The innate immune sensor IFI16 recognizes foreign DNA in the nucleus by scanning along the duplex
Figures
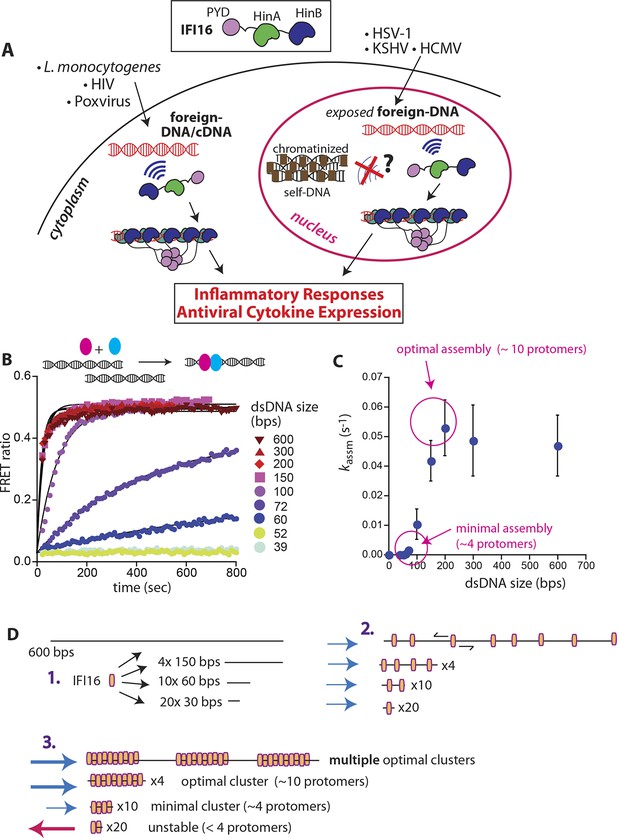
IFI16 assembles faster on longer dsDNA.
(A) Top: IFI16 is composed of three functional domains flanked by unstructured linkers, namely one pyrin domain (PYD) and two dsDNA-binding Hin domains (HinA and HinB; Hin: hematopoietic interferon-inducible nuclear antigen). Bottom: IFI16 detects foreign dsDNA from invading pathogens in both the host nucleus and cytoplasm. (B) Top: a cartoon scheme for FRET experiments. The two differentially colored ovals represent fluorescently (Dylight-550 and Dylight-650) labeled IFI16. Bottom: The time-dependent changes in the emission ratio between FRET donor and acceptor labeled IFI16 (50 nM) were monitored at 33 µg/ml of each dsDNA (e.g. sixfold higher than the dissociation constant for 39-bp dsDNA [Morrone et al., 2014]). Lines are fits to a first-order exponential equation (see Figure 1—figure supplement 1 for 25 nM protein). All shown representative experiments were performed at least three times. (C) A plot of observed assembly rates (kassms) vs. dsDNA-sizes (see also Figure 1—figure supplement 1). (D) 1D-diffusion assisted assembly mechanism can explain the observed assembly profile of IFI16. 1. At the same mass-concentrations, the number of individual dsDNA fragments present in each assay is inversely proportional to the length of dsDNA. 2. Individual IFI16 molecules initially bind dsDNA at random positions and diffuse one-dimensionally while searching for other respective protomers; the number of IFI16 residing on the same dsDNA fragment should be proportional to the length of dsDNA (e.g. there are four times more individual 150-bp fragments than 600-bp fragments) 3. IFI16 fails to assemble into an oligomer on dsDNA shorter than 60 bp (indicated by a red arrow pointing left). The saturating rates can be explained if the final FRET signals arise from formation of distinct optimal oligomers.
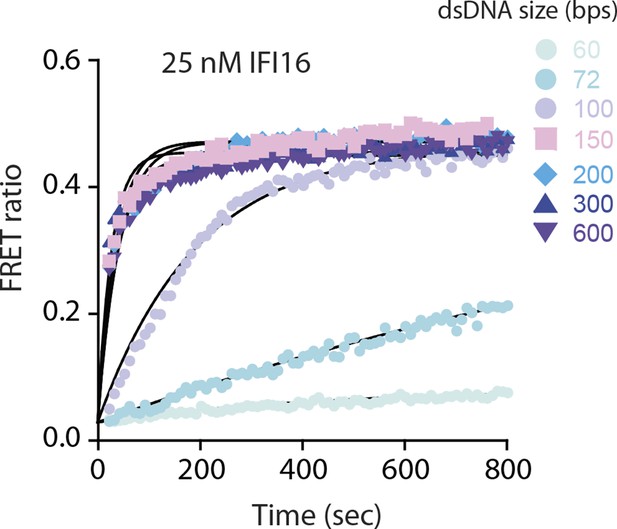
FRET assembly assays using 25 nM donor and acceptor labeled IFI16 compared to 50 nM in Figure 1B.
