Guanylate binding proteins directly attack Toxoplasma gondii via supramolecular complexes
Figures
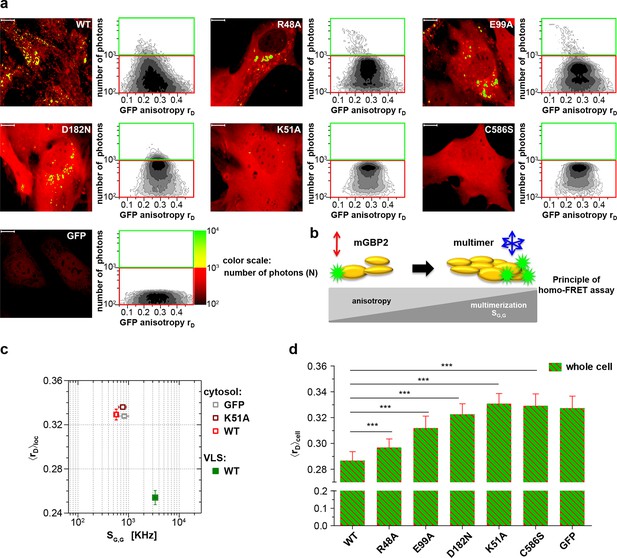
Intracellular homo-multimerization of WT and mutant mGBP2.
All cells were pre-treated with IFNγ for 16 hr prior investigation (a) Left panel. GFP fluorescence intensity (SG,G) images of GBP2-/- MEFs expressing G-mGBP2-WT (G-mGBP2 MEFs), mutants (R48A, K51A, E99A, D182N, C586S) or GFP highlighted with selections of pixels within different cellular compartments. Right panel. MFIS 2D-histograms of GFP anisotropy (rD) on x axis vs. photon number per pixel on y axis, the frequency of pixels color coded from white (lowest) to black (highest). This allows the identification and selection of pixel populations with unique fluorescence properties for a detailed subsequent pixel integrated analysis. The pixels with low photon numbers (below 1000) are selected in red boxes (defined as cytosol) and those with more than 1000 photons in green boxes (defined as VLS). Bars, 10 µm. (b) Scheme of the principle of homo-FRET assays. Compared to G-mGBP2 monomers, rD in G-mGBP2 multimers decreases due to depolarization of GFP fluorescence while GFP SG,G increases. (c) For specific compartments (cytosol and VLS, respectively), the anisotropy values are averaged over all cells generally denoted as <rD>loc. <rD>loc and SG,G in cytosol and VLS were plotted for G-mGBP2-WT, and the K51A mutant and GFP in the cytosol. (d) Mean anisotropy of averages over whole cells <rD>cell for G-mGBP2 WT and mutant proteins. GFP expressing cells served as controls (***p<0.0001).
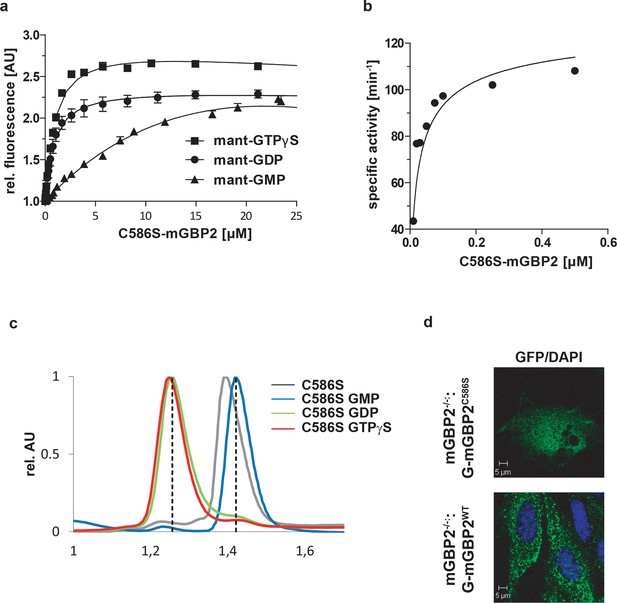
Biochemical properties and intracellular localization of the C586S mutant of mGBP2.
(a) Nucleotide binding. A solution containing 0.5 µM mant-GTPγS, mant-GDP and mant-GMP was titrated with C586S mutant of mGBP2. The fluorescence was excited at 355 nm and detected at 448 nm. The values were normalized to the fluorescence of the nucleotide alone. Dissociation constants are calculated from the fit of the binding curves as in (Kravets et al., 2012). The results averaged over two to four experiments each are given in the Table 1. (b) GTP-hydrolysis. Concentration-dependent GTP-hydrolysis catalyzed by the C586S mutant was measured with a fixed concentration of GTP (1 mM) at 37°C. The initial rates were measured (< 30% GTP hydrolyzed) from the linear parts of time-course experiments and normalized to the protein concentrations used (specific activity). The specific activities were then plotted against protein concentrations. The data were fitted to a model describing the interaction of two molecules of mGBP2, yielding KD (µM) and the maximal specific activity Kmax (min-1). The maximum specific GTPase activitiy, the dimer dissociation constant and the amount of GMP production are summarized in the Table 1. (c) Nucleotide-dependent multimerization. Size-exclusion chromatography of the C586S mutant of mGBP2 bound to GTPγS, GDP, GMP and in the nucleotide free state at 4°C. Elution of all proteins was followed using absorbance by 280 nm. The protein size was estimated by appropriate standard proteins and the absorbance values were normalized to the peaks of the curves. (d) Intracellular localization of WT and C586S-mGBPs was analyzed by transduction of the GFP fusion constructs in mGBP2-/- MEFs. Cells were stimulated with IFNγ for 16 hr. Glass slides were analyzed by confocal microscopy. Bars, 5 µm.
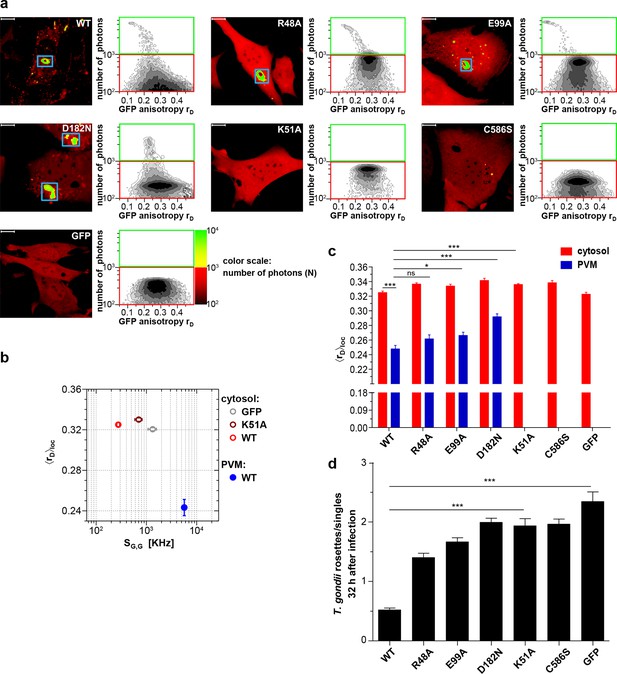
Intracellular homo-multimerization of WT and mutant mGBP2 at the PVM of T. gondii and parasite inhibition.
