The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation
Peer review process
This article was accepted for publication as part of eLife's original publishing model.
History
- Version of Record published
- Accepted Manuscript published
- Accepted
- Received
Decision letter
-
Ivan DikicReviewing Editor; Goethe University Medical School, Germany
In the interests of transparency, eLife includes the editorial decision letter and accompanying author responses. A lightly edited version of the letter sent to the authors after peer review is shown, indicating the most substantive concerns; minor comments are not usually included.
Thank you for submitting your work entitled "The IBD-associated autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation" for consideration by eLife. Your article has been reviewed by two peer reviewers, and the evaluation has been overseen by Ivan Dikic as the Senior Editor.
The following individuals involved in review of your submission have agreed to reveal their identity: Vojo Deretic and Janneke Samsom (peer reviewers).
The reviewers have discussed the reviews with one another and the Reviewing Editor has drafted this decision to help you prepare a revised submission.
Summary:
The manuscript describes the role for autophagy in orchestrating intestinal T-cell responses. The authors convincingly show that autophagy is required for maintenance of Foxp3+ Treg in vivo while, on the contrary, autophagy inhibits expansion of TH2 cells. In consequence, defects in autophagy in the CD4 T-cell compartment in vivo cause TH2 pathology by the lack of immune regulation due to reduced numbers of Foxp3 Treg and is aggravated by a direct boosting of TH2 responses. These findings are novel in the field of autophagy and intestinal homeostasis.
Overall, the manuscript is nicely structured, convincing and guides the reader very well.
Essential revisions:
1) From the results describing Figure 5 it does not become clear why reconstitution with WT CD4+ T cells does not partially rescue the phenotype in Atg16l1ΔCD4 mice. On the basis of Figure 5B one would expect that 70% of the WT CD4 are Treg. Why are these cells unable to suppress the expanding Atg16l1 deficient TH2 cells? Are Atg161 deficient TH2 cells not sensitive to suppression by Foxp3+ Treg? This should be investigated in more detail. For example, is this expansion independent of IL-2 and can the Foxp3 Treg therefore not suppress as easily?
2) As noted in the Abstract the role of autophagy in limiting mucosal TH2 expansion was unexpected. Although the authors suggest in the Discussion that Gata3 may promote peripheral T-cell proliferation and maintenance they do not explain how they think this works mechanistically. Are TH2 cells independent of autophagy for their metabolism? Is it possible to study the metabolic function of in vitro differentiated Atg16l1ΔCD4 TH2 and control TH2 cells in a similar experiment as has been performed in Figure 8? How does autophagy inhibit? This should be better explained as expansion of TH2 in Atg16l1ΔCD4 is a major finding in the study.
3) The effects of autophagy on fatty acid metabolism seems to be the best available explanation for the TH2 and Treg disregulation but the experiments are a bit correlative here. The intestinal-specific defect seems to argue against that, although this reviewer does agree with the elegant thinking and correlations by the authors.
[Editors' note: further revisions were requested prior to acceptance, as described below.]
Thank you for resubmitting your work entitled "The IBD-associated autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation" for further consideration at eLife. Your revised article has been favorably evaluated by Ivan Dikic (Senior editor and Reviewing editor). The manuscript has been improved but there are some remaining issues that need to be addressed before acceptance, as outlined below:
The title should provide a clear indication of the biological system under investigation and it should avoid specialist abbreviations and acronyms where possible.
It is recommended to delete IBD from the title if you agree and this does not affect the main message of your paper:
"The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation"
https://doi.org/10.7554/eLife.12444.024Author response
Essential revisions: 1) From the results describing Figure 5 it does not become clear why reconstitution with WT CD4+ T cells does not partially rescue the phenotype in Atg16l1 δ
CD4 mice. On the basis of Figure 5B one would expect that 70% of the WT CD4 are Treg. Why are these cells unable to suppress the expanding Atg16l1 deficient TH2 cells? Are Atg161 deficient TH2 cells not sensitive to suppression by Foxp3+ Treg? This should be investigated in more detail. For example, is this expansion independent of IL-2 and can the Foxp3 Treg therefore not suppress as easily?
