Synchronization of endothelial Dll4-Notch dynamics switch blood vessels from branching to expansion
Figures
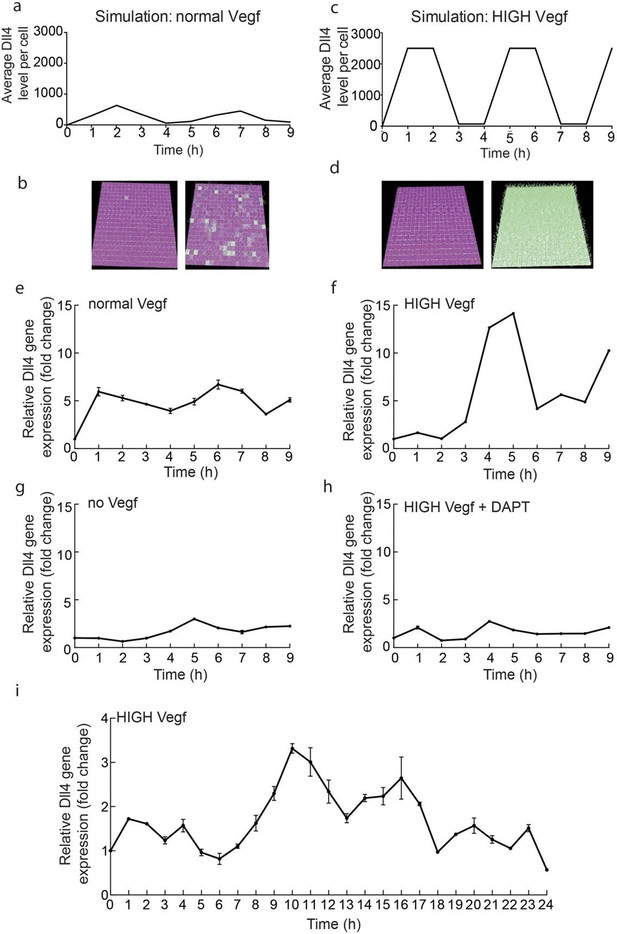
Endothelial Dll4 mRNA levels fluctuate dynamically in a Vegfa and Notch dependent manner.
(a–d) Simulation of Dll4 dynamics in a monolayer of 20×20 cells using the memAgent-Spring model under normal Vegfa and high Vegfa (20x normal level). (a) The total Dll4 level across all cells fluctuated overtime with a regular periodicity even though normal levels (b) generated a transient salt and pepper pattern with many non-adjacent cells high in Dll4 at high points in the fluctuation and few in the low phase. (c) High Vegf caused the cells to synchronise with the two phases of behavior showing either (d) most adjacent cells low, or most high in Dll4. Dll4 level low to high are represented purple-green. (e–i) Quantitative real time PCR analysis of dll4 mRNA levels in bEND5 cell monolayer. Cells were starved for four hours with serum-depleted medium and then stimulated with medium supplemented with either 50 ng/ml (e), 1 μg/ml (f, i), 0 Vegf (g), or 1 μg/ml Vegf and 50 µM DAPT (h). Cell lysates were collected every hour for the times indicated in the graphs. Values represent means ± S.D of technical replicates.
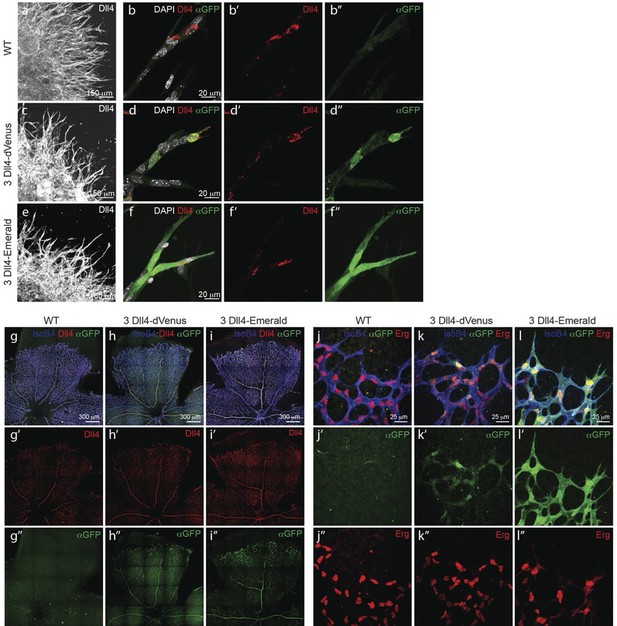
Dynamic and stable genomic reporters of Dll4 expression show differential distributions.