Cells were pre-treated with IFNγ for 16 hr prior infection with T. gondii ME49 (a) Left panel. GFP fluorescence intensity images of G-mGBP2-WT, mutants MEFs or GFP highlighted with selections of pixels with low and high numbers of photons. Blue boxes mark the PVM area. Bars, 10 µm. Right panel. MFIS 2D-histograms of GFP rD on x axis vs. photon number per pixel on y axis. The pixels with low photon numbers (below 1000) are selected in red boxes and the pixels containing more than 1000 photons in green boxes. (b) Mean values of <rD>loc and mean GFP SG,G were plotted for G-mGBP2-WT in the cytosol and at the PVM of T. gondii and for the K51A mutant and GFP in the cytosol. (c) Mean anisotropy <rD>loc of WT and mutants in the cytosol and at the PVM (blue boxes in (a)). GFP expressing cells served as controls (ns=not significant; *p<0.05; **p<0.01; ***p<0.0001). (d) Replication inhibitory capacity of G-mGBP2-WT and mutants. After fixation T. gondii were stained with the α-SAG1 antibody and the cell nuclei with DAPI. Slides were analyzed by confocal microscopy. Replication inhibition was calculated by the ratio of T. gondii single parasites versus replicative units (rosettes) in at least 100 infected cells (***p<0.0001).
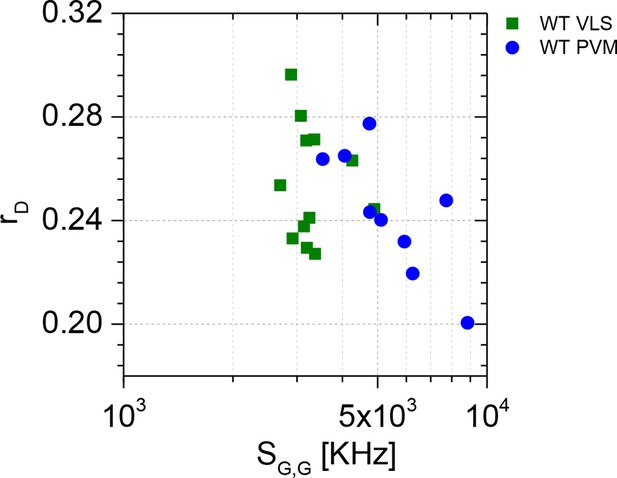
Spectroscopic characterization of G-mGBP2 WT in VLS in non-infected cells and at the PVM in T. gondii infected cells via homo-FRET assay.
Average values of GFP fluoresecnce anisotropy (rD) and signal intensity (SG,G) over single-cell measurements are plotted, in which SG,G values are proportional to protein concentration. A much wider distribution of SG,G can be observed for G-mGBP2 localizing at the PVM (blue circles) comparing to the SG,G values for G-mGBP2 localizing in the VLS (green squares).
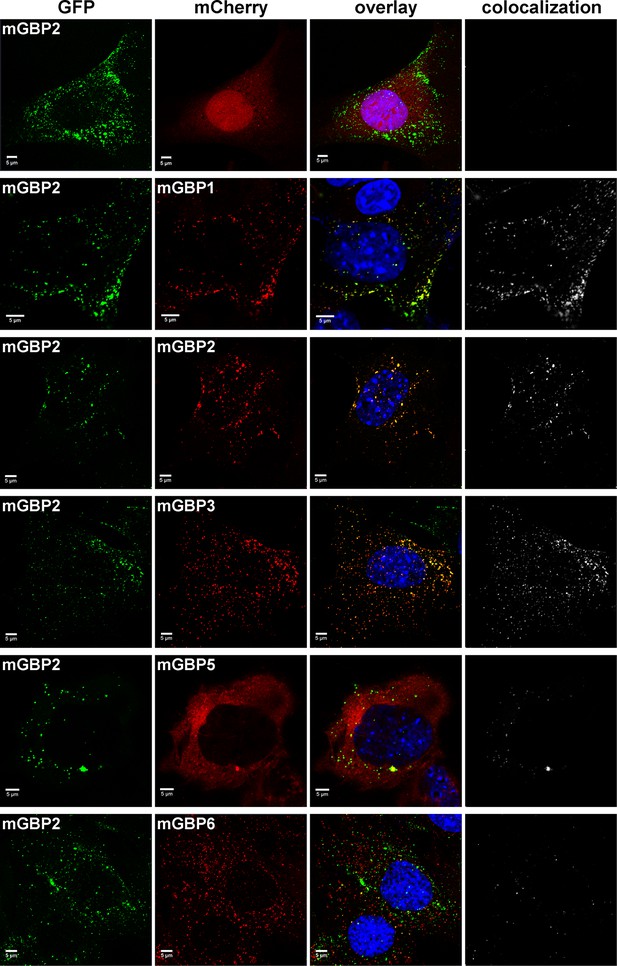
Intracellular colocalization of mGBP proteins.
Subcellular localization of mGBPs was analyzed in G-mGBP2 coexpressing one of the mCh-mGBPs (1, 2, 3, 5 or 6). mCherry expressing cells served as controls. Cells were pre-treated with IFNγ for 16 hr. After fixation, nuclei were stained with DAPI. Glass slides were analyzed by confocal microscopy. Bars, 5 µm. Colocalization analysis was performed with Imaris (Bitplane).
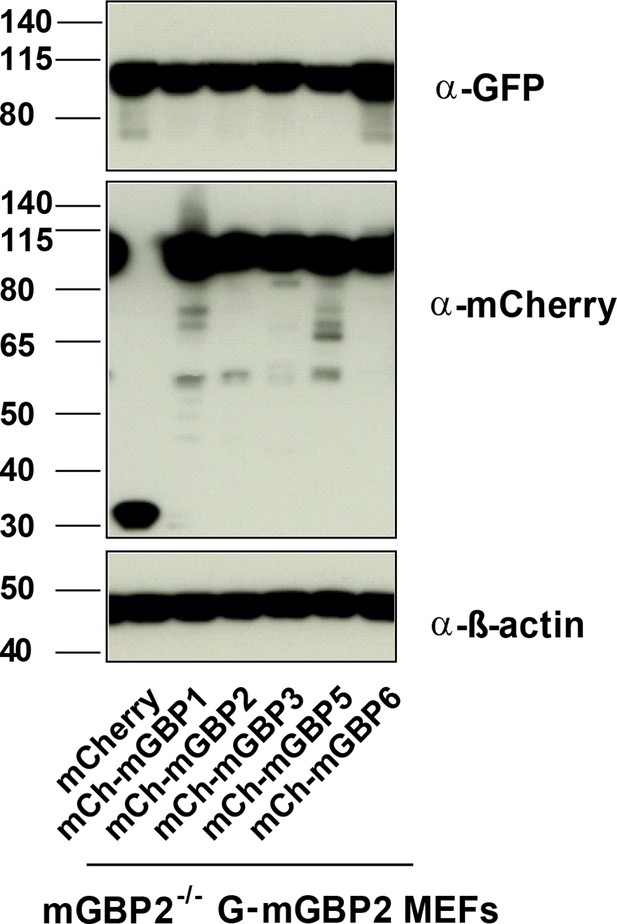
Expression analysis of coexpressed mGBP proteins.