The presentation of the data in the Figure 5 did not make clear to the reader what are the total levels of different T cell subsets in transferred Atg16l1ΔCD4 mice. We have now provided this information (Figure 5—figure supplement 1A-C, and in the subsection “Autophagy regulates intestinal TH2 responses in a cell-intrinsic manner“), which shows that the frequencies and numbers of Treg cells in reconstituted Atg16l1ΔCD4 mice are equivalent to those found in control Atg16l1fl/fl mice. Therefore, although 70% of the total Treg cells in the adoptively transferred Atg16l1ΔCD4 mice are of WT origin, the total Treg cell levels are comparable with control Atg16l1fl/fl mice.
To address whether autophagy-deficient TH2 cells were resistant to Treg cell suppression, we examined whether there was any correlation between the frequencies of Atg16l1-deficient-TH2 cells and Treg cells in the cLP of adoptively transferred Atg16l1ΔCD4 mice. This revealed a negative correlation, i.e. mice with higher levels of intestinal Treg cells had the lowest levels of Atg16l1-deficient-TH2 cells (data not shown), suggesting that autophagy-deficient TH2 cells may be restricted to some extent by WT Treg cells. However, the key conclusion from the data presented in Figure 5 (together with the supplemental data described above), is that despite efficient reconstitution of the Treg cell compartment, the adoptively transferred Atg16l1ΔCD4 mice still develop increased frequencies of autophagy-deficient TH2 cells. The simplest explanation for this result is that autophagy regulates TH2 cell accumulation in cell-intrinsic manner. This is supported by the results in Figure 4 showing that autophagy-deficient TH2 cells exhibit increased survival compared to WT TH2 cells. However, we cannot completely rule out the reviewer's suggestion that autophagy-deficient TH2 cells might be somewhat resistant to inhibition by Treg cells in the cLP in vivo and we therefore discuss this possibility in the Discussion.
To investigate whether autophagy-deficient TH2 cells might proliferate in an IL-2-independent manner, we assessed expression of IL-2Ra (CD25) on TH2 cells in the cLP of Atg16l1ΔCD4 mice and control mice. As shown below, we found comparable levels of CD25 on Atg16l1-deficient and WT TH2 cells in the cLP, as well as on polarized WT and Atg16l1-deficient TH2 cells in vitro. Furthermore, IL-2-driven STAT5 activation has been shown to contribute to TH2 lineage commitment (Zhu et al., Immunity, 2003; Liao et al., Nat. Immunol., 2008). Therefore, we think it unlikely that Atg16l1-deficient TH2 cells would develop or expand in an IL-2-independent manner.
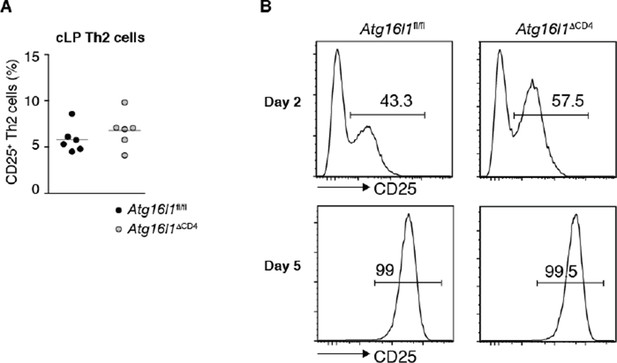
Autophagy deficiency does not influence expression of IL-2 receptor on TH2 cells (A) Expression of IL-2Ra (CD25) by TH2cells in the cLP of young Atg16l1ΔCD4 and Atg16l1fl/fl littermates (gated on CD4+ TCRβ+ Foxp3- Gata3+ T cells), data are representative of two independent experiments.
(B) Expression of CD25 was measured in naïve Atg16l1ΔCD4 or Atg16l1fl/fl CD4+ T cells cultured with anti-CD3 (5μg/ml) and anti-CD28 (1μg/ml) in TH2 polarizing conditions for 2 or 5 days, data are from one experiment.
