Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states
Figures
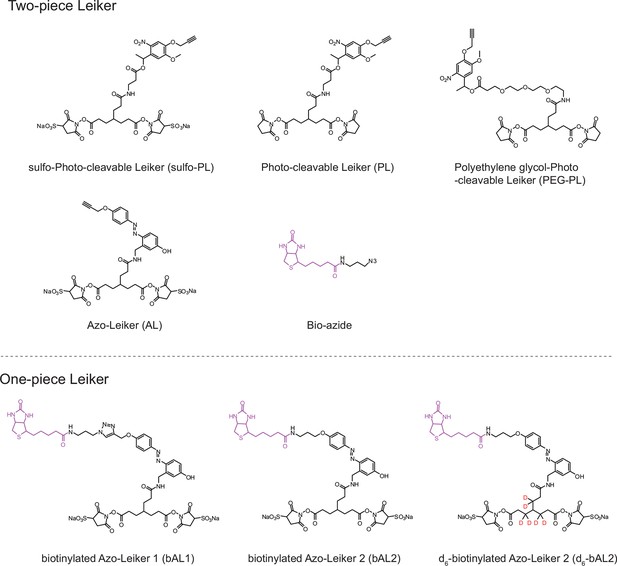
Chemical structures of different designs of Leiker.
The top panel shows four designs of two-piece Leiker with a photo-cleavage site (sulfo-PL, PL, and PEG-PL) or an azobenzene-based cleavage site (AL). Biotin is attached via click chemistry by reacting with bio-aizde. The bottom panel shows two unlabeled (bAL1, bAL2) and deuterium-labeled ([d6]-bAL2) one-piece Leiker molecules. The biotin moiety is colored magenta.
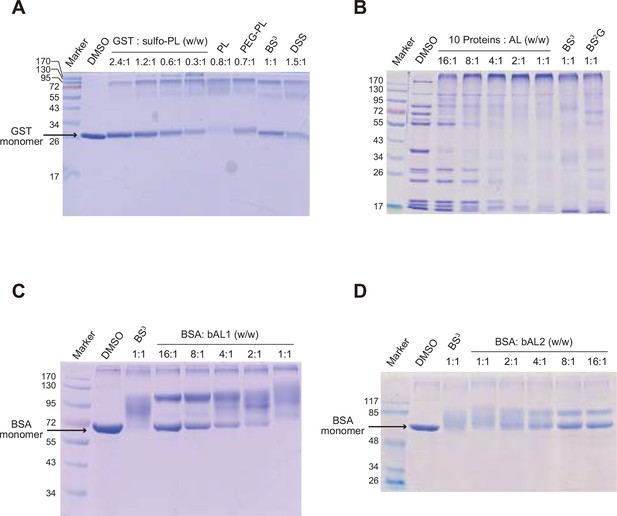
Optimization of protein-to-cross-linker ratio (w/w) for (A) sulfo-PL, (B) AL, (C) bAL1, and (D) bAL2.
https://doi.org/10.7554/eLife.12509.004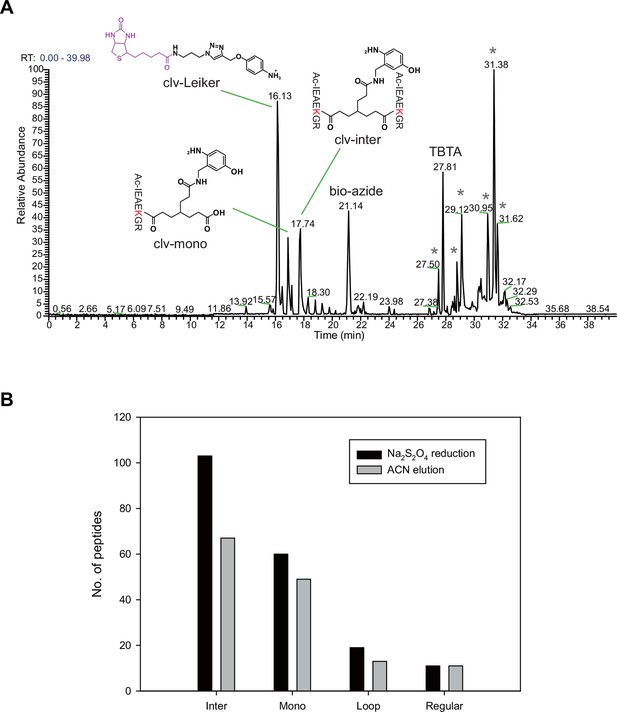
Evaluation of azobenzene-based chemical cleavage.