Expression levels of mGBPs in postnuclear supernatants of mGBP2-/- MEFs reconstituted with G-mGBP2 and coexpressing one of the mCh-mGBPs (mGBP1, mGBP2, mGBP3, mGBP5, mGBP6) were analyzed by Western Blotting. mCherry protein expressing cells served as controls. Cells were stimulated with IFNγ for 16 hr. Blots were stained with the α-mCherry antibody.
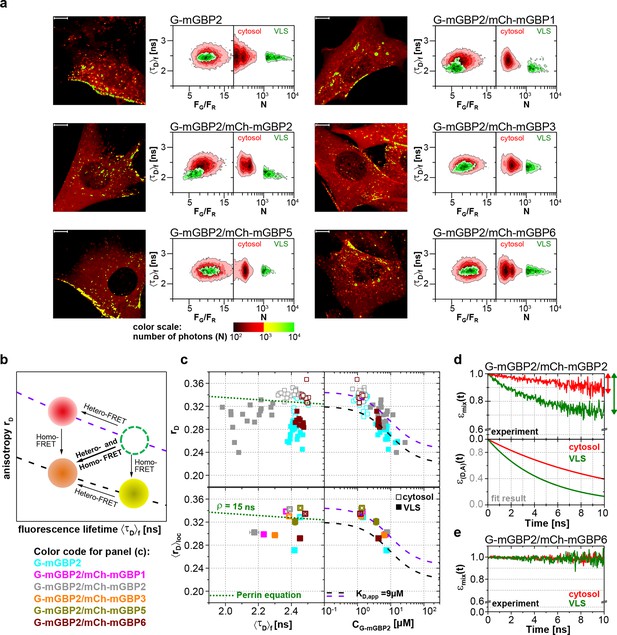
Intracellular homo- and hetero-multimerization of mGBPs.
All cells were pre-treated with IFNγ for 16 hr prior investigation (a) Left panels. GFP fluorescence intensity images of G-mGBP2 or G-mGBP2/mCh-mGBP(1,2,3,5,6) MEFs highlighted with selections of pixels with different intensities. Bars, 10 µm. Right panels. Two MFIS 2D-histograms of GFP fluorescence lifetimes (<τD>f) on y axes, GFP/mCherry fluorescence intensity ratios (FG/FR) or photon number per pixel (N) on x axes. The pixel populations locating in cytosol (N < 1000: red island) and VLS (N > 1000: green island) were separated according to photon numbers. (b) Schematic 2D MFIS plot detailing the effects of hetero- and/or homo-FRET on a reference data set (green circle). The average GFP <τD>f is plotted on the x axis from short to long, while the average steady-state rD is plotted on the y axis. For detailed explanation refer to results section. (c) Upper panel. For individual G-mGBP2, G-mGBP2/mCh-mGBP2or G-mGBP2/mCh-mGBP6 MEFs, mean values of rD in the cytosol (empty squares) and in the VLS (solid squares) were plotted against <τD>f and G-mGBP2 concentrations (CG-mGBP2). Lower panel. Mean anisotropy <rD>loc values (average over all cells weighted by CG-mGBP2) were plotted against <τD>f or CG-mGBP2. The two left panels contain an overlay calculated according to the Perrin equation: with GFP fundamental anisotropy r0 = 0.38 and rotational correlation time ρglobal= 15 ns. The two right panels are overlaid with function curves plotting which assumes a mGBP2 Langmuir binding model with an apparent dissociation constant KD,app. In all donor-only experiments the formation of mGBP2 homo-multimers could be described by KD,app = 9 μM, rmax = 0.32 and rmin = 0.22 (black curve). If other interaction processes interfere with homo-FRET between G-mGBP2 proteins, this curve is shifted upwards (violet curve) while keeping KD,app invariant (rmax = 0.345 and rmin = 0.245). (d, e) εmix(t) and ε(D,A)(t) diagrams of a representative G-mGBP2/mCh-mGBP2 MEF (d) and G-mGBP2/mCh-mGBP6 MEF (e). The drop in εmix(t) curves, as marked by the arrows, represents the species fractions of FRET-active complexes (xFRET) in the VLS (green) and in the cytosol (red). In (d), the FRET rate constant (kFRET) in the cytosol is 0.09 ns-1 and in the VLS 0.20 ns-1.
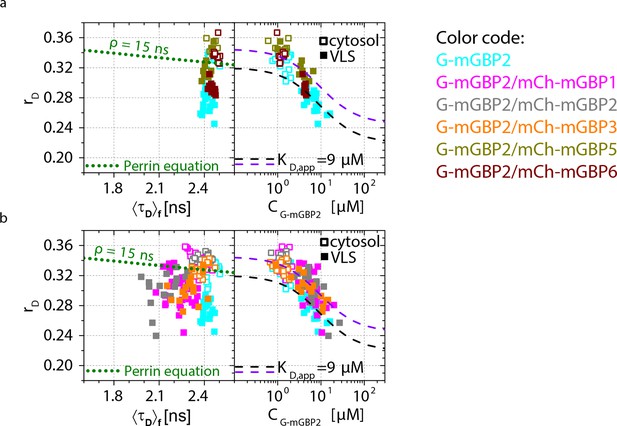
Intracellular homo- and hetero-multimerization of mGBPs in cells.
(a) For single IFNγ stimulated mGBP2-/- MEFs expressing G-mGBP2 alone or coexpressing G-mGBP2/mCh-mGBP5, and G-mGBP2/mCh-mGBP6, average values of rD in the cytosol (empty) and in the VLS (solid) were plotted against <τD>f or G-mGBP2 concentrations (CG-mGBP2). See the legend of Figure 4c for the description of the overlay curves in both panels. (b) Corresponding plots as in (a) for single cells expressing G-mGBP2 alone or coexpressing G-mGBP2/mCh-mGBP1, G-mGBP2/mCh-mGBP2 and G-mGBP2/mCh-mGBP3.