Shown is a representative of three experiments, and the calculated rates are listed in Supplementary file 1A.
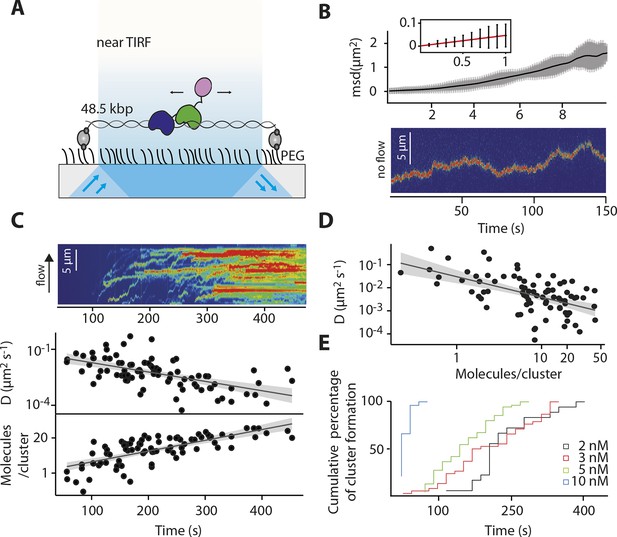
IFI16 scans dsDNA.
(A) Illustration of the TIRF setup. λdsDNA is anchored to the pegylated coverslip surface by either one or two biotinylated oligonucleotide linkers. Single-biotinylated λdsDNA is constantly stretched by flow during measurements and used for the clustering and nucleosome experiments, whereas double-biotinylated λdsDNA is stably attached whilst being stretched and used without flow for single-molecule diffusion coefficient analysis. (B) Double-biotinylated λdsDNA is anchored to the surface and Cy5-labeled IFI16 molecules (800 pM) are visualized in near TIRF while bound to DNA. Top: Mean-square displacement (msd) trajectories are fitted within their linear regime to calculate the 1D-diffusion coefficient. Bottom: A sample kymograph of a single molecule stably bound for tens of second to DNA and exerting Brownian motion. (C) Elevated IFI16 concentrations result in clustering along λdsDNA. Top: A sample kymograph of multiple IFI16 molecules (3 nM) diffusing along λdsDNA. IFI16 molecules display a net motion along the flow direction. Bottom: Time-resolved clustering is accompanied by a decrease in diffusion coefficients and increase in the number of molecules per cluster. (D) The diffusion coefficient inversely correlates with the number of IFI16 per cluster, resulting in immobile, stable oligomers. (E) Cluster formation is IFI16-concentration dependent.
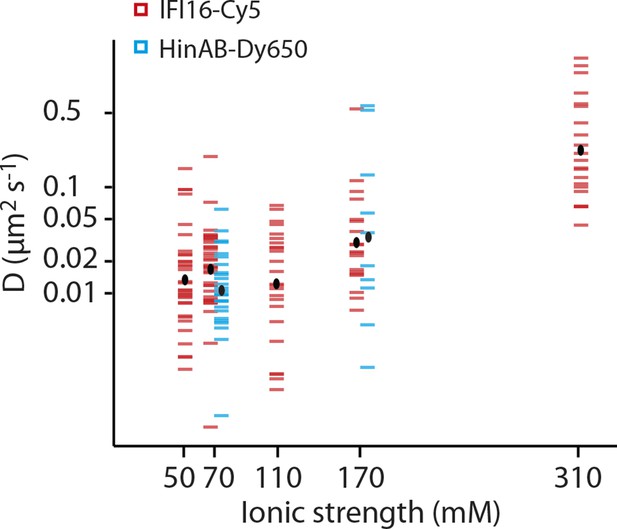
Dependence of the diffusion coefficient of IFI16 and IFI16HinAB mutant on salt concentration.
