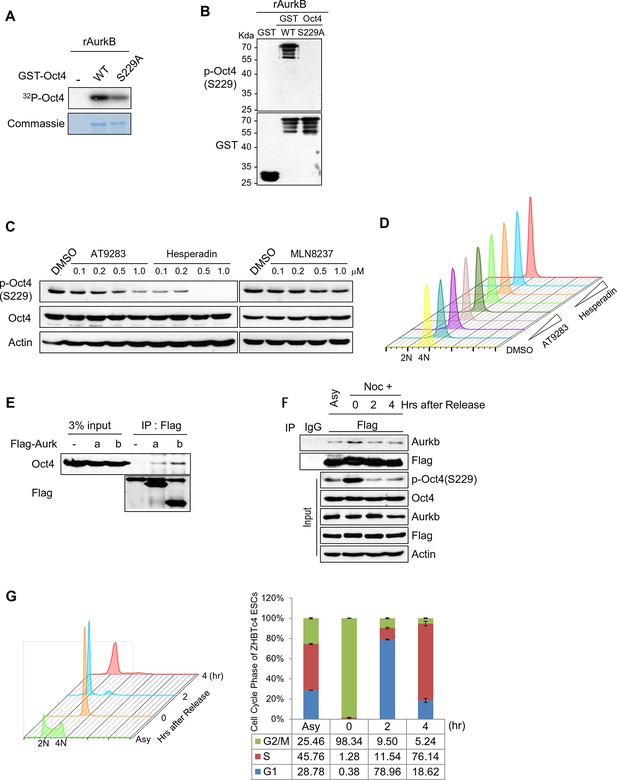
Aurkb binds and phosphorylates Oct4 at serine 229 during G2/M phase.
(A) Radioactive in vitro kinase assay using recombinant Aurkb to phosphorylate GST-Oct4 WT and S229A mutant. Coomassie staining of purified proteins and autoradiogram showing incorporation of γ-32P ATP. (B) Cold in vitro kinase assay reactions using recombinant Aurkb with purified GST, GST-Oct4 WT, and S229A mutant as substrate followed by western blot. (C and D) Nocodazole-arrested E14 ESCs (200 ng/ml for 10 hr) were treated with the Aurora kinase inhibitors AT9283 (inhibits Aurka and Aurkb), hesperadin (inhibits Aurkb), and MLN8237 (inhibits AurkA). Gradual decreases in p-Oct4(S229) levels with increasing concentrations of Aurkb inhibitors in E14 ESCs were seen by western blot (C). FACS analysis was performed under the same condition (D) (1x104 cells/sample). (E) Coimmunoprecipitation of Oct4 with Aurka and Aurkb from E14 ESCs stably expressing Flag-tagged Aurora kinases. (F) Changes in Oct4 interaction with Aurkb during cell cycle progression. Whole-cell lysates from Flag-Oct4-expressing ZHBTc4 ESCs were pulled down with anti-Flag beads. Bound proteins were immunoblotted with the indicated antibodies. (G) DNA content analysis of Flag-Oct4 expressing ZHBTc4 ESCs by FACS. Flag-Oct4-expressing ZHBTc4 ESCs, treated with nocodazole (200 ng/ml) for 6 hr, were released for 2 and 4 hr and DNA contents were counted (1x104 cells/sample).
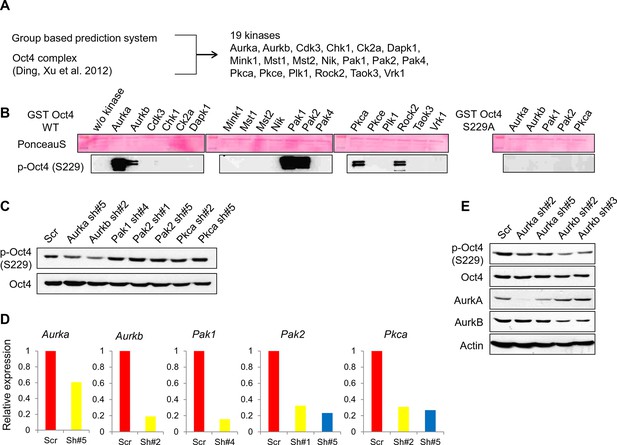
Screening of kinases responsible for phosphorylation on Oct4 at S229.