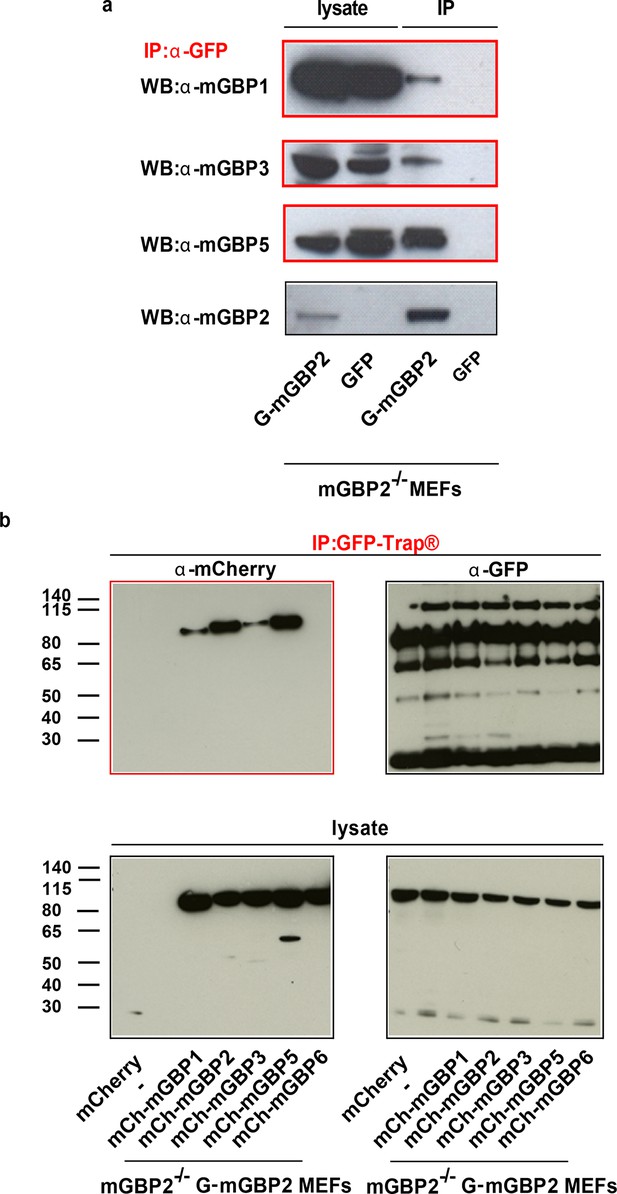
Immunoprecipitation analysis of mGBP proteins.
(a) mGBP2-/- MEFs reconstituted with G-mGBP2 or GFP were stimulated with IFNγ for 16 hr, subsequently lysed and postnuclear supernatants were incubated o/n with G-sepharose beads and the α-GFP antibody at 4°C. IP probes were subjected to Western Blotting. Blots were stained with the α-mGBP2, α-mGBP1, α-mGBP3, α-mGBP5 antibodies. (b) Postnuclear supernantants of mGBP2-/- MEFs reconstituted with G-mGBP2 and coexpressing mCherry protein or one of the mCherry fused mGBPs (mGBP1, mGBP2, mGBP3, mGBP5, mGBP6) were incubated o/n with GFP-Trap beads at 4°C. IP probes were subjected to Western Blotting. Blots were stained with the α-GFP and α-mCherry antibodies.
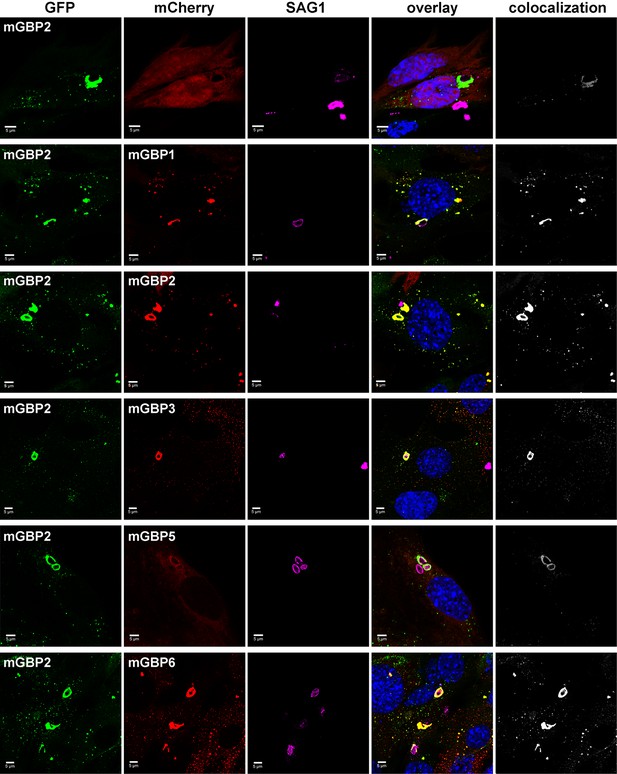
Intracellular colocalization at the PVM of T. gondii and enrichment of mGBP proteins.
Recruitment and colocalization of mGBPs was analyzed in G-mGBP2/mCh-mGBP(1,2,3,5,6) MEFs. mCherry expressing cells served as controls. Cells were stimulated with IFNγ for 16 hr and subsequently infected with T. gondii for 2 hr. After fixation, T. gondii were stained with an α-SAG1 antibody and cell and T. gondii nuclei with DAPI. Glass slides were analyzed by confocal microscopy. Bars, 5 µm. Colocalization analysis was performed with Imaris (Bitplane).
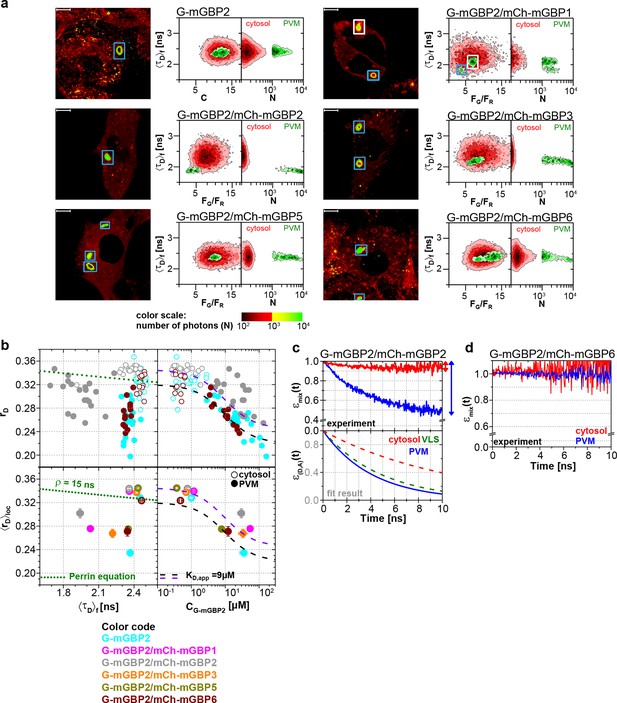
Intracellular homo- and hetero-multimerization of mGBPs at the PVM of T. gondii.