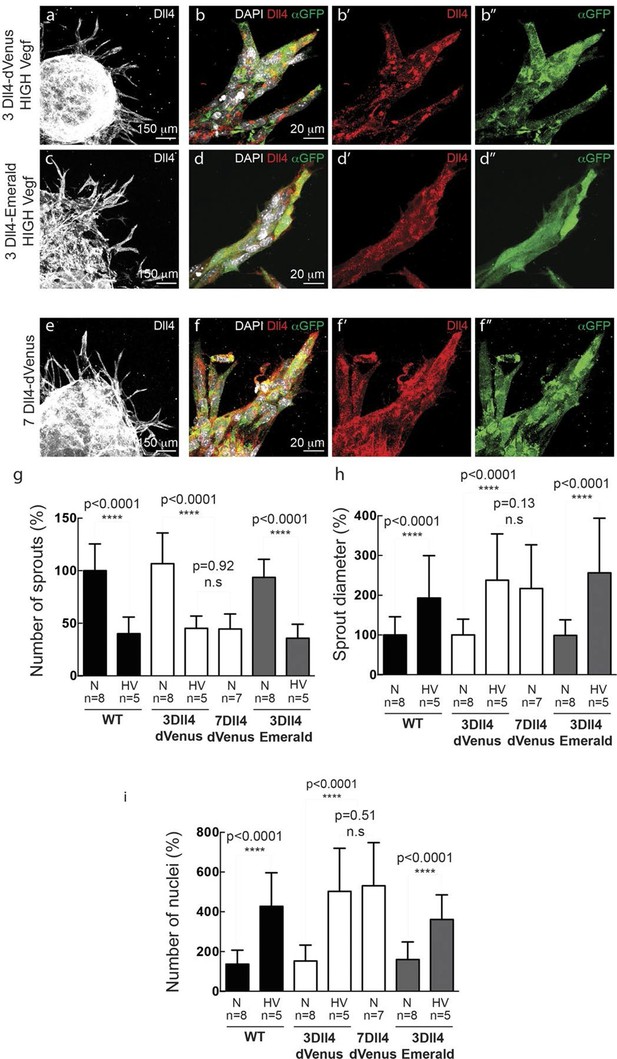
(a–f) Representative confocal overview (a, c, e) and high magnification (b, d, f) images of vascular sprouts in WT (a, b), 3Dll4-dVenus (c, d), 3Dll4-Emerald (e, f) embryoid bodies immunolabelled with antibodies specific to Dll4 (white in a, c, e and red in b, d, f) and GFP (green). Cell nuclei labeled with DAPI (white; only shown in b, d, f). (g–l) Isolectin B4 (blue), Dll4 (red in g–i), anti-GFP (green) and endothelial nuclear marker ERG (red in j–l) protein staining in representative overview tile-scan (g–i) and high magnification (j–l) images of whole-mounted WT (g, j), 3Dll4-dVenus (h, k) and 3Dll4-Emerald (i, l) P5 retinas.
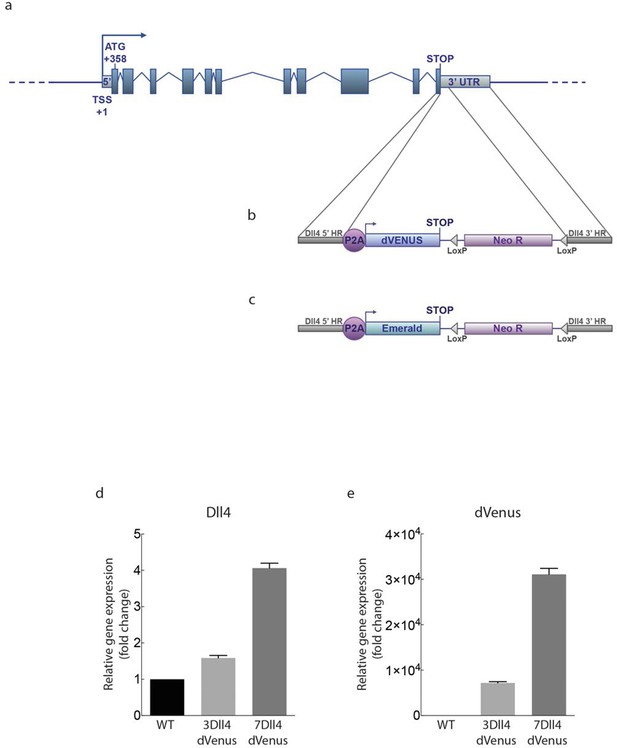
Generation of dynamic dVenus and stable Emerald Dll4 reporters.
https://doi.org/10.7554/eLife.12167.007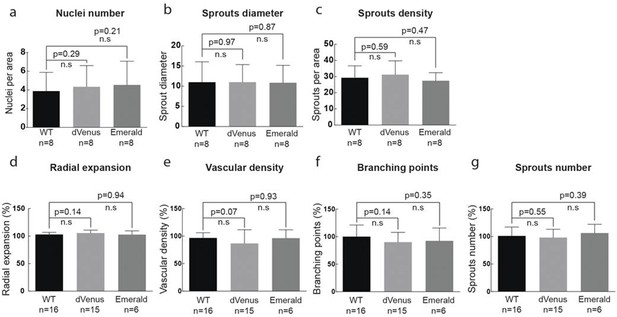
(g–i) Quantification of sprout density (g), sprout diameter (h), and nuclei number (i) in WT, 3Dll4-dVenus and 3Dll4-Emerald embryoid bodies (See Materials and methods for details).