(i) IFI16: The median of D increases from 0.017 to 0.18 μm2s-1 with increasing ionic strength from 0.07 to 0.31 M concomitant with decreasing binding times to DNA. (ii) HinAB: The median increases from 0.010 to 0.027 μm2s-1 with increasing ionic strength from 0.07 to 0.017 M.
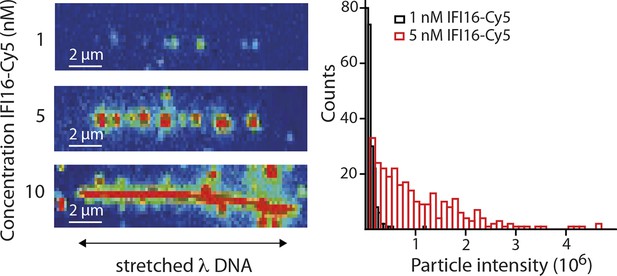
(Left) Representative fluorescence images of IFI16-Cy5 on DNA molecules: Concentrations of 1 and 5 nM result in the resolution of single IFI16 molecules and distinguishable Ifi16 clusters, respectively, whereas a higher concentration of 10 nM eventually leads to DNA congestion with protein clusters or filaments.
(Right) Intensities per particle are shown for 1 nM (single molecules) and 5 nM (clusters of IFI16). The mean particle intensity for single molecules serves as basis for calculating the number of IFI16 molecules within clusters (Figure 2—figure supplement 3).
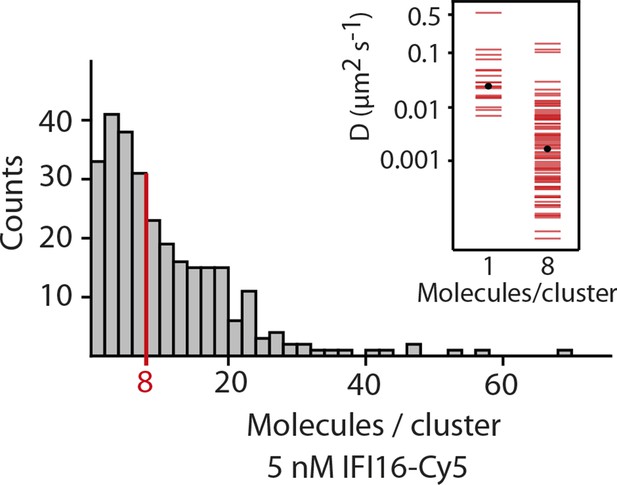
Fluorescence data of 5 nM IFI16 are converted to cluster sizes (see Figure 2—figure supplement 2).
The mean cluster size amounts to 8 IFI16 molecules within a cluster. Clusters are almost immobile, represented by a significant reduction in the diffusion coefficient D compared to that of single molecules of IFI16.
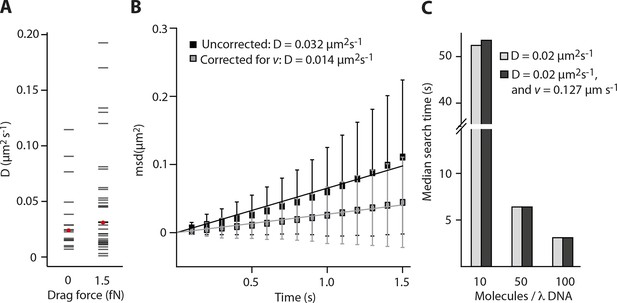
Impact of the flow-induced drift on diffusion and clustering is minimal.
(A) Constant flow (20 µl/min) is applied for the clustering experiments with elevated IFI16 concentrations, corresponding to a drag force of 1-2 fN on the DNA substrate and bound IFI16 molecules. The median measured diffusion coefficient D for single molecules increases from 0.026 µm2s-1 (no flow) to 0.031 µm2s-1 (with flow). (B) As described in the supplementary material, we calculated the drag velocity v on single molecules, with which we can define the effect of flow on D. Exemplarily shown here is a single diffusion trace, uncorrected and corrected for v. (C) Monte-Carlo simulation of IFI16 dimerization on λDNA that is occupied by varying amounts of 1D-diffusing IFI16s. The presence of a drag velocity v does not significantly alter the median search time.
