Community effects in regulation of translation
Figures
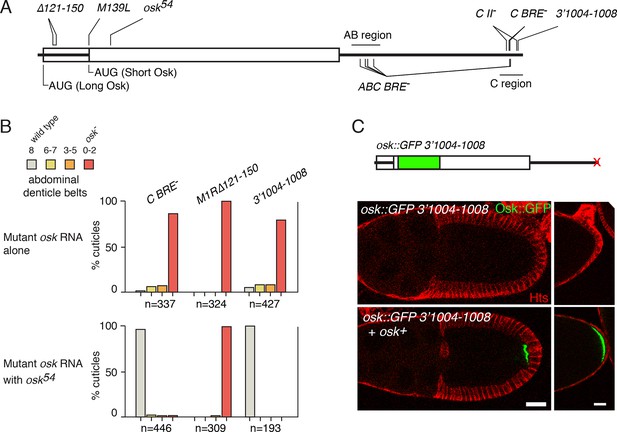
Rescue of osk expression in trans is not limited to Bru-dependent regulation.
(A) Diagram of the osk mRNA showing sites of mutations discussed in the text. The UTRs are shown as thick lines and the coding region as a rectangle. Because two different start codons are used, portions of the 5’ region can be both UTR and coding region. Not all the mutations at the 3’ end of the mRNA are shown. (B) Embryonic patterning assays to monitor osk activity. For the upper panel, the only source of osk mRNA was a genomic osk transgene with the indicated mutation. For the lower panel, osk54 mRNA was also present. Embryos from these mothers were were scored for cuticular patterning defects. Wild-type embryos have eight abdominal segments. Lower osk activity results in fewer abdominal segments. n values are the number of embryos scored. (C) Rescue in trans of the Osk expression defect caused by mutation 3’1004–1008. At top is a diagram of the mRNA from the genomic osk transgene whose activity was monitored. All sequences are from osk, except for the inserted GFP. Below are images of stage 10 egg chambers (left) and the posterior ends of late stage egg chambers (right). Scale bars, 25 µm.
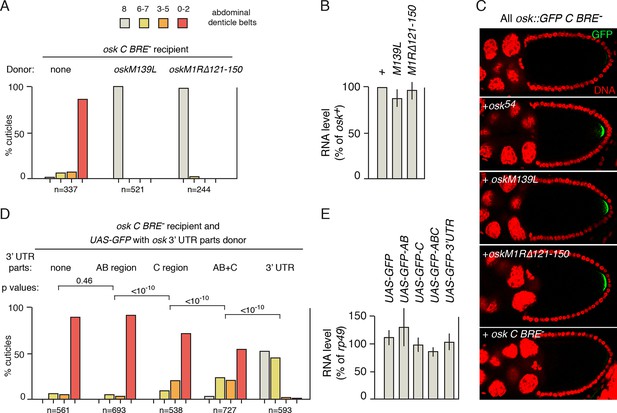
Identification of sequences required in donor mRNAs for rescue in trans.
(A) Embryonic patterning assays to monitor rescue of the translational activation defect of osk C BRE-. The osk C BRE- mRNA was expressed as the only osk mRNA, or in combination with donor osk mRNAs as indicated. (B) Levels of transgenic osk mRNAs. Values are relative to an osk+ transgene that fully rescues an osk mutant. Error bars indicate standard deviations. The osk C BRE- mRNA used in panel C is not included but has similar abundance (Reveal et al., 2010). (C) Detection of GFP signal from osk::GFP C BRE-, expressed as the only osk mRNA or in combination with the donor osk mRNAs indicated. (D) Embryonic patterning assays to monitor rescue of the translational activation defect of osk C BRE-. The osk C BRE- mRNA was expressed as the only osk mRNA (in the oskA87/osk0 background), together with a version of UAS-GFP expressed under GAL4 transcriptional control. Each UAS-GFP transgene contains osk 3’ UTR sequences as indicated (positions of the AB and C regions are shown in Figure 1A). p values from student’s t tests were determined as described in 'Experimental Procedures', with comparison sets highlighting the incremental increases in rescuing activity resulting from the presence of the C region together with additional parts of the 3’ UTR. Values for effect size and power were: AB vs C, 0.444 and 1.0; C vs AB+C, 0.448 and 1.0; AB+C vs osk 3’ UTR, 2.21 and 1.0. Additional student’s t tests for comparison of GFP alone to GFP plus the C, AB+C and 3’ UTR parts all had p values of <10–10. (E) Levels of mRNAs from D. Error bars indicate standard deviations.