Values represent means ± S.D. n= number of individual embryoid bodies analysed. p values calculated using two-tailed unpaired t-test. (p–s) Quantifications of radial expansion (p), vascular density (q), branching points (r) and sprouts number (s) in WT, 3Dll4-dVenus and 3Dll4-Emerald P5 retinas (See Materials and methods for details). Values represent means ± S.D. n=number of mice analysed. p values calculated using a two-tailed, unpaired t test.
In vitro high Vegfa or Dll4 levels lead to a switch from branching and elongation to sprout expansion.
(a-d) Dll4 (white in a, c; red in b, d) and reporter protein (anti-GFP, green) staining in representative confocal overview (a, c) and high magnification (b, d) images of vascular sprouts in 3Dll4-dVenus (a, b) and 3Dll4-Emerald (c, d) embryoid bodies cultured under high Vegf concentration. Cell nuclei are labeled with DAPI (white; only shown in b, d). (e, f) Representative confocal overview (e) and high magnification (f) images of 7Dll4-dVenus embryoid bodies immunolabelled with antibodies specific to Dll4 (white in e, red in f) and GFP (green). Cell nuclei labeled with DAPI (white; only shown in f). (g-i) Quantification of sprout density (g), sprout diameter (h), and nuclei number (i) in WT, 3Dll4-dVenus, 7Dll4-Venus and 3Dll4-Emerald embryoid bodies treated with either normal (N) or high Vegfa levels (HV). (See Materials and methods for details). Values represent means ± S.D. n= number of individual embryoid bodies analysed. P values calculated using two-tailed unpaired t-test.
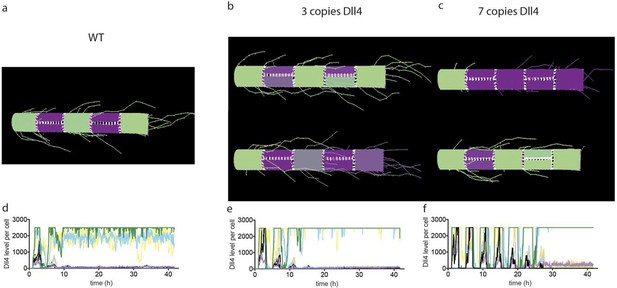
Simulated Dll4 dynamics with 2, 3 and 7 copies of Dll4.
https://doi.org/10.7554/eLife.12167.010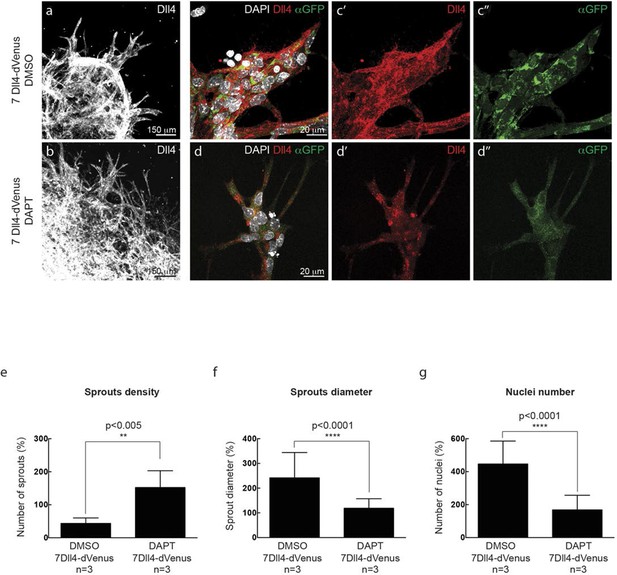
In vitro synchronization of cell signaling and behavior driven by Dll4 overexpression is Notch dependent.