(A) A schematic diagram represents strategy for kinase screening. (B) Cold in vitro kinase assay was performed using predicted 19 kinases with purified GST-Oct4 WT and S229A mutant. 50nM of kinases were used in all reactions. Kinase responsible for Oct4 phosphorylation at S229 was detected by the p-Oct4(S229) antibody and PonceauS staining was used as a loading control. (C and D) Identification of kinase responsible for phosphorylation at S229 on Oct4 in vivo Each kinase was stably knocked down in E14 ESCs by infection of indicated lentiviral shRNA and only Aurkb-knockdown ESCs harbored decreased p-Oct4(S229) level. p-Oct4(S229) level was detected by Western blot after treatment of nocodazole for 10 hr in each E14 ESCs and Oct4 was used as an internal control (C). Stable knockdown of indicated kinases in E14 ESCs were analyzed by qRT-PCR (D). (E) Transient knockdown of Aurkb in E14 ESCs reduced p-Oct4(S229) level. By infection of lentiviral shRNAs targeting Aurka and Aurkb in E14 ESCs, knockdown levels were detected with indicated antibodies 2 days after infection. p-Oct4(S229) level in each E14 ESCs was detected by Western blot after treatment with nocodazole for 10 hr and Actin was used as an internal control.
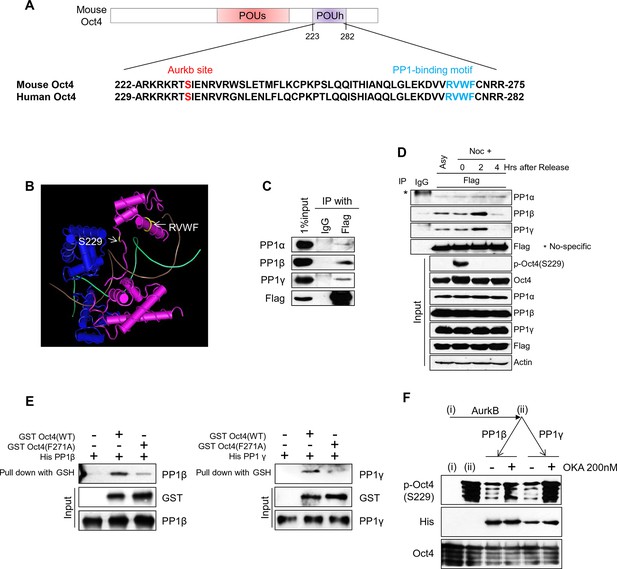
PP1 binds and dephosphorylates Oct4 at serine 229 during G1 phase.
(A) Sequence alignment of Oct4. Oct4 contains a conserved PP1 docking motif (RVXF). (B) Three-dimensional structure of Oct4 and DNA complex (MMDB ID: 87311) was adapted from the Molecular Modeling Database (MMDB) of NCBI. Each yellow region indicates S229 and an RVWF PP1-binding domain. (C) Coimmunoprecipitation assay revealing the endogenous interaction between Oct4 and PP1 catalytic subunits. Proteins were immunoprecipitated from Flag-Oct4-expressing ZHBTc4 ESCs with Flag antibody, followed by western blot. (D) Changes in Oct4 interaction with PP1 catalytic subunits during cell cycle progression. Whole-cell lysates from Flag-Oct4-expressing ZHBTc4 ESCs were pulled down with anti-Flag beads. Immunoprecipitated proteins were immunoblotted with the indicated antibodies. (E) Purified GST-Oct4(WT) or GST-Oct4(F271A) mutant was incubated with purified (His)6-PP1β and PP1γ and then pulled down with GST beads. Immunoblot shows that PP1β and γ directly bind GST-Oct4(WT). PP1β and PP1γ show weaker interaction with GST-Oct4(F271A) than wild-type Oct4. (F) In vitro phosphatase assay using PP1β or PP1γ with phosphorylated Oct4 as substrate. Okadaic acid (OKA) treatment decreased PP1-mediated dephosphorylation of Oct4.