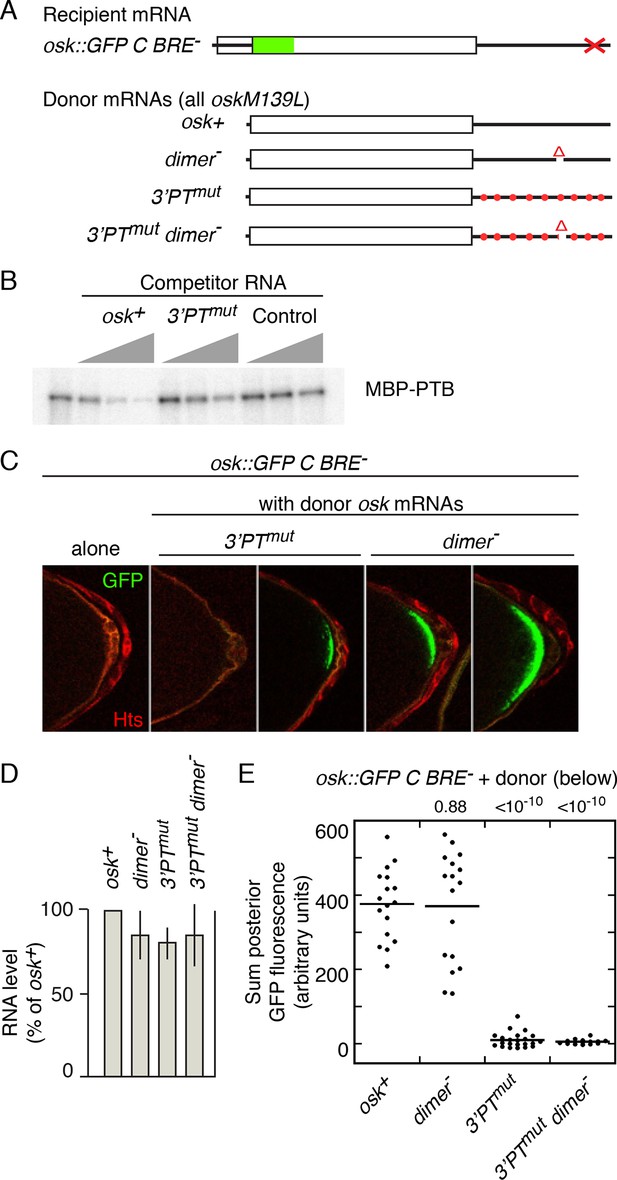
Mutation of pyrimidine tracts in the osk mRNA 3’ UTR disrupts rescue in trans.