https://doi.org/10.7554/eLife.12167.011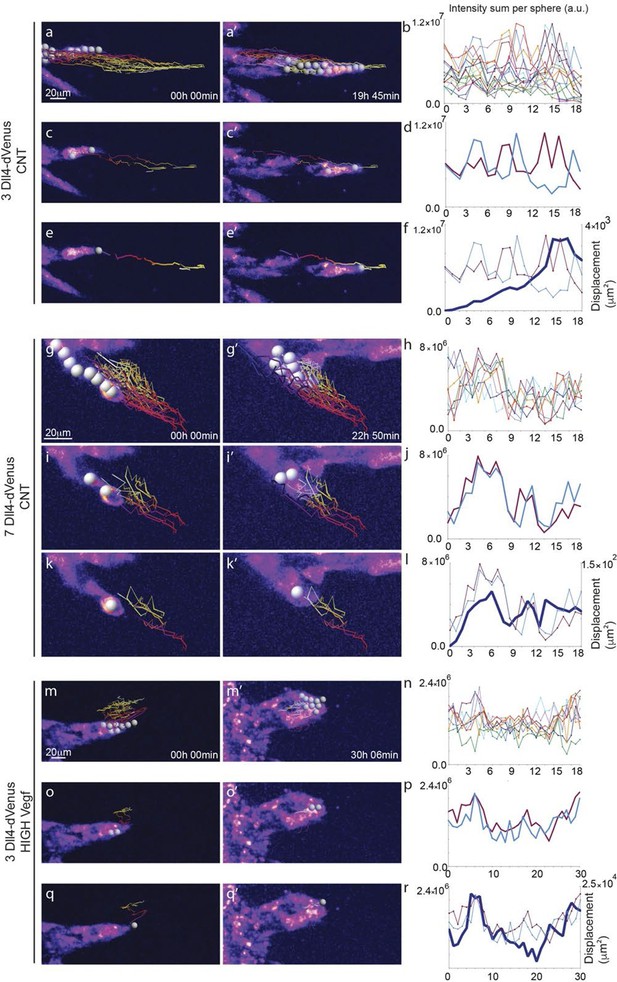
Time-lapse analysis of Dll4 reporter levels identify Dll4/Vegf dose- dependent shift from ‘individual cell’ to ‘synchronous cell’ signalling and behavior.
Start (a, c, e, g, i, k, m, o, q) and end (a’, c’, e’, g’, i', k’, m’, o’, q’) point of confocal time-lapse acquisitions of 3Dll4-dVenus (a-f) and 7Dll4-dVenus (g-l) embryoid bodies cultured in normal Vegf conditions (50 ng/ml), and 3Dll4-dVenus embryoid bodies treated with high Vegf concentration (500 ng/ml) (m-r). A heatmap intensity range color was used to represent dVenus expression levels. (See supplementary information; Videos 7, 8, 9). (a-a’, g-g’, m-m’) Arbitrary 10 μm spheres were employed to fill the sprout volume, using the Imaris ‘Spot’ cell tracking tool. (c-c’, i-i’, o-o’) Sprout volumes covered by two neighboring spheres are monitored overtime. (e-e’, k-k’, q-q’) A single sphere placed at the sprout tip is used to monitor the tip advance and retraction. (b, h, n) Quantification by time-lapse microscopy of dVenus signal levels relative to each sphere volume covering the sprout. dVenus intensity sum is represented by arbitrary units (a. u.). For technical reasons absolute intensity sum values are not comparable between experiments. (d, j, p) Quantification by time-lapse microscopy of dVenus signal intensity sum in two neighboring sphere volumes. (f, l) Quantification by time-lapse microscopy of the sprout tip displacement along the three axes x, y, z is shown together with dVenus intensity sum from (d, j). (r) To counteract the effect of a sprout drift along the x axis on x-y-z displacement (See supplementary information; Video 9) only the y-displacement is quantified by time-lapse microscopy and shown together with dVenus intensity sum from (p). (For details on the tracking technique see Materials and methods)
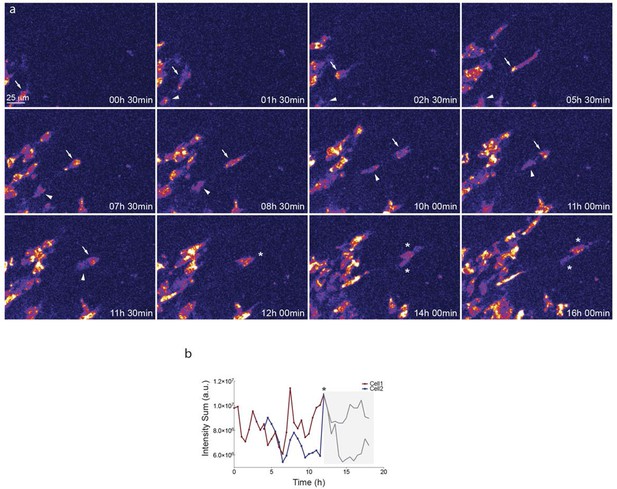
Dll4-dVenus reporter expression is dynamically and differentially regulated in vitro in individual endothelial cells.
https://doi.org/10.7554/eLife.12167.013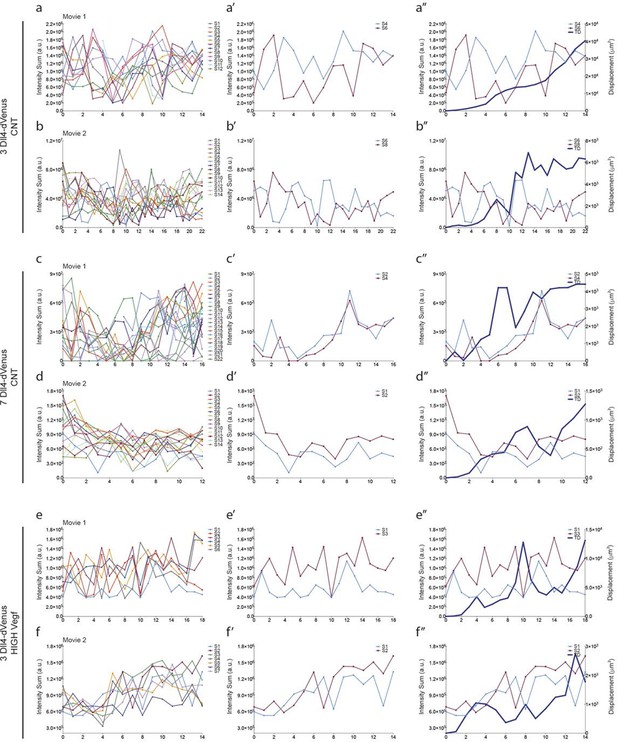
Additional examples of dynamic Dll4-reporter intensity profiles and sprout tip displacement illustrating a Dll4- and Vegfa-level dependent switch from individual cell to synchronous cell signaling and behaviour.