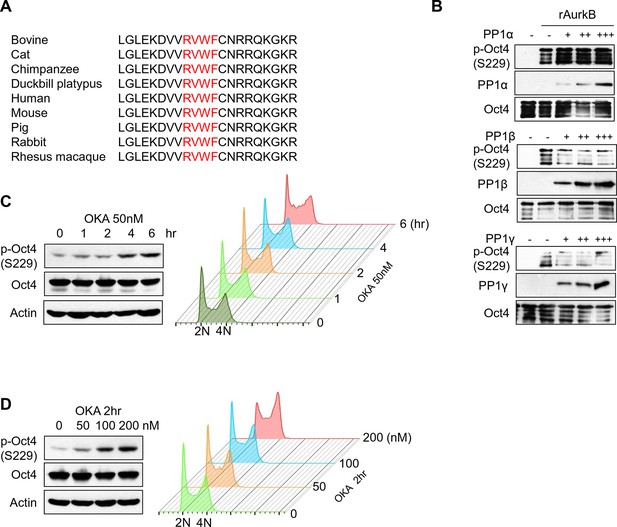
PP1 dephosphorylates Oct4 at S229 in vitro and in vivo .
(A) Sequence alignment of PP1 binding motif in Oct4 between species. (B) In vitro phosphatase assay using purified PP1 isoforms with GST Oct4 (WT). rAurkb-mediated phosphorylation of Oct4 at S229 was decreased by incubation with PP1β and PP1γ, but not by PP1α. Reduced p-Oct4(S229) levels were detected by Western blot with indicated antibodies. (C and D) Treatment of okadaic acid (OKA) in E14 ESCs increased p-Oct4(S229). E14 ESCs were incubated with 50nM of OKA for indicated time and increased p-Oct4(S229) level was detected by Western-blot with indicated antibodies (Left panel). DNA contents were analyzed by FACS (right panel) (C). E14 ESCs were treated with indicated concentrations of OKA for 2 hr and p-Oct4(S229) level was increased by dose-dependent of OKA. p-Oct4(S229) was detected by Western blot (left panel) and DNA contents were analyzed by FACS (right panel) (D).
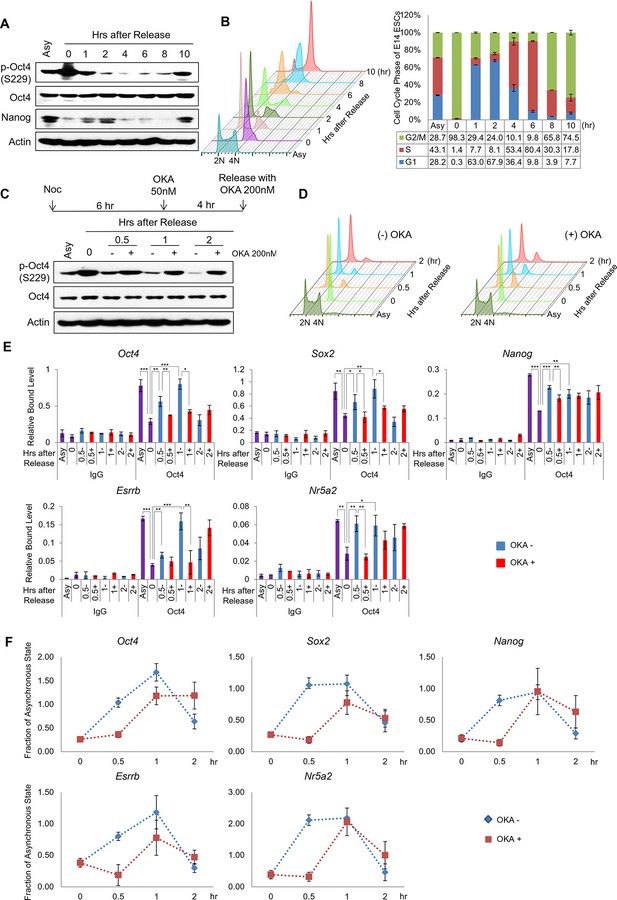
PP1-mediated dephosphorylation of Oct4(S229) correlates with the resetting of pluripotency genes in the next G1 phase.