2) As noted in the Abstract the role of autophagy in limiting mucosal TH2 expansion was unexpected. Although the authors suggest in the Discussion that Gata3 may promote peripheral T-cell proliferation and maintenance they do not explain how they think this works mechanistically. Are TH2 cells independent of autophagy for their metabolism? Is it possible to study the metabolic function of in vitro differentiated Atg16l1 δ CD4 TH2 and control TH2 cells in a similar experiment as has been performed in Figure 8? How does autophagy inhibit? This should be better explained as expansion of TH2 in Atg16l1 δ
CD4 is a major finding in the study.
The reviewers bring up a very interesting point and we have now performed additional experiments to compare the metabolic status of Atg16l1-deficient and WT Th2 cells. Our data in Figure 8 show increased expression of glycolytic genes in Atg16l1-deficient Treg cells and recent studies reported similar glycolytic changes in autophagy-deficient T cells (Puleston et al., 2014; Wei et al., 2016). Thus, a consistent theme emerges whereby autophagy perturbation results in increased glycolytic metabolism in T cells. We therefore investigated the hypothesis that this glycolytic shift in autophagy-deficient CD4+ T cells negatively impacts on their survival, with the exception of TH2 cells, as previous publications suggested that TH2 cells have higher levels of glycolysis (Michalek et al., 2011). As it was not possible to isolate viable intestinal TH2 cells to perform comparative analysis of metabolic gene expression (as we were able to do with the Treg cells that expressed a YFP marker), we analysed in vitro polarised TH2 cells. We confirmed that TH2 cells were highly glycolytic and found that this was independent of autophagy (Figure 8—figure supplement 3, and the last two paragraphs of the Results section). Mechanistically, we suggest that this highly glycolytic state is orchestrated by Gata3, which may enhance glycolysis through direct effects on c-Myc (Wang, 2013) and we found high levels of c-Myc expression in TH2 cells (Figure 8—figure supplement 3C). As discussed in the Discussion section, we believe that TH2 cells are uniquely able to cope with prolonged high levels of glycolysis, and this may explain why they have a clear survival advantage over other T cells in the context of autophagy-deficiency.
3) The effects of autophagy on fatty acid metabolism seems to be the best available explanation for the TH2 and Treg disregulation but the experiments are a bit correlative here. The intestinal-specific defect seems to argue against that, although this reviewer does agree with the elegant thinking and correlations by the authors.
We would like to argue that intestinal-specific defect fits with our observations and hypothesis. We saw the most striking defect in Treg populations in the colonic LP in both Atg16l1ΔCD4 and Atg16l1ΔFoxp3 mice (Figure 2B, C) and this is the same site where we observed that control Treg cells had increased expression of genes implicated in lipid metabolism (Figure 8D). To further support this observation we used an in vivo assay to measure lipid uptake by intestinal and systemic Treg cells. We also measured their expression of the fatty acid translocase CD36. As shown in Figure 8—figure supplement 2 (described in the subsection “Differential survival of autophagy-deficient Treg cells and TH2 cells is associated with an altered metabolic profile”), we found increased uptake of the fluorescent C16 lipid analogue and increased CD36 expression by colonic LP Treg cells when compared with Treg cells from the mLN and spleen. This further suggests that colonic Treg cells are more adapted for lipid metabolism. Interestingly, short chain fatty acids (SCFA) produced by commensal bacteria were shown to facilitate pTreg induction and to increase proliferation of intestinal Treg cells. Although this was attributed to regulation of Foxp3 expression 8,9, our results tempt us to speculate that some of the beneficial effects could be due to SCFA acting as a metabolic fuel for intestinal Treg cells.
[Editors' note: further revisions were requested prior to acceptance, as described below.]
The manuscript has been improved but there are some remaining issues that need to be addressed before acceptance, as outlined below: The title should provide a clear indication of the biological system under investigation and it should avoid specialist abbreviations and acronyms where possible. It is recommended to delete IBD from the title if you agree and this does not affect the main message of your paper: "The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation"
I can confirm that we have now made this change to the title, which now reads,
“The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation”.
https://doi.org/10.7554/eLife.12444.025