https://doi.org/10.7554/eLife.12167.014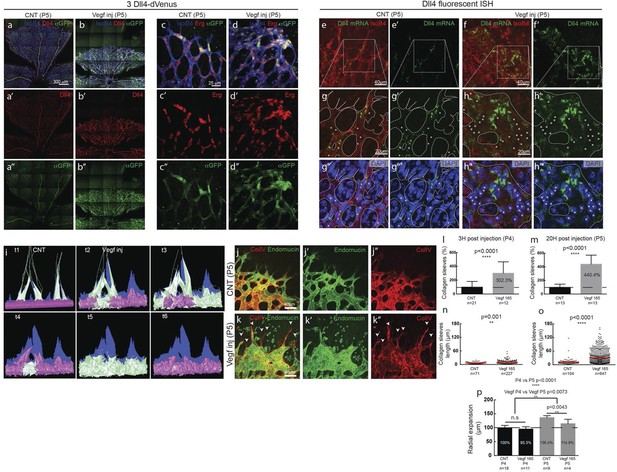
Endothelial cell Dll4 expression synchronization under high Vegfa in vivo.
(a–d) Dll4 (red in a, b), ERG (red in c, d), anti-GFP (green) and isolectin B4 (blue) staining in representative overview tile-scan (a, b) and high magnification (c, d) images of whole-mounted 3Dll4-dVenus P5 retinas not injected (CNT; a, c) and injected with mVegfa165 (Vegf inj; b, d). For anti-GFP (dVenus) and ERG signal a median filter of 3 and 5 pixel, respectively, was used. (e–h) Representative confocal images of whole-mounted WT P5 retina not injected (e) and mVegfa165 injected (f), labeled for dll4 mRNA detected by fluorescent ISH (green) and isolectin B4 (red). White dashed boxed areas in each panel (e, f) are magnified in (g,h) images. To facilitate endothelial cell nuclei visualization (DAPI; blue) together with dll4 mRNA only one stack is shown in panels g and h. White dashed lines delimit endothelial cells (Iso B4). Asterisks represent EC negative for dll4 mRNA expression. (i) Computational simulation of collagen IV deposition (blue) after high Vegfa stimulation (for details on simulation, see Materials and methods). At t1 a normal Vegfa condition with a linear gradient extending above the sprout is simulated. High uniform Vegfa levels are simulated from t2 through t6. Cells with high Dll4 expression and 'tip cell phenotype' are represented in green; cells with low Dll4 expression and 'stalk cells phenotype' are shown in purple. (j, k) Representative confocal images of the collagen IV distribution at the sprouting front of WT P5 retinas not injected (j) and injected with mVegfa165 (k). Collagen IV is shown in red; Endomucin in green. Arrows indicate empty collagen IV sleeves. (l–o) Quantification of the total number (l, m) and length of empty collagen IV sleeves (n, o) in WT retinas three (l, n) and twenty (m, o) hours post-injection. Mean ± S.D values are indicated (n, o). n= number of retinas analyzed (l, m); n= total number of collagen sleeves observed (n, o). P values calculated using a two-tailed, unpaired t test. (p) Quantification of the radial expansion in P4 (0H post injection) and P5 (20H post injection) WT retinas not injected and injected with mVegfa165. n= number of retinas analyzed. Values represent mean ± S.D. p values calculated using a two-tailed, unpaired t test.