All cells were pre-treated with IFNγ for 16 hr prior investigation (a) Left panels. GFP fluorescence intensity images of living G-mGBP2 or G-mGBP2/mCh-mGBP(1,2,3,5,6) MEFs infected with T. gondii highlighted with selections of pixels within different intracellular localizations. Right panels. Two MFIS 2D-histograms of GFP <τD>f on y axes, GFP/mCherry FG/FR and photon number per pixel (N) on x axes. The pixel populations locating in cytosol (N < 1000: red island) and at the PVM (N > 1000: green island) were separated according to photon numbers. (b) Upper panel. For individual G-mGBP2, G-mGBP2/mCh-mGBP2 or G-mGBP2/mCh-mGBP6 MEFs pixel averages of rD in the cytosol and at the PVM were plotted against <τD>f or CG-mGBP2. Lower panel. Averages of <rD>loc were plotted against <τD>f and CG-mGBP2. Please refer to Figure 4c for further information on the legend and overlaid curves. (c, d) εmix(t) and ε(D,A)(t) diagrams of a representative T. gondii infected G-mGBP2/mCh-mGBP2 MEF (c) and G-mGBP2/mCh-mGBP6 MEF (d). The drop in εmix(t) curves, as marked by the arrows, represents xFRET at the PVM (blue) and in the cytosol (red). The dashed curves representing the ε(D,A)(t) diagrams of G-mGBP2/mCh-mGBP2 interactions in the cytosol (red) and VLS (green) in uninfected cells are inserted for comparison from Figure 4d. In (c), kFRET at the PVM is 0.24 ns-1.
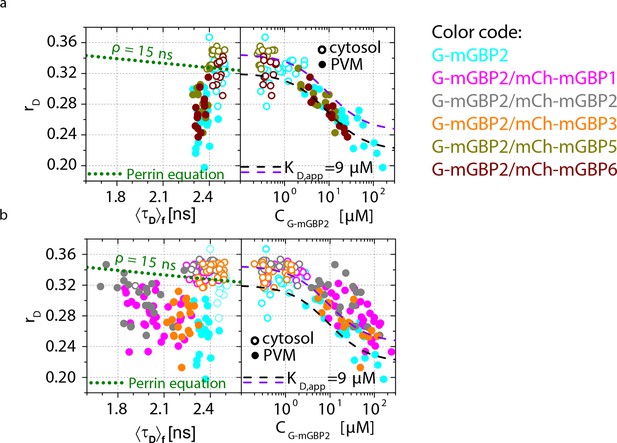
Intracellular homo- and hetero-multimerization of mGBPs in T. gondii infected cells.
(a) For single IFNγ stimulated and T. gondii infected mGBP2-/- MEFs expressing G-mGBP2 alone or coexpressing G-mGBP2/mCh-mGBP5, and G-mGBP2/mCh-mGBP6, average values of rD in the cytosol (empty) and at the PVM (solid) were plotted against <τD>f or G-mGBP2 concentrations (CG-mGBP2). See the legend of Figure 4c for the description of the overlay curves in both panels. (b) Corresponding plots as in (a) for single cells expressing G-mGBP2 alone or coexpressing G-mGBP2/mCh-mGBP1, G-mGBP2/mCh-mGBP2 and G-mGBP2/mCh-mGBP3.
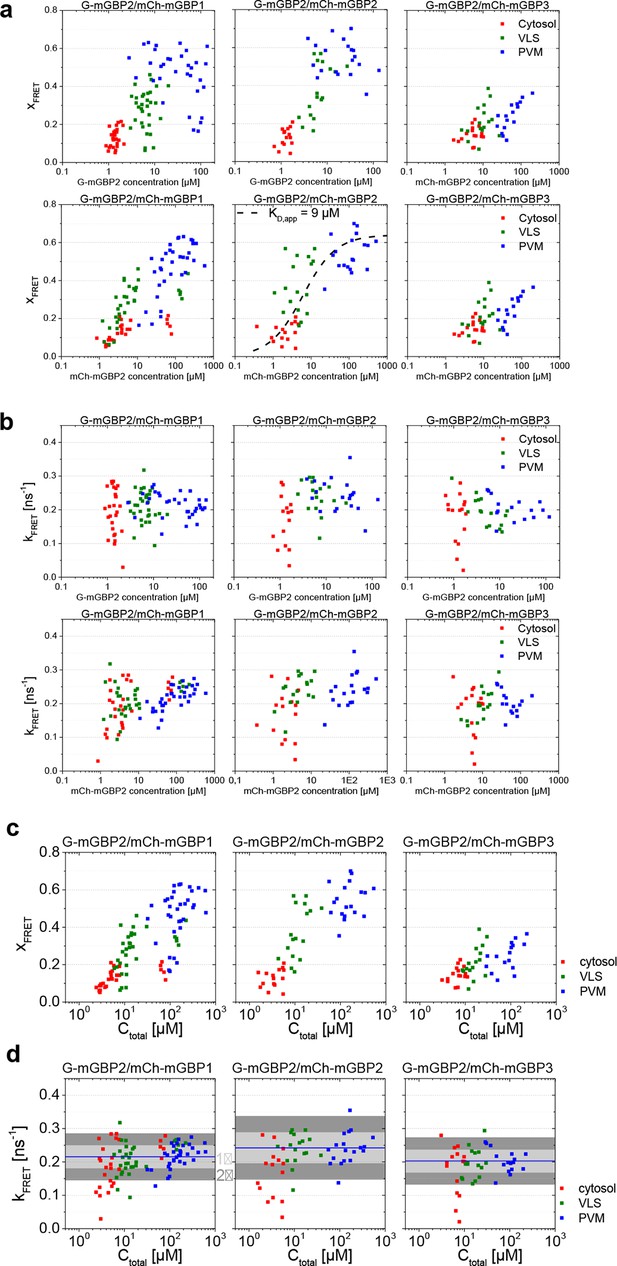
Quantitative MFIS-FRET analysis of mGBP2 hetero-multimerization in living IFNγ stimulated cells.
(a) All the experiments on G-mGBP2/mCh-mGBP1, G-mGBP2/mCh-mGBP2 and G-mGBP2/mCh-mGBP3 interactions were formally analyzed according to Equations 1–5. Fit results of species fraction of FRET-active complex (xFRET) is plotted against G-mGBP2 and mCh-mGBPs concentrations determined in cytosol (red), in VLS (green) and at PVM (blue). The overlaid fuction curve plotting assumes a mGBP2 Langmuir binding model with apparent dissociation constant, KD,app = 9 μM, the same value as applied in Figure 4c and 6b. The scaling factor S = 0.64 was adjusted according to the saturation level of xFRET. (b) For the same experiments as in (a), FRET rate constants (kFRET) are plotted versus G-mGBP2 and mCh-mGBPs concentrations. (c) xFRET in (a) is plotted versus total protein concentration. (d) kFRET in (b) is plotted versus total protein concentration.