(A) Diagram of osk mRNAs used in the assays. The dimer- mRNA has a deletion of positions 665–685 of the 3’ UTR. The 3’PTmut mRNA has multiple mutations (Figure 3—figure supplement 1), indicated figuratively by the red dots. Each donor osk transgene for rescue has the M139L mutation to prevent translation of the Short Osk isoform. Only Short Osk has patterning activity (Vanzo and Ephrussi, 2002), and we eliminated this isoform to address the possibility that some mutations might cause inappropriate Osk expression and dominant maternal-effect lethality from excess osk patterning activity. (B) PT mutations disrupt PTB binding. UV crosslinking assay of MBP-PTB binding to the osk 3’ UTR. The left lane shows binding to the radiolabeled osk 3’ UTR RNA in the absence of competitor RNA. For the remaining lanes, increasing amounts (3, 10, 30x molar excess) of unlableled competitor RNAs were included in the binding assays. The competitor RNAs were the wild type osk 3’ UTR (osk+), the osk 3’ UTR with the PT mutations included in the osk 3’PTmut transgene (3’PTmut), or a nonspecific control RNA corresponding to a portion of the bicoid mRNA 3’ UTR (segment 3R: 6756590–6756463, r6.08) with no strong predicted PTB binding sites (Control). (C) Examples of oocytes expressing osk::GFP C BRE- alone (and thus strongly defective in translational activation), or in combination with a donor osk transgene as indicated. Late stage oocytes were used because the translational activation defect caused by the C BRE- mutations is not fully penetrant at earlier stages. Although only a small fraction of stage 9/10 osk C BRE-oocytes have detectable Osk protein (Reveal et al., 2010), we wanted to remove this variability for experiments in which there could be small differences between different genotypes. Note that the imaging conditions used to provide these examples of different levels of rescue are not the same as those used for quantitation in panel E. The conditions for these images were chosen to highlight trace levels of GFP, and as a consequence the signal in some samples is saturated. (D) Levels of donor mRNAs. Values are in comparison to a rescuing osk transgene as in Figure 2. Error bars indicate standard deviations. (E) Posterior GFP fluorescence from osk::GFP C BRE- when expressed in combination with donor osk mRNAs bearing the indicated mutations. Each dot represents one late stage oocyte, with averages indicated by horizontal lines. All samples were from flies grown, fixed, processed and imaged in parallel (see 'Experimental Procedures' and Figure 3—figure supplements 2 and 3 for details of quantitation). The student’s t test was used to test for significance of differences relative to the osk+ donor mRNA (left), with the p values shown above.
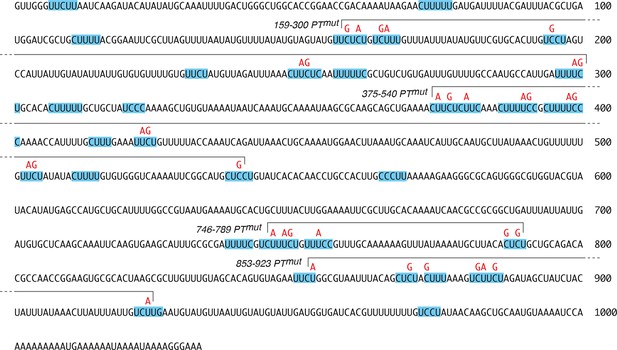
Predicted PTB-binding sites in the osk mRNA 3’ UTR.
The complete osk 3’ UTR sequence is shown, with runs of 4 or more mixed pyrimidines (Pyrimidine Tracts, or PTs) highlighted with blue shading. The mutations included in the osk3’PTmut transgene are indicated in red. The subsets of these mutations included in the derivative transgenes are shown using overlying brackets, with each mutant named to the left of the bracket.
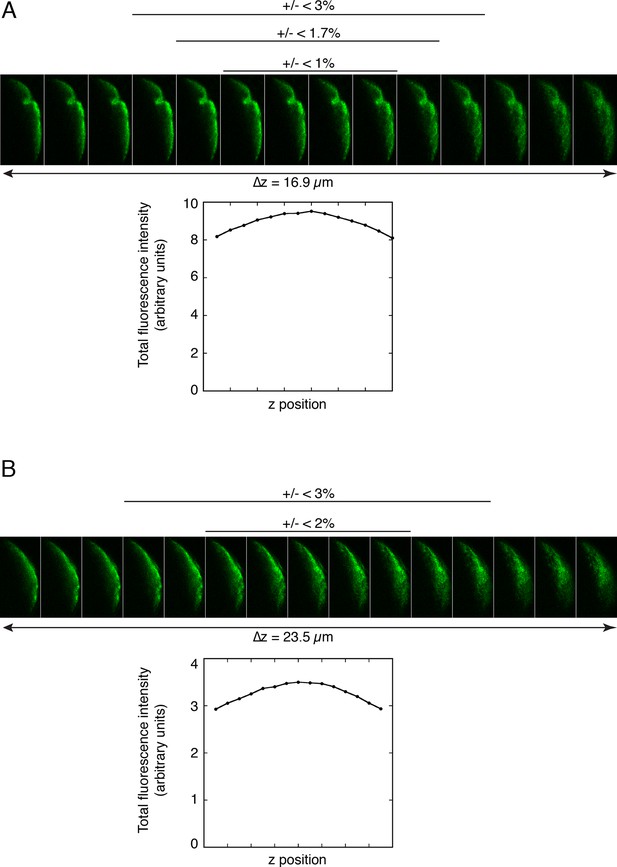
Quantitation of fluorescence at the posterior pole of oocytes.