(A) p-Oct4(S229) levels after release of E14 ESCs from M-phase arrest. Shown are immunoblots for the indicated proteins. (B) Nocodazole-treated E14 ESCs were released and analyzed for DNA content by FACS (1x104 cells/sample). (C) OKA treatment retards dephosphorylation of p-Oct4(S229) during the M/G1 phase transition. The experimental strategy is shown (upper panel). The same strategy was applied to (D–F). Whole-cell lysates from E14 ESCs were collected and assessed by western blot. (D) Histogram shows cell cycle state of E14 ESCs without (upper panel) or with (lower panel) OKA treatment. (E) ChIP-qPCR analysis of E14 ESCs with anti-Oct4 in regions of pluripotency-associated Oct4 target genes during the M/G1 phase transition with or without OKA treatment. IgG was used as a control. Values represent mean ± standard deviation (n≥3). t-test was used to calculate the statistical significance of differences in enrichment levels of Oct4 at pluripotency-associated Oct4 target genes in ESCs during the M/G1 transition with or without OKA. (*p<0.05, **p<0.01, ***p<0.001) (F) Nascent RNA of pluripotency-associated Oct4 target genes from E14 ESCs were collected and analyzed by real-time qPCR during the M/G1 phase transition with or without OKA. Levels of each nascent RNA were normalized by those in asynchronous E14 ESCs.
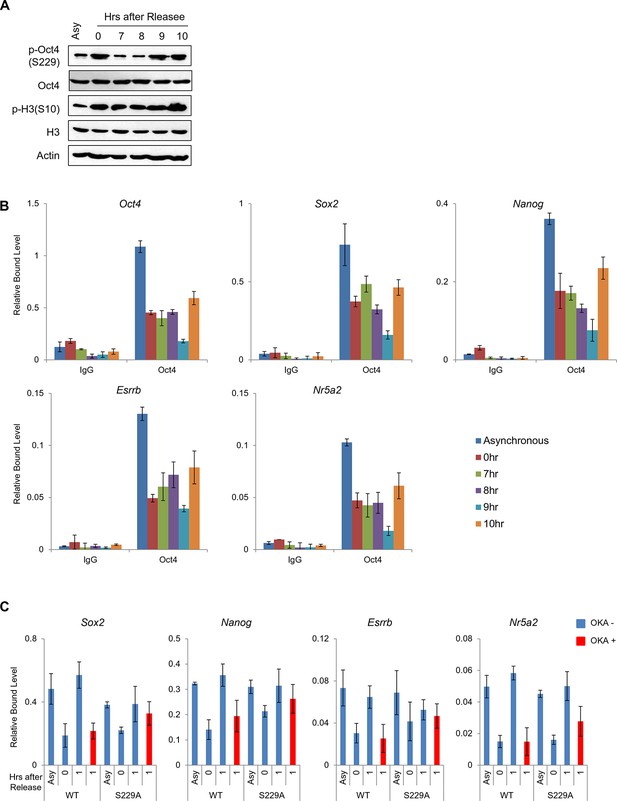
Dissociation of p-Oct4(S229) from chromatin occurs independent to chromatin status.
(A) p-Oct4(S229) levels after release of E14 ESCs from M-phase arrest. Shown are immunoblots for the indicated proteins. (B) ChIP-qPCR analysis of E14 ESCs with anti-Oct4 in regions of pluripotency-associated Oct4 target genes after release of E14 ESCs from M-phase arrest. IgG was used as a control. Values represent mean ± standard deviation (n≥3). (C) Anti-Oct4 ChIP-qPCR of ZHBTc4 ESCs that express exogenous Oct4 in regions of pluripotency-associated Oct4 target genes during the M/G1 phase transition with or without OKA treatment. Values represent mean ± standard deviation (n≥3).
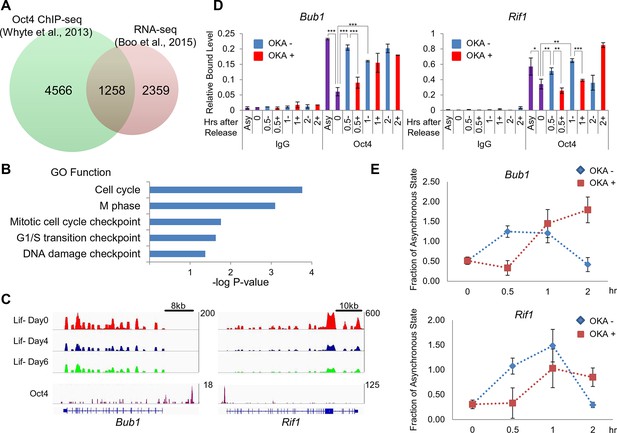
Oct4 regulates cell cycle related genes by direct targeting and resetting during the M/G1 transition.