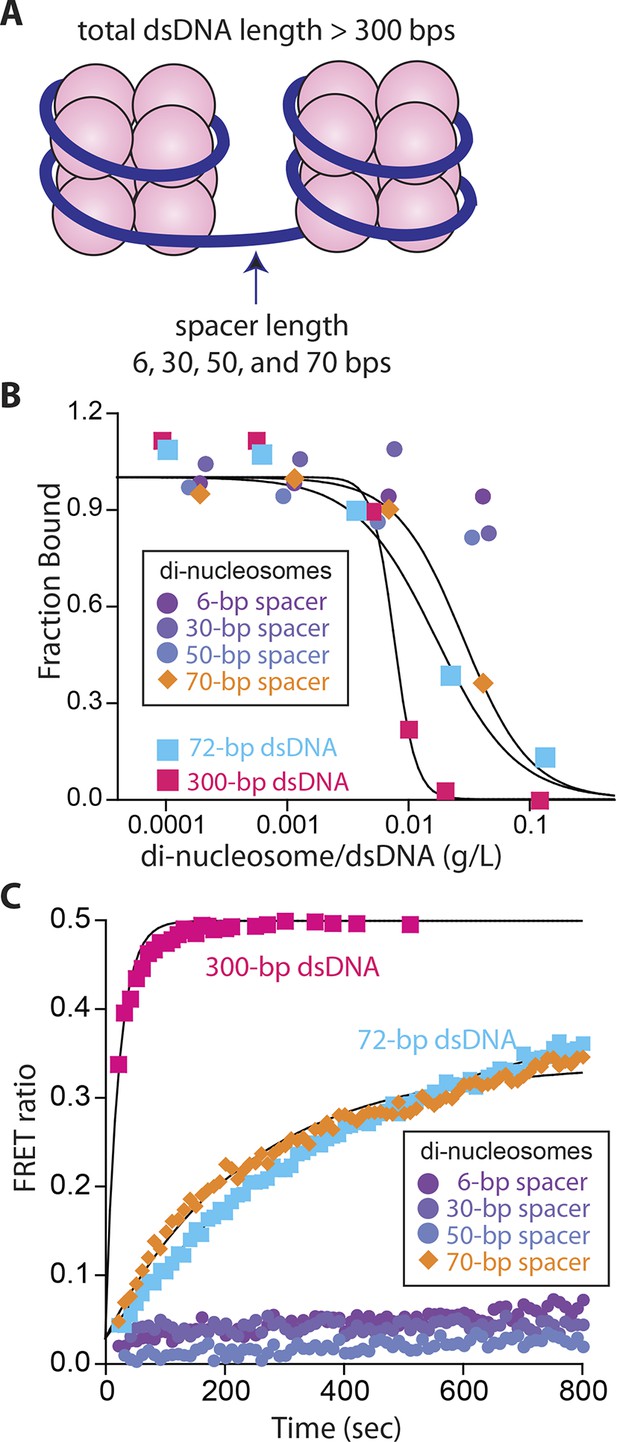
Nucleosomes inhibit oligomerization.
(A) A cartoon of di-nucleosome constructs with varying dsDNA-spacers. (B) Competition binding assays using IFI16 bound FAM-labeled 70-bp dsDNA against various di-nucleosomes and naked dsDNA. The lines are fits to: 1/(1+([DNAcompetitor]/IC50)^Hill constant), where IC50 indicates the concentration of competitor at 50% efficiency. The mass-concentration of each competitor was calculated using dsDNA, but not histones. (C) The time-dependent changes in the emission ratio between the FRET donor and acceptor labeled IFI16 (50 nM) were monitored at 33 µg/ml of each nucleosome or naked dsDNA. The lines are fits to a first-order exponential equation.
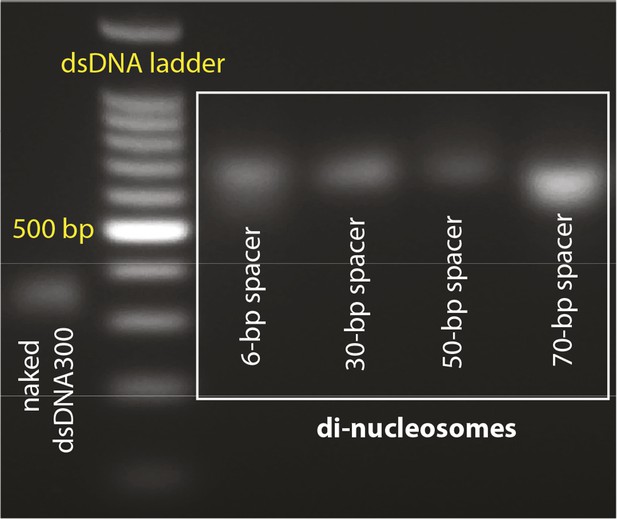
Agarose gels with nucleosome preparations.