A and B. Measuring the amounts of Osk::GFP fluorescence in different z positions at the posterior pole of an oocyte to determine the variability introduced by selecting a single focal plane for analysis. A series of images were captured using the Series function of the Leica SP software, and sums of posterior fluorescence were determined with FIJI ('Experimental Procedures') and plotted with Kaleidagraph (below the images). Deviations from the maximal values are shown (above the images). For both A and B, the z position selected visually as being brightest had an intensity less than 2% different than the true maximum. Thus, there is only a small potential error from choice of focal plane for imaging, with the magnitude being much less than the differences observed in Figures 3–4.
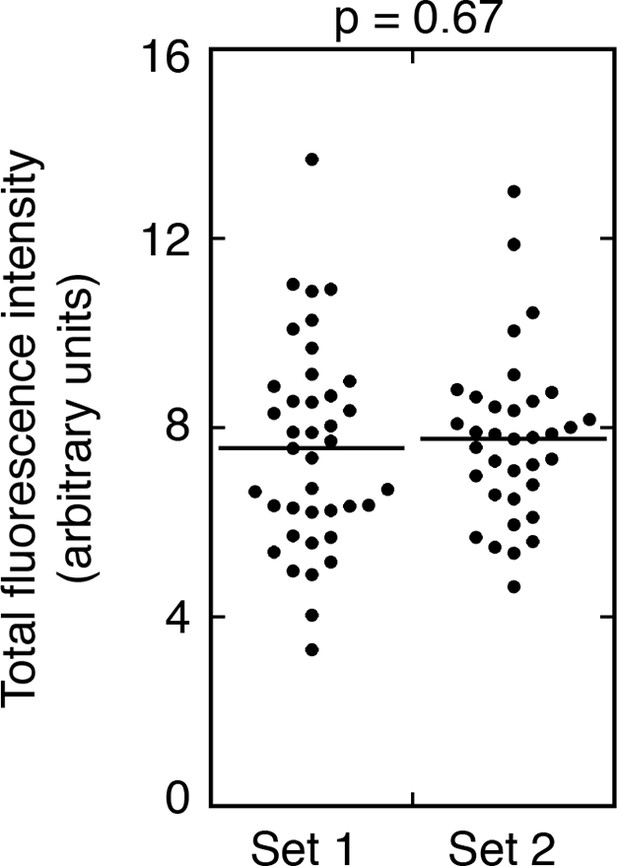
Consistency of rescue of Osk::GFP expression.
Measuring the amounts of Osk::GFP fluorescence at the posterior pole of different oocytes of the same genotype. Samples from a single experiment (with osk::GFP C BRE- in combination with an osk+ donor) were mounted on two slides. Set 1 images were from one slide, and Set 2 images from the other slide. The student’s t test was used to test for significance of differences between the two data sets, with the p value shown above. The results are statistically indistinguishable, indicating that sampling errors are unlikely to explain to differences observed for different genotypes in Figures 3–4.