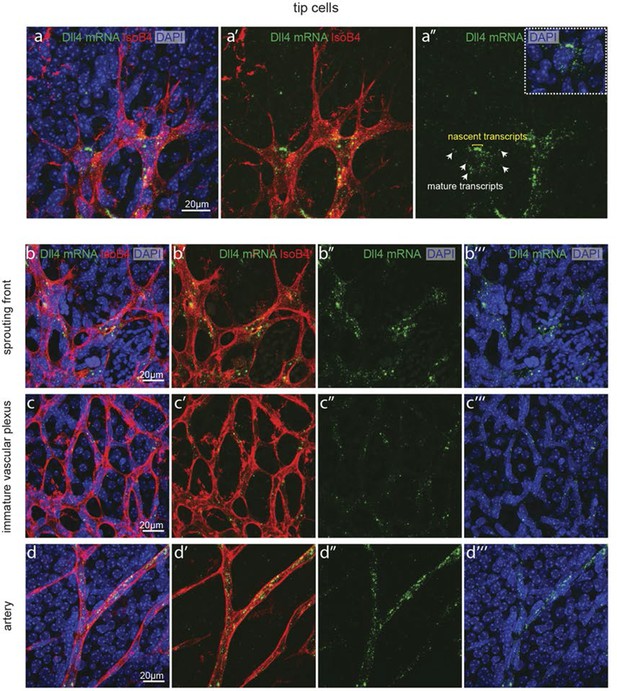
Detection of mature and nascent dll4 mRNA transcripts using whole mount fluorescent ISH.
https://doi.org/10.7554/eLife.12167.029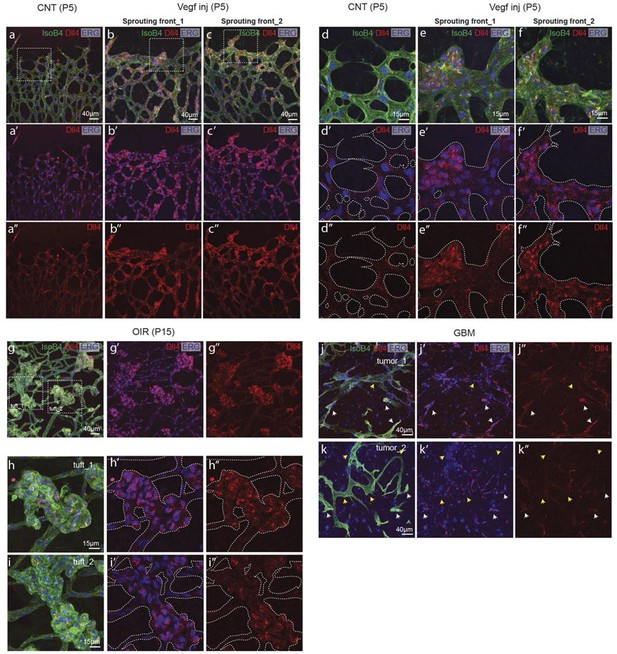
Dll4 protein expression is synchronized under pathological Vegfa concentration.
https://doi.org/10.7554/eLife.12167.030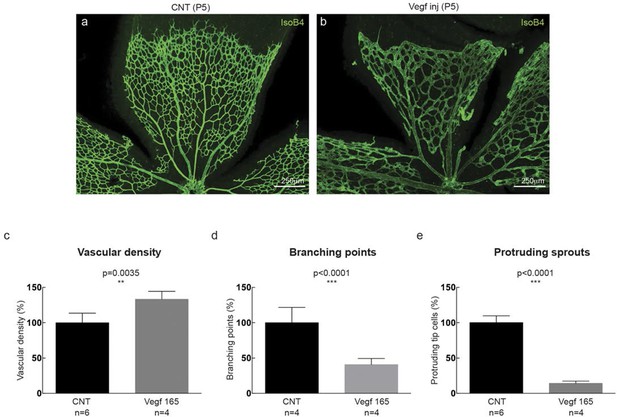
High Vegf concentrations during retinal angiogenesis result in aberrant vascular patterning.
https://doi.org/10.7554/eLife.12167.031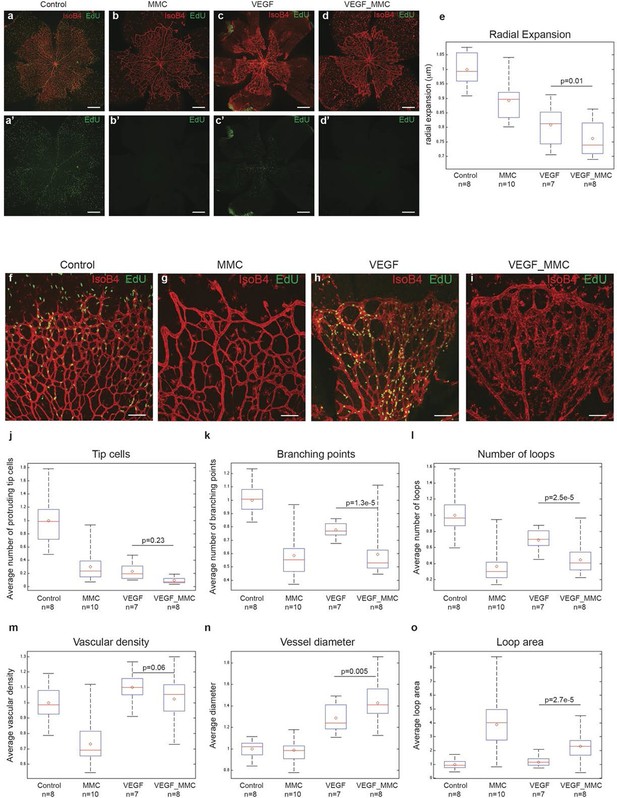
High levels of Vegfa induce vessel expansion even in absence of EC proliferation.