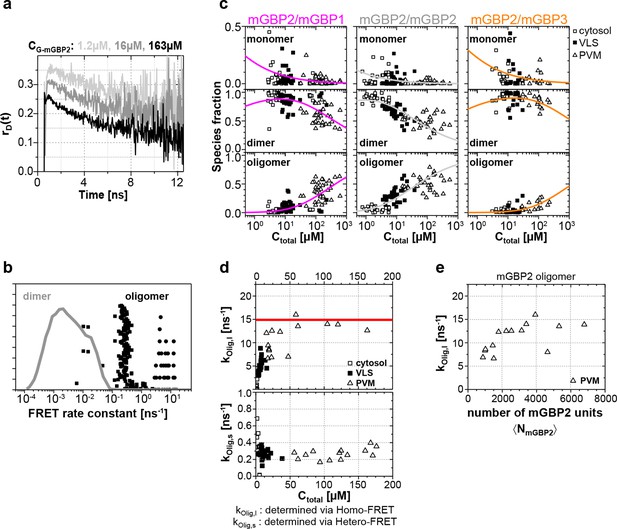
Species-resolved analysis of mGBP2 homo- and hetero-complexes.
(a) G-mGBP2 MEFs with higher concentration exhibited larger quasi instantaneous drop of rD(t) from its initial value of ~0.35, which proves the appearance of a very fast depolarization process due to homo-FRET in mGBP2 oligomers. (b) Distribution of FRET rate constants (kFRET) for mGBP2 dimer (gray curve) and oligomer species (black symbols). Small (black squares) and large (black dots) oligomers, as formally differentiated in the pattern-based MFIS-FRET analysis, show generally higher kFRET than that of the mGBP2 dimer estimated by the MC simulation. (c) Concentration dependence of the three mGBP species (monomer, dimer and oligomer) obtained by the global pattern fit (Equations 6 and 7) of rmix(t) and εmix(t) for two localizations VLS and PVM. The line depicts the fit ('Pattern based pixel-integrated MFIS-FRET analysis' and 'Determination of dissociation constants', Materials and methods section) to the corresponding binding equilibrium with KD,dim, and KD,app-oligo (values are given in the main text). (d) Concentration dependence of FRET rate constants for mGBP2 oligomers which formally differentiated as small (kOlig,s) and large (kOlig,l). (e) kOlig,l versus the number of monomer units in mGBP2 multimers at the PVM determined by scanning FIDA (see 'Scanning fluorescence intensity distribution analysis (FIDA) for determination of oligomer size', Materials and methods section).
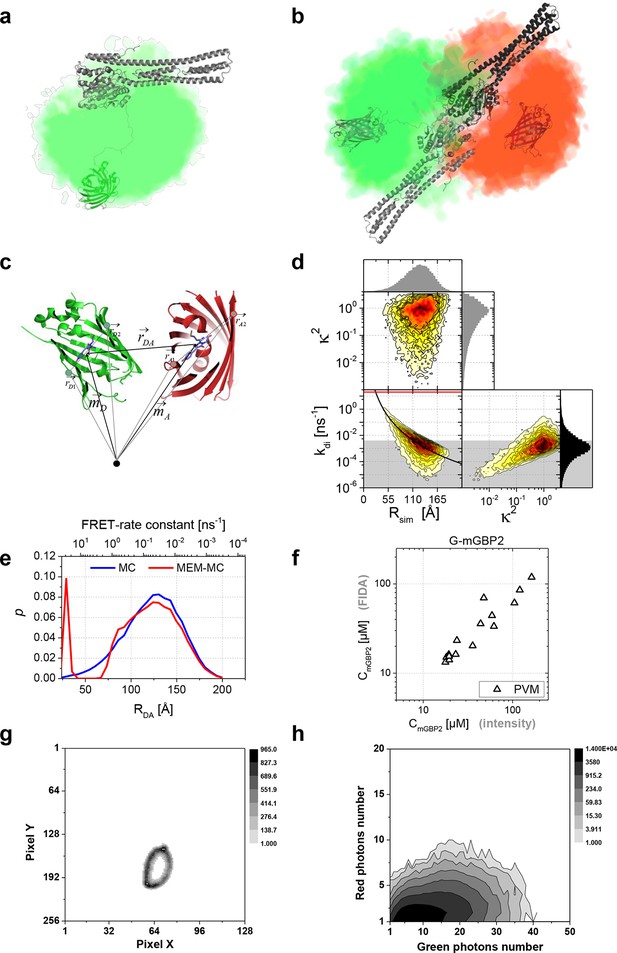
Sample mGBP2 dimer conformations by MC molecular simulation.
(a) Conformational space sampled by the MC simulations of free mGBP2 is illustrated by the density of the GFP-chromophore, one conformation is shown using cartoon representation. (b) Structural properties of a predicted mGBP2 dimer based on the crystal structure of the hGBP1 dimer (PDB-ID 2BC9). The characteristic FRET features of the dimer with flexibly linked fluorescent proteins can be predicted by calculating inter fluorophore distances from the space that is sterically accessible to the fluorescent proteins. The accessible space of attached fluorescent proteins (green (GFP) and red (mCherry) is depicted as fuzzy cloud; ≥ 60% of all D-A configurations are FRET-inactive due to their large distances between the fluorophores, 'Monte Carlo sampling of the donor-acceptor conformational space of mGBP2 dimer', Materials and methods section). (c) Illustration of FRET parameter calculation on each sampled G-mGBP2/mCh-mGBP2 dimer conformation in the MC simulation. Vectors and coordinates in the figure are listed in supplementary file 1a. GFP: green, mCherry: red. (d) Donor-acceptor orientation factor (κ2), spatial distance (Rsim) and FRET rate (kdi) were computed for each sampled mGBP2 dimer conformation, and their relation is plotted in the histogram. In the left panel, the overlay curve in black assumes that the Förster radius between GFP and mCherry is 52 Å, unquenched GFP fluorescence lifetime is 2.6 ns and <κ2> is 2/3. The red line indicates the maximum resolvable FRET rate constant for our detection system (20 ns-1). The area shade in grey indicates the irresolvable low FRET rate constant (E < 1%, kdi < 0.004 ns-1), in which the conformations constitute ~73% of the whole population. (e) The donor-acceptor distance distribution (RDA) obtained from the Monte Carlo (MC) simulation of mGBP2 dimer (blue) and its optimized distance distribution according to the experimental data using maximum entropy method (MEM-MC, in red), see the subsection 11 of Materials and methods for details. (f) mGBP2 concentration determined by 2D FIDA analysis is plotted versus that directly derived from G-mGBP2 fluorescence intensity. (g) A typical image showing the pixels at the PVM area which were analysed by scanning FIDA. The grey scale indicates acquired photon count per pixel. (h) The corresponding 2D FIDA matrix analysing the fluorescence intensity in the green and red detection channel of (g) (the details of FIDA are given in 'Scanning fluorescence intensity distribution analysis (FIDA) for determination of oligomer size', Materials and methods section).