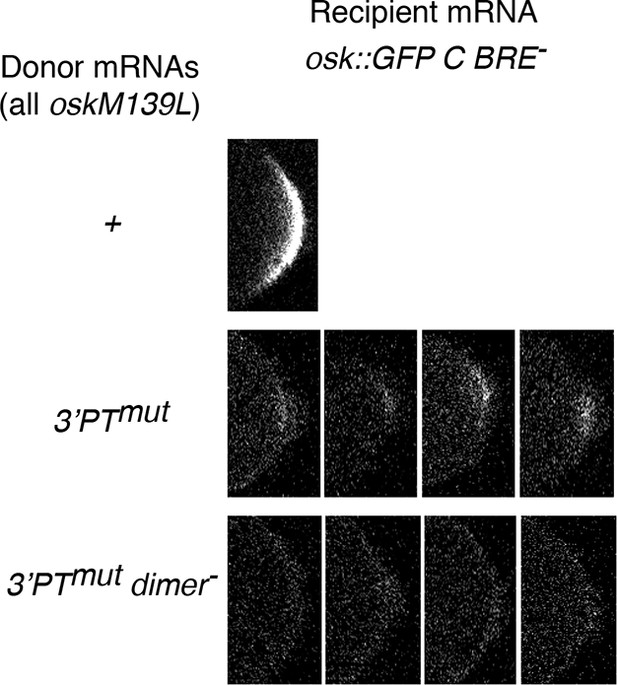
Qualitative differences in weak rescue of osk::GFP C BRE- expression.
Examples of posterior GFP signal from osk::GFP C BRE- recipient mRNA in combination with the donor mRNAs indicated at left. The examples show the tract levels of posterior GFP sometimes found with the 3’PTmut donor, but never seen with the 3’PTmut dimer-donor.
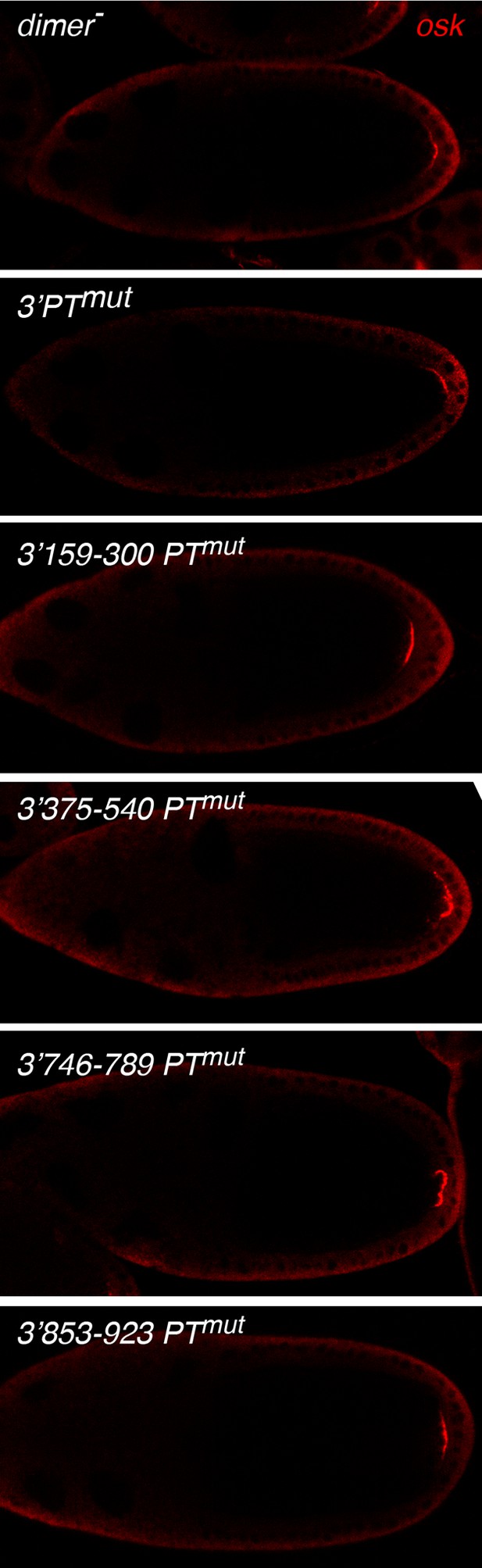
PT and dimer-mutations in the osk mRNA 3’ UTR do not disrupt posterior localization.
Transgenes with the indicated mutations were expressed in flies lacking any other osk mRNA, and the transgenic mRNAs were detected by in situ hybridization. In all cases, the transgenic mRNAs appear in a crescent at the posterior pole of the oocyte.