https://doi.org/10.7554/eLife.12509.005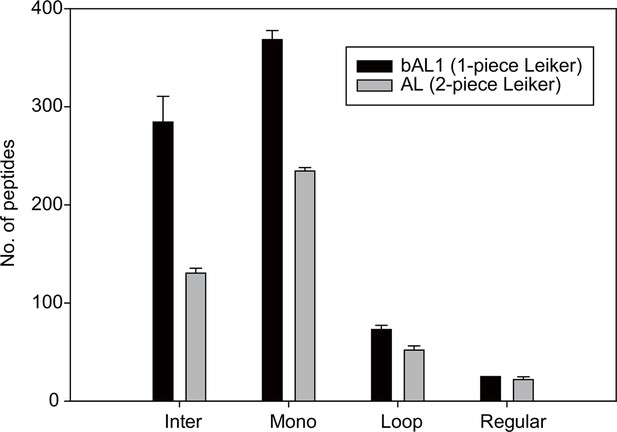
The one-piece Leiker (bAL1) outperformed the two-piece Leiker (AL) in the CXMS analysis of a mixture of ten standard proteins.
https://doi.org/10.7554/eLife.12509.006
Evaluation of the two piece Azo-Leiker (AL).
https://doi.org/10.7554/eLife.12509.007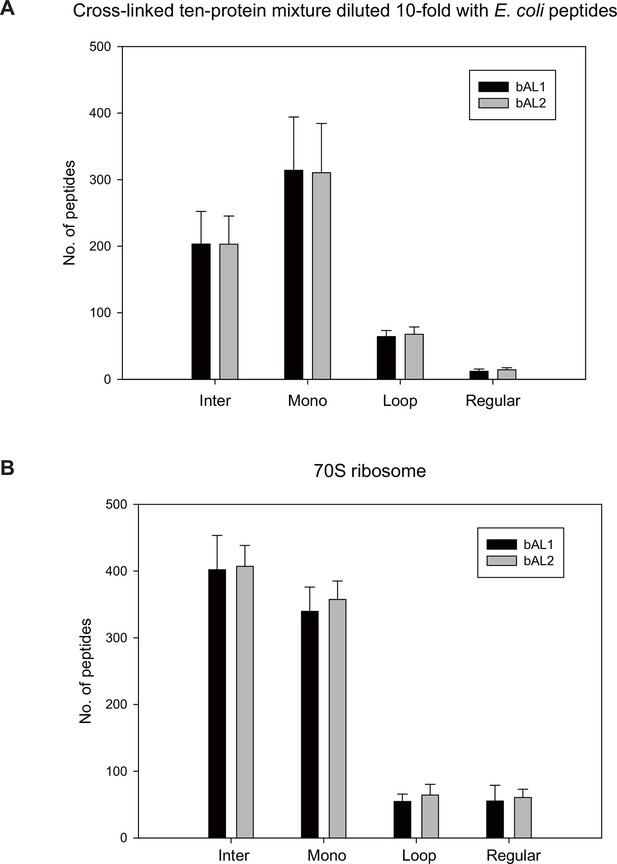
bAL1 and bAL2 performed similarly.