(A) A Venn diagram shows overlapped genes between proximal genes of Oct4 binding sites (green, n=5824; (Whyte et al., 2013)) and downregulated genes (fold changes≤0.75) in ZHBTc4 ESCs after Oct4 depletion by doxycycline treatment for 2 days (red, n=3617; [Boo et al., 2015]). (B) Gene ontology (GO) functional categories for putative Oct4 target genes. Cell cycle related GO functional categories are enriched. (C) RNA-seq reads of Bub1 and Rif1 of E14 ESCs during ESC differentiation upon LIF withdrawal (upper panel; [Xiao et al., 2012]) and ChIP-seq binding profiles of Oct4 at the Bub1 and Rif1 locus in undifferentiated E14 ESCs (lower panel; [Whyte et al., 2013]). (D) ChIP-qPCR analysis of E14 ESCs with anti-Oct4 in regions of Bub1 and Rif1 during the M/G1 phase transition with or without OKA treatment. IgG was used as a control. Values represent mean ± standard deviation (n≥3). t-test was used to calculate the statistical significance of differences in enrichment levels of Oct4 in ESCs during the M/G1 transition with or without OKA treatment. (*p<0.05, **p<0.01, ***p<0.001) (E) Nascent RNA levels of Bub1 and Rif1 in E14 ESCs were nalyzed by real-time qPCR during the M/G1 phase transition with or without OKA treatment. Levels of nascent RNA were divided by those in asynchronous state of E14 ESCs.
-
Figure 5—source data 1
Identification of putative Oct4 target genes.
- https://doi.org/10.7554/eLife.10877.013
-
Figure 5—source data 2
Cell-cycle related genes in putative Oct4 target genes.
- https://doi.org/10.7554/eLife.10877.014
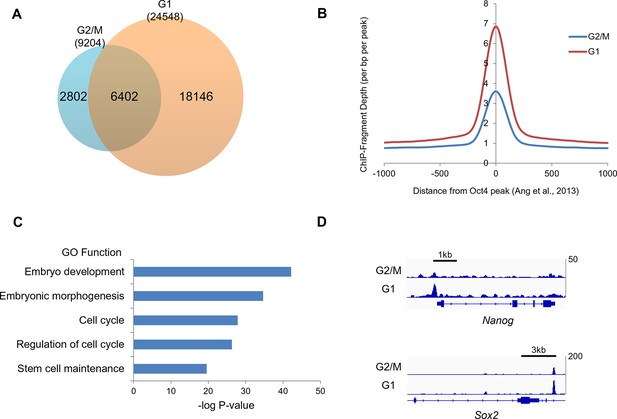
Oct4 ChIP-seq at G2/M and G1 phase of cell cycle.
(A) Venn diagram of overlap between G2/M and G1 ChIP-seq peaks. (B) Mean ChIP-seq density of Oct4 around previously published Oct4-occupied regions (Ang et al., 2011) between G2/M and G1 phase. The level of Oct4 increased after release into M/G1 transition compared to G2/M phase. (C) Gene ontology (GO) functional categories for genes which are reset by Oct4. Pluripotency and cell cycle related GO functional categories are significantly enriched. (D) Integrated genomics viewer (IGV) screenshots for ChIP-seq data of Nanog and Sox2.
-
Figure 6—source data 1
Identification of G2/M or G1 specific Oct4 binding peaks.
- https://doi.org/10.7554/eLife.10877.016
-
Figure 6—source data 2
Candidate genes reset by Oct4.