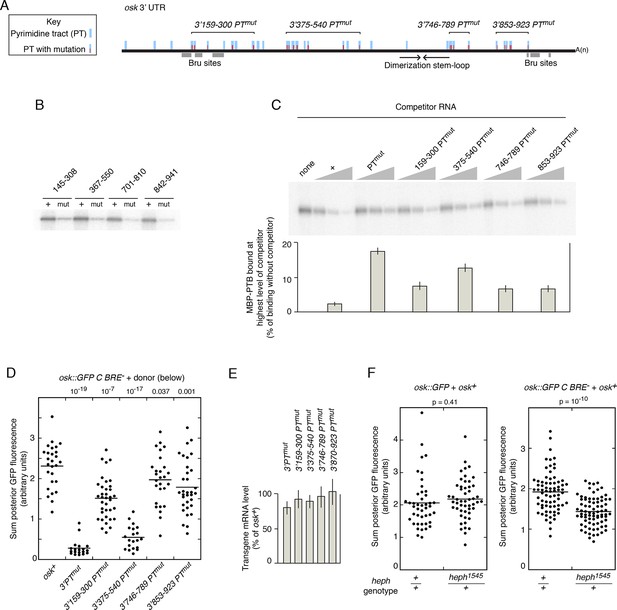
Evidence that PTB is required for rescue in trans.
(A) Diagram of the osk mRNA 3’ UTR. Above the black line (the 3’ UTR) are indicated candidate PTB-binding sites (blue), consisting of any tract of mixed pyrimidines at least 4 nt in length. Mutations within pyrimidine tracts (PTs) that are present in the osk 3’PTmut transgene are indicated in red. Subsets of PT mutations, incorporated into genomic osk transgenes used in panel B, are indicated above (numbers correspond to positions in the osk 3’ UTR). Below the black line, the Bru sites and position of the stem-loop structure containing the RNA dimerization motif are indicated. (B) Subsets of PT mutations disrupt PTB binding to short segments of the osk mRNA 3’ UTR. Short segments of the osk 3’ UTR, each encompassing a subset of PT sites noted in panel A, were used as RNA substrates for UV crosslinking assays with MBP-PTB. The extent of each RNA is indicated above (numbers correspond to positions in the osk 3’ UTR). Radiolabeled RNAs were prepared in wild type form (+), or with the PT mutations from that region (i.e. the same mutations as indicated in red in A)(mut). (C) Subsets of PT mutations disrupt PTB binding to the complete osk mRNA 3’ UTR. UV crosslinking assays with unlabeled competitor RNAs were performed as in Figure 3E, again using the complete osk 3’ UTR as a radiolabeled-binding substrate. The competitor RNAs are indicated above, and consist of the complete osk 3’ UTR with the designated PT mutations. The bar graph below shows the level of residual binding to the radiolabeled substrate RNA at the highest level of competitor (30x molar excess). Smaller values indicate stronger binding of the competitor RNA. Assays were performed three times, and the error bars indicate standard deviations. The 375–540 PTmut RNA was a significantly less effective competitor than the other RNAs with subsets of mutations (p<0.05 for all). (D). Posterior GFP fluorescence from osk::GFP C BRE- when expressed in combination with donor osk mRNAs bearing the indicated mutations. Data are presented as in Figure 3. The student’s t test was used to test for significance of differences relative to the osk+ donor mRNA (left), with the p values shown above. Additional statistical tests evaluated effect size (Cohen’s d) and power. Effect size and power, respectively, for the different donors relative to osk+ were: 3’PTmut, 5.01 and 1.0; 3’159–300 PTmut, 1.53 and >0.99; 3’375–540 PTmut, 4.12 and 1.0; 3’746–789 PTmut, 0.58 and 0.56; 3’853–923 PTmut, 0.88 and 0.91. The statistical significance of the reduced rescuing activity of the 3’746–789 PTmut donor is less compelling than for the other mutants, but this does not invalidate the overall conclusion that the PT mutations have an additive effect on loss of rescuing activity. (E) Levels of donor mRNAs. Values are in comparison to a rescuing osk transgene as in Figure 2. Error bars indicate standard deviations. (F) Posterior GFP fluorescence from osk::GFP (left) or osk::GFP C BRE- (right) when expressed in combination with osk+. Flies were either heph+ or heterozygous for heph1545, as indicated below. The student’s t test was used to test for significance of differences between the two samples in each panel, with the p values shown above. Additional statistical tests evaluated effect size (Cohen’s d) and power. Effect size and power, respectively, were 0.169 and 0.134 (left panel) and 1.12 and >0.99 (right panel).