Representative image of a tile scan showing IsoB4 (red) and EdU staining (green) in non treated (control; a–a’), MMC (bb’), Vegfa 165 (cc’) and MMC_Vegfa 165 (d–d’) treated retinas. (e) Quantification of the radial expansion in control (a), MMC (b), Vegfa 165 (c) and MMC_Vegfa 165 (d) treated retinas. High magnification images showing IsoB4 (red) and EdU staining (green) in non-treated (control; f), MMC (g), Vegfa 165 (h) and MMC_Vegfa 165 (i) treated retinas. Quantification of the number of tip cells (j), branching points (k), number of loops (l), vascular density (m), vessel diameter (n) and loop area (o) in non treated (control), MMC, Vegfa 165 and MMC_Vegfa 165 treated retinas. Scale bar correspond to 400 μm (a–d’) and 100 μm (fi) respectively. Statistical comparison and number of animals analyzed are indicated in the graphs.
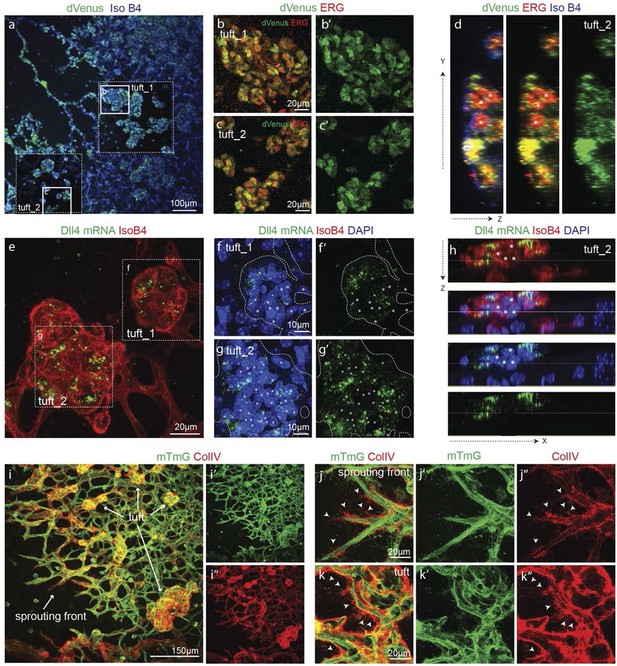
Epiretinal tufts in oxygen induced retinopathy show Dll4 expression sychronization.
(ac) Representative overview tile-scan of a whole-mounted P15 3Dll4-dVenus OIR retina (a). White dashed boxes highlight two different tuft regions; to facilitate tufts visualization only one optical section is shown in the boxed area. Full line boxes are magnified in b and c. For dVenus and ERG signals (b, c) a median filter of 3 and 5 pixel, respectively, was used. dVenus (anti-GFP) is shown in green, endothelial nuclei are labeled in red (ERG) and endothelial cells in blue (Iso B4). (d) Y–Z confocal section of (c). dVenus (anti-GFP) expression is shown in green, endothelial cell are labeled in blue (Iso B4) and endothelial nuclei in red (ERG). Asterisks represent EC negative for dVenus expression. (e–g) Representative confocal overview image (e) and high magnification images (f, g) of two different tufts in a WT P15 OIR retina. dll4 mRNA, detected by fluorescent ISH, is shown in green, endothelial cells (Iso B4) are shown in red and endothelial nuclei in blue (DAPI). White dashed boxes in panel (e) highlight the regions of the tufts analyzed in f and g. In f and g, white dashed lines delineate endothelial cells on each panel to help visualization. Asterisks represent EC negative for dll4 mRNA expression. (h) X-Z confocal section of (g). Stainings for dll4 mRNA (green), IsoB4 (red) and DAPI (blue) are shown. Asterisks represent EC negative for dll4 mRNA expression. (i) Overview image of collagen IV distribution in a mTmG WT P15 OIR retina, labeled with collagen IV (red). New sprouting front and tuft regions are highlighted in the merged image. (j, k) Representative images of collagen IV empty sleeves protruding ahead of the new sprouting front (j) and of tufts region (k) in the mTmG WT p15 OIR retina. Collagen IV is labeled in red. Arrows indicate empty collagen IV sleeves.