(A) Purified dinucleosomes for the bulk FRET assays.
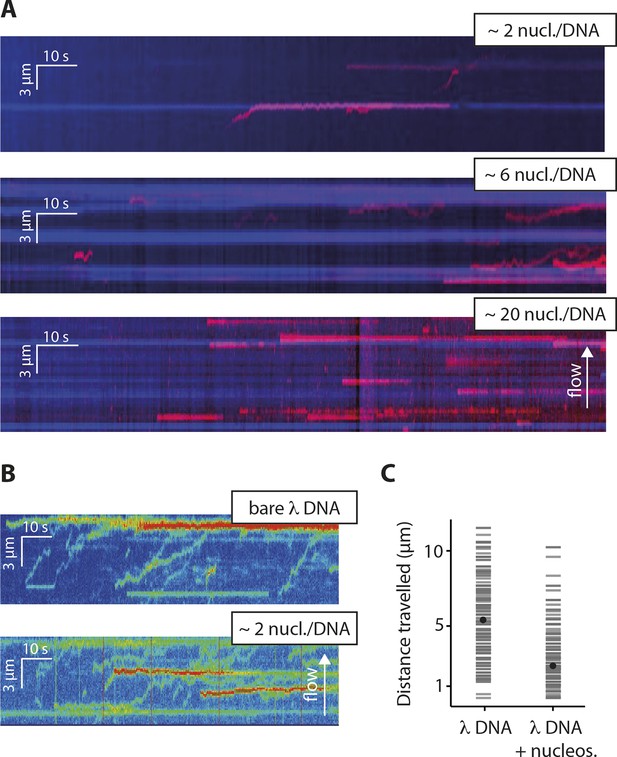
Nucleosomes inhibit 1D-diffusion.
(A) Kymographs of Cy5-labeled IFI16 (magenta) binding to λdsDNA with varying numbers of nucleosomes tagged with anti-H4-Atto488 (blue). The number of nucleosomes per λdsDNA were estimated by quantifying IFI16 clustering sites for the lowest nucleosome/λdsDNA ratio (Figure 4—figure supplement 3C), yielding ~2 nucleosomes/ λdsDNA. At low nucleosome concentrations (~2 to 6 nucleosomes/DNA), IFI16 binds to λdsDNA and diffuses with the flow direction, until encountering a nucleosome. At higher concentrations (~20 nucleosomes per λdsDNA), individual IFI16 show only very short diffusive movements upon binding. (B) On naked λdsDNA, IFI16 travels with the flow to the free tip (top), whereas it oligomerizes along the path on nucleosome-loaded λdsDNA (bottom). (C) The overall travel distance of single IFI16 on nucleosome-loaded λdsDNA is reduced compared to bare λdsDNA (nucleosomal λdsDNA: N = 167, bare λdsDNA: N = 141).
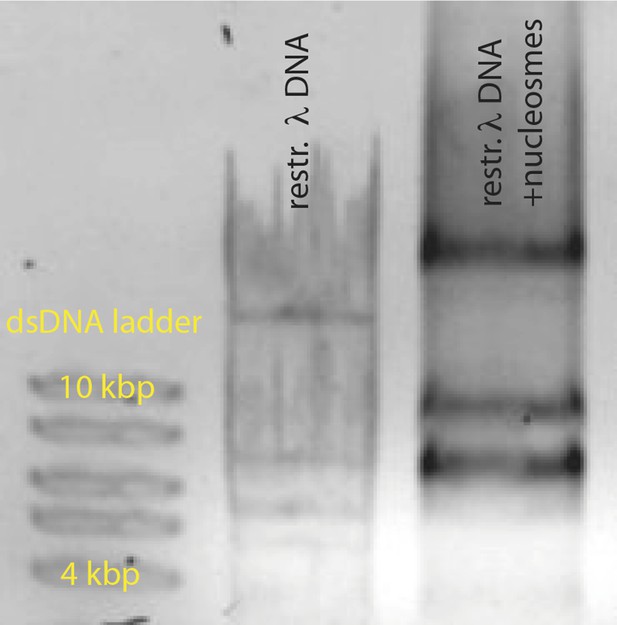
Reconstituted nucleosomes on restricted λdsDNA: EcoRI digestion generated l-DNA fragments of 21 kbp, 7.5 kbp, 5.8 kbp, 5.6 kbp, 4.8 kbp, and 3.5 kbp.