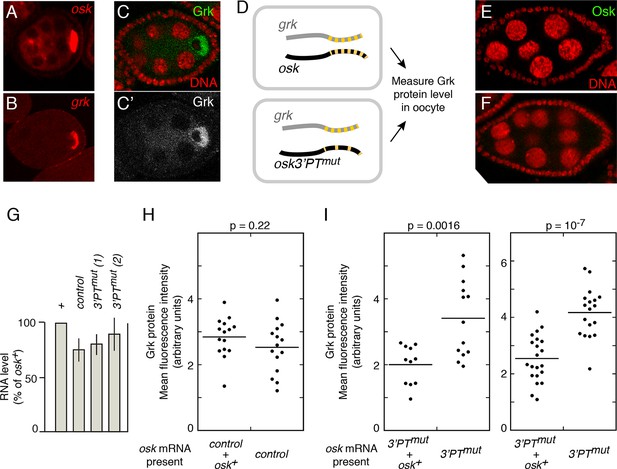
osk mRNA influences the level of Grk protein.
A and B. Distribution of osk (A) and grk (B) mRNAs in egg chambers early in oogenesis. mRNAs were detected by in situ hybridization. For each egg chamber, the cell at right with intense signal is the oocyte. (C) Distribution of Grk protein at stage 6 of oogenesis. C shows both Grk (green) and nuclei (red), and C’ shows only Grk (white). Grk is most strongly expressed in the oocyte, and is excluded from the oocyte nucleus. (D) Experimental design. Each rectangle represents an oocyte, showing the grk and osk mRNAs present. Both have multiple PTs, indicated by the yellow dashes. The osk 3’PTmut mRNA has many PTs mutated, and thus fewer yellow dashes. (E and F) Immunodetection of Osk protein, showing no detectable precocious expression of Osk protein from either the control osk mRNA (E) or the osk3’PTmut mRNA (F). Nuclei are red. (G) Levels of donor mRNAs. Values are in comparison to a rescuing osk transgene as in Figure 2. Error bars indicate standard deviations. (H) Levels of Grk protein in oocytes when expressed in the presence of both wild type and control osk mRNAs (left) or only the control osk mRNA (right). The control osk mRNA is from the osk 11-13- transgene (Figure 6) which has no defect in Osk expression or rescue in trans. See 'Experimental Procedures' and Figure 5—figure supplement 1 for details of quantitation. (I) Levels of Grk protein in oocytes when expressed in the presence of osk mRNAs as indicated at bottom. In the left panel, osk 3’PTmut line 1 was used, and in the right panel osk 3’PTmut line 2 was used. Additional statistical tests evaluated effect size (Cohen’s d) and power, respectively: 1.51 and 0.94 (left panel) and 1.91 and >0.99 (right panel). For comparison, the values for the control experiment in panel H were 0.45 (effect size) and 0.22 (power).
Quantitation of fluorescence in Stage 5/6 oocytes.
Measuring the amounts of Grk immunofluorescence in different z positions within an oocyte to determine the variability introduced by selecting a single focal plane for analysis. A series of images were captured using the Series function of the Leica SP software, covering the region identified visually as having the maximum brightness, and average fluorescence levels were determined with FIJI ('Experimental Procedures') and plotted with Kaleidagraph (below the images). Variation in levels across the entire z series was less than 5%, much less than the differences observed in Figure 5.