- https://doi.org/10.7554/eLife.10877.017
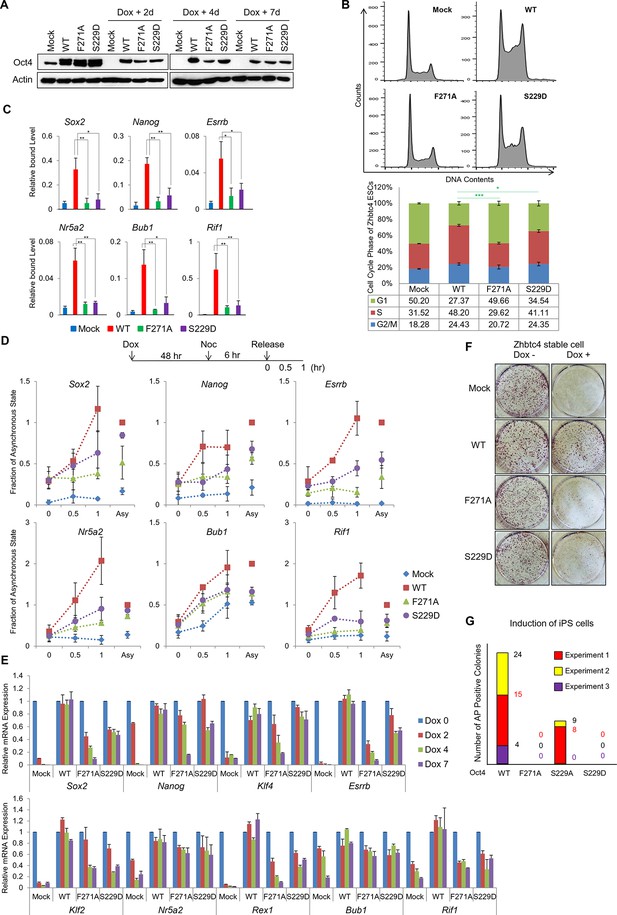
Oct4 mutants—Oct4(S229D) and Oct4(F271A)—effect the loss of pluripotency and alter the cell cycle by impeding gene expression.
(A) Flag Oct4 WT and mutants were stably incorporated into the genome of ZHBTc4 ESCs. These cells were treated with doxycycline for the indicated days. Stable expression of exogenous Oct4 was confirmed by western blot. (B) After 2 days of doxycycline treatment in ZHBTc4 ESCs (Mock, wild-type Oct4(WT)-, Oct4(F271A)-, and Oct4(S229D)- backup cells), DNA content (1x104 cells/sample) were analyzed in each ESC by FACS. Values represent mean ± standard deviation (n≥3). t-test was used to calculate the statistical significance of differences in G1 phase of ZHBTc4-Oct4(WT) versus -Oct4(S229D) and -Oct4(F271A). (*p<0.05, ***p<0.001) (C) Anti-Oct4 ChIP-qPCR of ZHBTc4 ESCs that express exogenous Oct4 at 2 days after doxycycline treatment. F271A and S229D mutants showed decreased binding to target genes compared with Oct4 WT. Oct4-depleted ZHBTc4 cells (Mock) were used as a control. Values represent mean ± standard deviation (n≥3). t-test was used to calculate the statistical significance of differences in enrichment levels of Oct4 in ZHBTc4-Oct4(WT) versus Oct4(S229D) and Oct4(F271A) ESCs. (*p<0.05, **p<0.01) (D) Levels of nascent RNA were measured by qRT-PCR and each nascent RNA levels were normalized by the levels in Oct4(WT) backup ZHBTc4 ESCs in the asynchronous state after doxycycline treatment for 2 days. The experimental scheme is shown (upper panel). (E) Relative expression of genes targeted by Oct4 related to pluripotency and cell cycle in ZHBTc4. mRNA levels of the indicated genes decreased significantly in Mock, Oct4(F271A), and Oct4(S229D) backup cells after doxycycline treatment but not in Oct4(WT) backup cells. (F) Indicated ZHBTc4 ESCs were stained for alkaline phosphatase (AP) activity after 7 days of doxycycline treatment. (G) Reprogramming of MEFs into iPS cells driven by Oct4, Sox2, and Klf4. Oct4 wild-type was replaced by F271A, S229A, and S229D mutants. Reprogrammed cells were identified by AP staining and counted. Results from 3 independent experiments are presented.