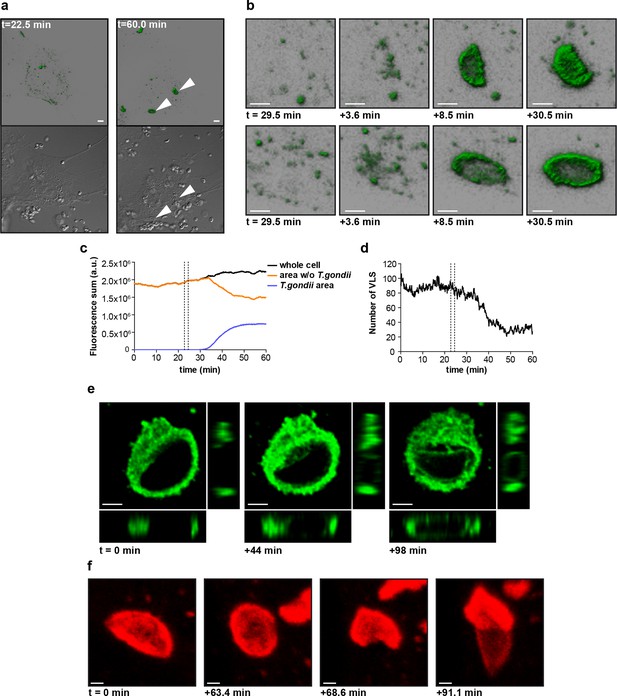
Live-cell imaging of mGBP2 in T. gondii infection.
(a) G-mGBP2 MEFs were treated o/n with IFNγ and infected with T. gondii ME49. Living cells were observed by confocal microscopy at 37°C and a z-stack was recorded every 5–10 s. 4D data were processed and rendered in normal shading mode (upper panels) and the DIC images are displayed (lower panels) for the indicated time points. One out of at least 3 similar experiments is shown. Bar = 5 µm. (b) Magnification from Video 1 and Figure 8a of G-mGBP2 accumulation around two T. gondii parasites at time points indicated. Bar = 2 µm. (c) Quantification of the total fluorescence intensity over the indicated voxels from Video 1. Vertical lines indicate the time points of T. gondii infection of MEFs. One representative analysis out of at least 3 similar experiments is shown. (d) Number of cytosolic VLS with at least approx. 0.25 µm diameter from Video 1 over time. Fluorescence signals close to the T. gondii area were excluded from the analysis. Vertical lines indicate the time points of T. gondii infection of MEFs. One representative analysis out of at least 3 similar experiments is shown (e) XY, XZ, and YZ projections of G-mGBP2 around one T. gondii PVM are shown for the indicated time points. Bar = 2 µm. (f) Maximum intensity projections of mCh-mGBP2 around one T. gondii are shown for the indicated time points. Bar = 1 µm.
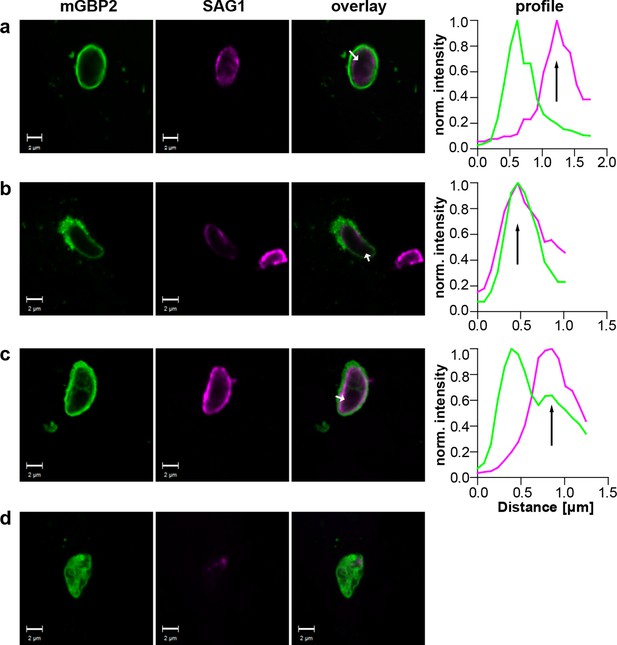
Localization of mGBP2 at the PVM, the plasma membrane, or the cytosol of T. gondii.
G-mGBP2 cells were stimulated with IFNγ for 16 hr and subsequently infected with T. gondii ME49 for 6 hr. After fixation, T. gondii were stained with an α-SAG1 antibody. Glass slides were analyzed by confocal microscopy. Bars, 2 µm. Profiles show individually normalized intensities of GFP (mGBP2, green) or Alexa633 (SAG1, magenta) fluorescence along the indicated white arrows. Black arrows indicate the localization of the T. gondii plasma membrane, as identified by the SAG1 staining. (a) Example of mGBP2 accumulation at the PVM of T. gondii without disruption or permeabilization of the PVM. (b) Example of mGBP2 accumulation at the plasma membrane of T. gondii with obvious disruption of the PVM. (c) Example of mGBP2 accumulation at the plasma membrane of T. gondii without apparent PVM disruption. (d) Example of T. gondii death and accumulation of mGBP2 in the cytosol of the parasite.
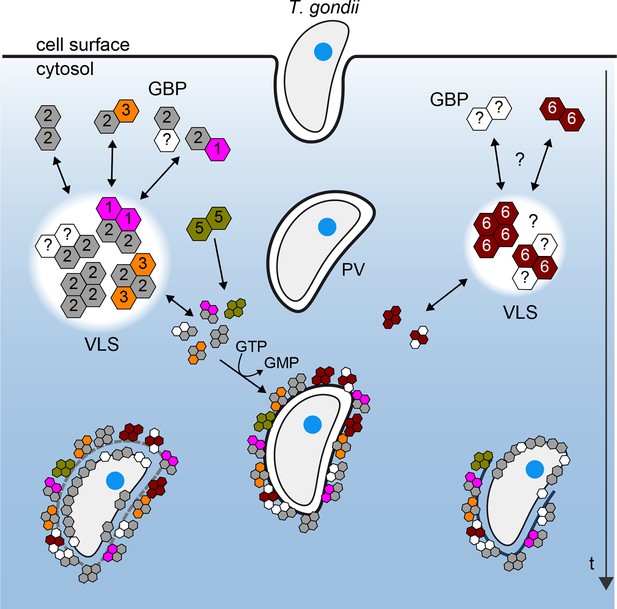
Schematic view of mGBP dynamics and multimerization in T. gondii infected cells.