https://doi.org/10.7554/eLife.11721.015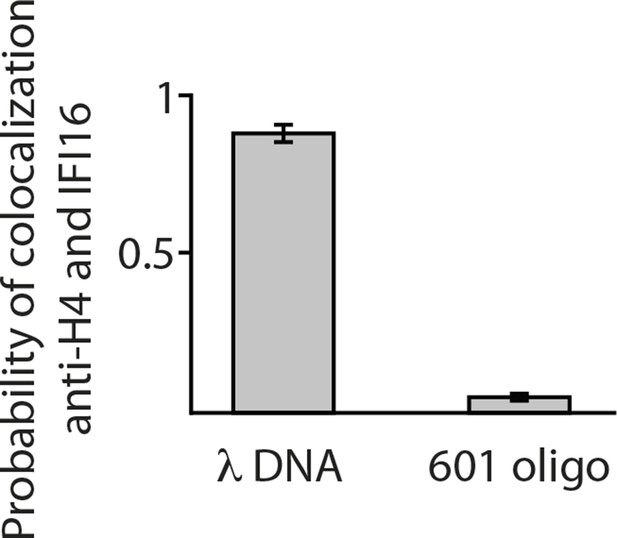
Single-molecule co-localization probability of anti-H4- Atto488 with IFI16-Cy5 on nucleosomal biotin-λdsDNA and on biotin-601-nucleosomes (N = 419 and 472, respectively).
601-nucleosomes do not provide sufficient exposed dsDNA for IFI16 binding, whereas on λdsDNA IFI16 molecules travel until encountering nucleosome obstacles.
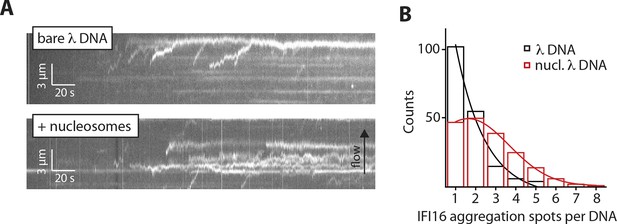
Nucleosomes interfere with the 1D-diffusion of IFI16.
(A) Example kymographs of IFI16-Cy5 on bare λdsDNA with long processivities to the free DNA tip and on nucleosomal λdsDNA with reduced run lengths. (B) Analysis of the number of IFI16 aggregation sites along DNA templates (including the free DNA tip) points to 1–3 nucleosomes per λdsDNA for the chosen reconstitution ratio DNA:histones, as it was used for run length measurements (Figure 4B,C).
Videos
Single Cy5-labeled IFI16 protein (1 nM) moving on double-biotinylated dsλ-DNA at 300 mM KCl (without flow).
The movie is played at 5x acceleration. This video is related to Figure 2.
Cy5-labeled IFI16 molecules (3 nM) moving and clustering on single-biotinylated dsλ-DNA at 160 mM KCl at constant flow from left to right.
This video is related to Figure 2.
Cy5-labeled IFI16 molecules (1 nM, red) moving on λ-DNA, with on average two nucleosomes per DNA, at constant flow from left to right.
Nucleosomes are tagged with fluorescent anti-H4 (Atto488, blue). Most nucleosomes co-localize with IFI16, indicating that they are efficiently blocking 1D-diffusion by IFI16. This video is related to Figure 4.
Additional files
-
Supplementary file 1
(A) dsDNA-mediated oligomerization rates of FRET-labeled IFI16. (B) Oligonucleotides used in this study.
- https://doi.org/10.7554/eLife.11721.019
-
Source code 1
Code for random walk simulation written in Python.
- https://doi.org/10.7554/eLife.11721.020