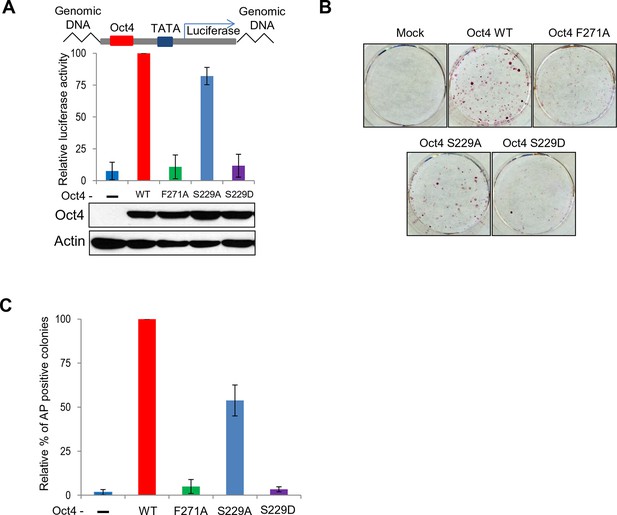
Both S229D and F271A mutants of Oct4 decrease Oct4 activity.
(A) Oct4-transcriptional activity was measured using NIH3T3 cells harboring ten copies of Oct4-responsive element (10X Oct4 RE)-driven luciferase reporter gene stably. These stable cells (NIH 3T3) were infected with retroviruses expressing Oct4 wild-type (WT), F271A, S229A and S229D mutants and luciferase activity was measured 4 days after infection. Both S229D and F271A mutants of Oct4 barely induced Oct4-driven luciferase activities. Values represent mean ± standard deviation (n≥3). (B and C) Endogenous Oct4 in ZHBTc4 ESCs was replaced by infection with indicated retroviral Oct4 mutants. AP staining was performed after selection of infected cells (2 days) in the presence of doxycycline 7 days later. AP-positive colony numbers were assessed as relative percent mean ± standard deviation (n≥3).
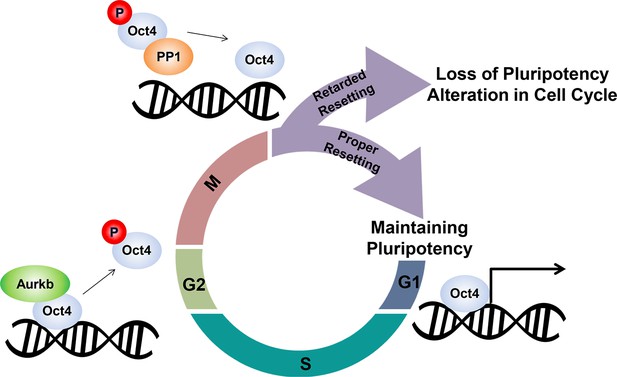
Schematic.
A model describing the dissociation and resetting of Oct4 on chromatin by Aurkb/PP1 during the cell cycle. Aurkb phosphorylates Oct4(S229), leading to dissociation of Oct4 from chromatin during G2/M phase. On mitotic exit, PP1 binds to Oct4 and dephosphorylates Oct4(S229), which resets Oct4-driven transcription to maintain pluripotency and cell cycle progression.
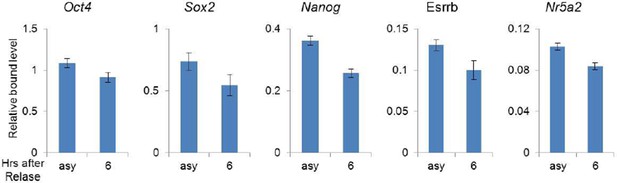
Oct4 bound weakly to target genes in the S phase.
https://doi.org/10.7554/eLife.10877.022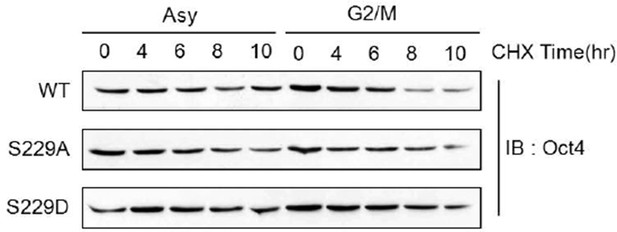
Serine 229 mutation didn’t affect Oct4 stability.
https://doi.org/10.7554/eLife.10877.023Additional files
-
Supplementary file 1
The lists of primers for nascent RNA, real-time qPCR, and ChIP-qPCR in this study.
- https://doi.org/10.7554/eLife.10877.021