For details see Results and Discussion
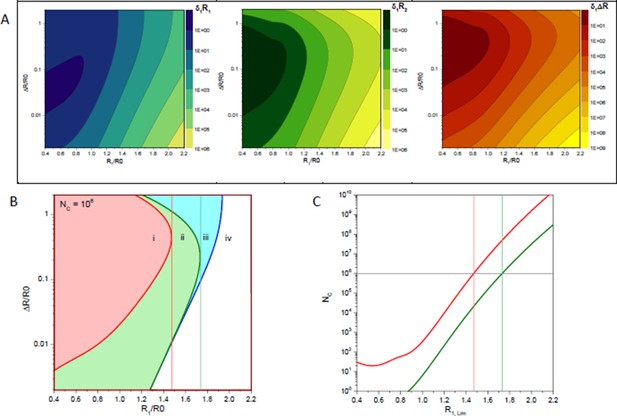
Statistical error estimates of a two distance model with a distance RDA,1 and a second distance RDA,1+ΔRDA; a Förster radius of 50 Å, a fluorescence lifetime of 4 ns and a time-window of 50 ns.
(A) Relative errors of per one counted photon of distances RDA,1, RDA,2 and their difference ΔRDA. (B) Isolines (blue line), (green line) and (red line) for 106 counted photons. Vertical lines indicate the limiting distances RDA,lim for parameters RDA,2 (green) and ΔRDA (red). (C) Limiting distances RDA,lim for a given number of detected photons for parameters RDA,2 (green) and ΔRDA (red).
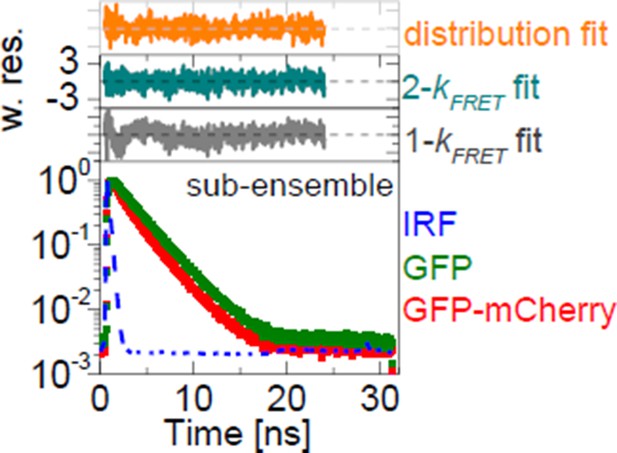
Comprehensive characterizations of FRET-FLIM data.
Two representative experiments of donor-only (GFP) and FRET (GFP-mCherry) samples (left). Fitting the sub-ensemble fluorescence decay containing 3.96×106 photons of the GFP-mCherry experiment (in red) with 1-kFRET, 2-kFRET and kFRET-distribution models resulted in reduced χr2 = 1.41 (in gray), 1.03 (in dark cyan) and 1.08 (in orange), respectively. Pre-determined donor-decay parameters were set as global restraints in all fits: xD0(1) = 0.854, xD0(2) = 0.146, τD0(1) = 2.747 ns, τD0(1) = 1.526 ns. Parameters obtained from 1-kFRET fit were xFRET = 0.303 and kFRET = 0.556 ns-1; from 2-kFRET fit: xFRET = 0.392, xFRET(1) = 0.561, xFRET(2) = 0.439, kFRET(1)= 0.225 ns-1 and kFRET(2) = 1.765 ns-1; and from distribution fit: xFRET = 0.652.
Videos
mGBP2-/- MEFs transduced with G-mGBP2 were treated o/n with IFNγ and infected with T. gondii.
The living cells were observed with a confocal microscope at 37°C and a z-stack was recorded every 5–10 s. 4D data were processed and rendered in normal shading mode. Bar = 5 µm.
mGBP2-/- MEFs transduced with G-mGBP2 were treated o/n with IFNγ and infected with T. gondii.
The living cells were observed with a confocal microscope at 37°C and a z-stack was recorded every 5–10 s. 4D data were processed and rendered as maximum intensity projection. Bar = 2 µm.
mGBP2-/- MEFs transduced with mCh-mGBP2 were treated o/n with IFNγ and infected with T. gondii.
The living cells were observed with a confocal microscope at 37°C and a z-stack was recorded every 5–10 s. 4D data were processed and rendered as maximum intensity projection. Bar = 1 µm.
mGBP2-/- MEFs transduced with G-mGBP2 and cytosolic mCherry were treated o/n with IFNγ and infected with T. gondii.
The living cells were observed with a confocal microscope at 37°C.
Tables
Dissociation constants KD of mant-nucleotides for mGBP2 WT and C586S mutant determined by fluorescence titrations and GTPase activity parameters obtained by protein concentration-dependent hydrolysis.
| Nucleotide binding | GTP-hydrolysis | ||||||
|---|---|---|---|---|---|---|---|
| mant-GTPγS | mant-GDP | mant-GMP | |||||
| KD (µM) | KD (µM) | KD (µM) | Kmax (min-1) | Dimer KD (µM) | GMP (%) | ||
| WT | 0.45 | 0.54 | 14.4 | 102 | 0.029 | 74 | |
| C586S | 0.50 | 0.45 | 15.5 | 133 | 0.026 | 72 | |
-
The % GMP indicates the relative amount of the two products, GDP and GMP
G-mGBP2 cells were stimulated with IFNγ for 16 hr and infected with T. gondii ME49 or BK strains for 2 hr. After fixation and permeabilization with the indicated amounts of saponin, T. gondii were stained with an α-SAG1 antibody and DAPI. T. gondii were counted and categorized according the indicated mGBP2 and SAG1 fluorescence. N.d = not detected.
| ME49 T. gondii | BK T. gondii | |||||
|---|---|---|---|---|---|---|
| mGBP2+ SAG1- | mGBP2+ SAG1+ | mGBP2- SAG1+ | mGBP2+ SAG1- | mGBP2+ SAG1+ | mGBP2- SAG1+ | |
| w/o Saponin | 50% | 38% | 12% | n.d. | n.d. | 3% |
| 0,15% Saponin | n.d. | 57% | 43% | n.d. | 1% | 99% |
Additional files
-
Supplementary file 1
(a) Amino-acid sequence settings in the MC molecular simulation.
The residues used to define the dipole of the chromophoric groups are indicated. (b) Calculations of donor-acceptor distances (Rsim) and orientation factors (κ2) from each sampled conformation from MC molecular simulation of G-mGBP2/mCh-mGBP2 dimer in steps. See Experimental procedures and Figure 7—figure supplement 1 for details.
- https://doi.org/10.7554/eLife.11479.027