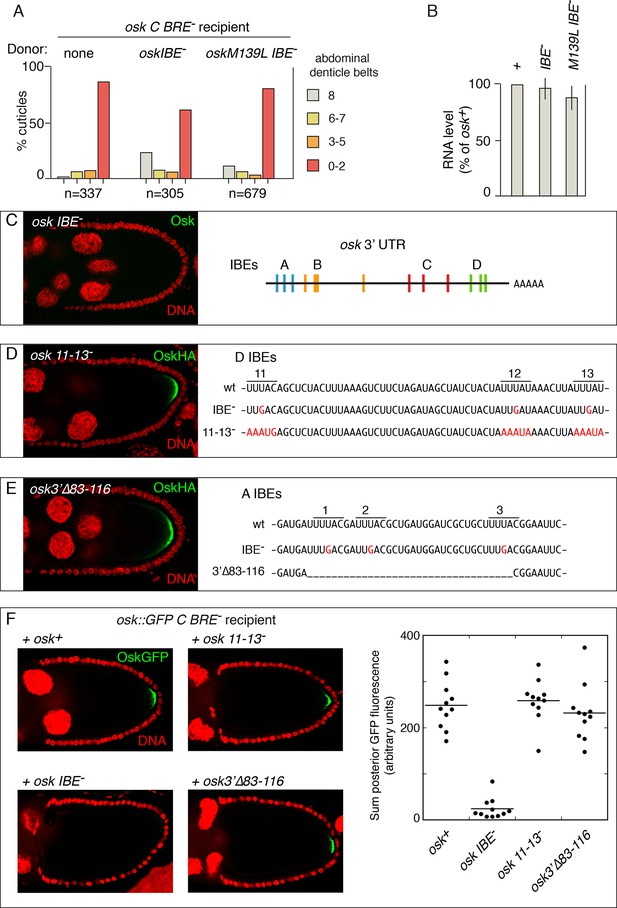
IBE mutations disrupt rescue in trans and have gain-of-function properties.
(A) Embryonic patterning assays to monitor rescue of the translational activation defect of osk C BRE-. (B) Levels of transgenic osk mRNAs. Values are relative to an osk+ transgene that fully rescues an osk mutant. Error bars indicate standard deviations. (C) Absence of Osk expression from the osk IBE- transgene (left)(stage 9 egg chamber) and positions of the IBEs in the osk 3’ UTR [diagram adapted from Figure 6A of (Munro et al., 2006)]. The osk IBE- transgene used has the A subset of mutations, and makes no detectable Osk protein (the same phenotype as osk mutants with the C or D subsets of IBE- mutations) (Munro et al., 2006). (D) Mutation and phenotype of IBEs 11–13. Left, Osk expression from the osk 11-13- transgene mRNA (stage 9 egg chamber). The osk 11-13- transgene has an HA epitope tag, which was used for immunodetection (an osk+ transgene with the same tag is expressed at similar levels; not shown). Right, sequences of the osk 3’ UTR region containing IBEs 11–13. Mutated bases are in red. The IBE- mutations are those used by Munro et al. (2006). (E) Mutation and phenotype of IBEs 1–3. Left, Osk expression from the osk3’∆83–116 transgene mRNA (numbering indicates position in the osk mRNA 3’ UTR)(stage 9 egg chamber). Right, sequences of the osk 3’ UTR region containing IBEs 1–3. Mutated bases are in red, and the region deleted is indicated by underscoring. The IBE- mutations are those used by Munro et al. (2006). (F) Osk::GFP expression in stage 10 egg chambers (posterior portion only) from females expressing osk::GFP C BRE- in combination with the donor osk mRNAs indicated. Fluorescence intensities were quantitated for the graph at right.
Additional files
-
Supplementary file 1
Lists of Drosophila genes expressed in oogenesis or in oocytes were obtained from Flybase.org using QuickSearch and searching for Stage: oogenesis or Tissue: oocyte.
Either the entire mRNA, or just the 3’ UTR, of the longest variant of each mRNA was scanned for the pyrimidine tracts indicated.
- https://doi.org/10.7554/eLife.10965.015