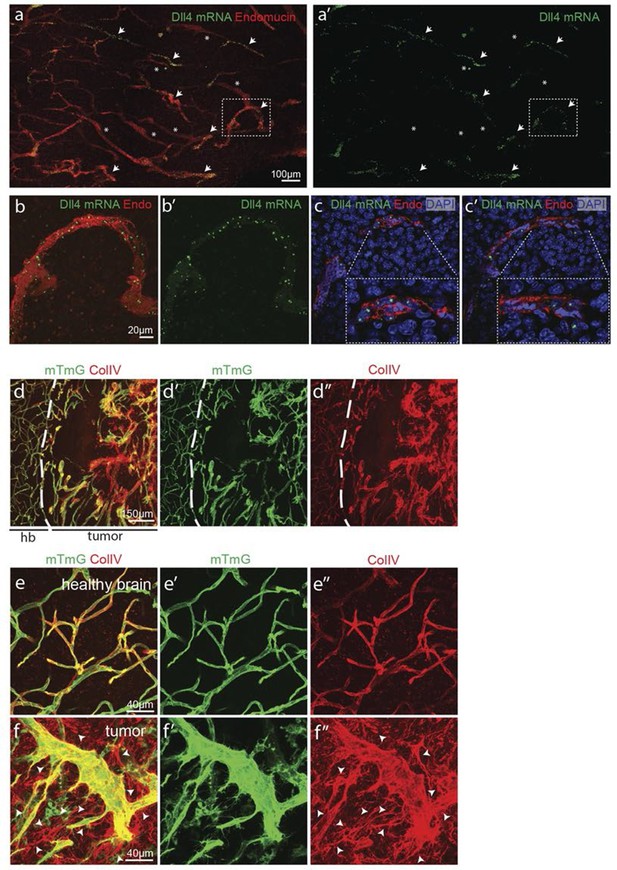
Synchronized Dll4 expression and sprouting behavior in tumour angiogenesis.
(a) Tile scan representative overview of dll4 mRNA expression detected using fluorescent ISH (green) in CT-2A glioblastoma tumor vessels labeled with endomucin (red). Asterisks and arrows indicate tumor vessels negative and positive for dll4 mRNA expression, respectively. White dashed boxes indicate the tumor region analyzed at high magnification on panel b and c. (b, c) High magnification of the positive tumor vessel for dll4 mRNA highlighted in panel (a). dll4 mRNA ISH is shown in green, cell nuclei stained with DAPI in blue and endothelial cells, detected using endomucin, in red. To facilitate endothelial cell nuclei visualization together with dll4 mRNA expression only one stack is shown in the panels where nuclear staining (DAPI; blue) is included (c). (d–f) Confocal overview image (d) and high magnification images (e, f) showing collagen IV (red) deposition around healthy (d, e) and tumor vasculature (d, f) in the glioblastoma tumor model (GBM) developed in a mTmG cre-reporter mouse brain. Endothelial cells express membrane eGFP by PdgfbiCreERT tamoxifen-induced recombination. Dashed line separates the healthy brain (hb) from the tumor region on d. Arrows indicate empty collagen IV sleeves.
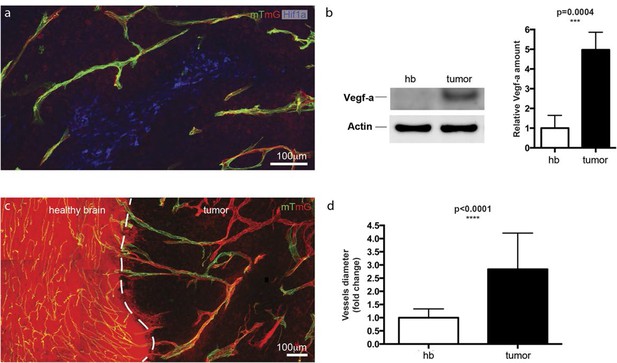
Hypoxic tumor cells induce high Vegfa concentrations leading to vessel expansion in mouse glioblastoma model.
https://doi.org/10.7554/eLife.12167.036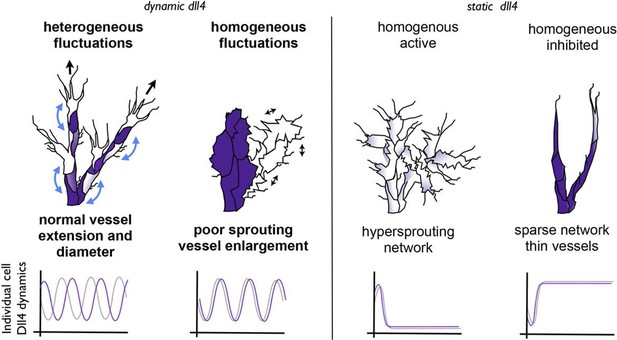
Schematic illustrating the important role of dynamic dll4 fluctuations in driving vessel expansion.
Individual cells Dll4 levels fluctuate asynchronously during normal vessel growth. Under high VEGF or high Dll4 these fluctuations become more synchronized leading to homogenous cellular dll4 levels that fluctuate between high and low levels together, driving vessel expansion rather than branching. In contrast, when Dll4 levels are homogeneous, but not fluctuating (termed 'static dll4' scenarios here for simplicity) the result is that cells either all remain activated and behave as tip cells (hypersprouting phenotype) or are all constantly inhibited, prohibiting vessel extension and driving a sparse branching phenotype. Thus the expansion phenotype is specifically driven by the dynamic fluctuations between homogeneous Dll4 high/low levels, rather than by the homogeneity of the Dll4 levels alone.