https://doi.org/10.7554/eLife.12509.008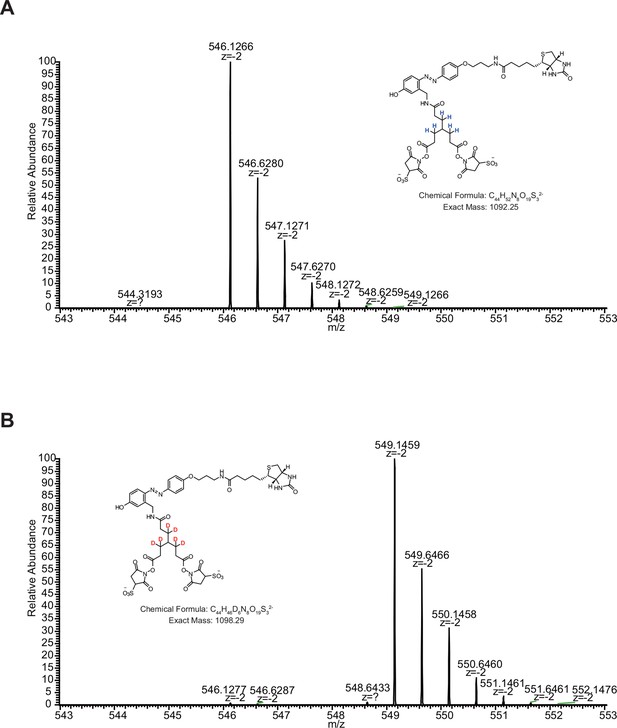
MS1 spectra of (A) [d0]-bAL2 and (B) [d6]-bAL2.
https://doi.org/10.7554/eLife.12509.009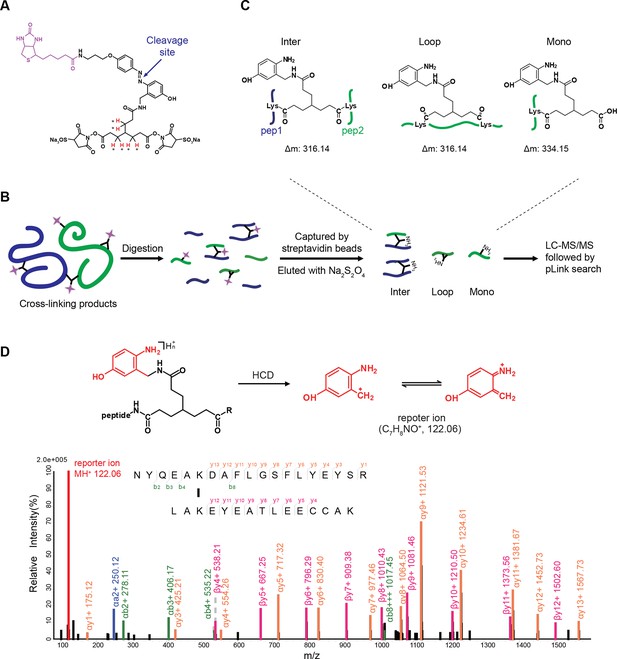
Scheme of the Leiker-based CXMS workflow.
(A) Leiker contains a biotin moiety (magenta), a cleavage site (arrows), and six hydrogen atoms that are accessible to isotope labeling (asterisks). (B) The workflow for purification of Leiker-linked peptides. (C) Three types of Leiker-linked peptides. (D) Leiker-linked peptides generate a reporter ion of 122.06 m/z in HCD, as shown in the spectrum of an inter-linked peptide NYQEAKDAFLGSFLYEYSR-LAKEYEATLEECCAK (+4 charged, MH+ 4433.0553), in which C denotes carbamidomethylated cysteine.
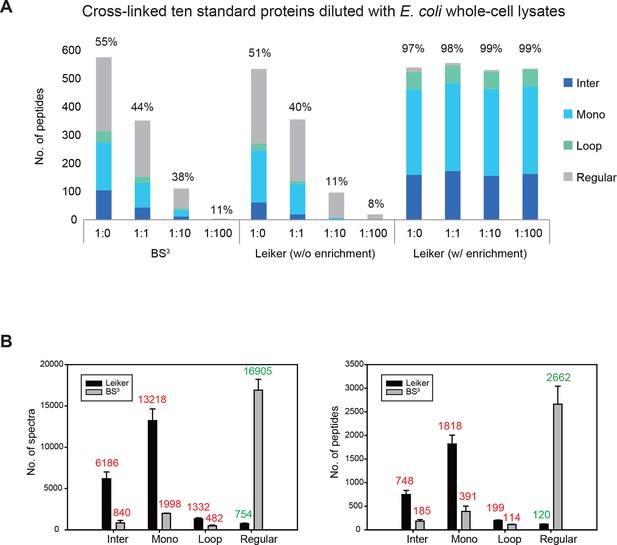
Evaluating the performance of Leiker.
(A) Leiker allowed near 100% enrichment of target peptides from a cross-linked ten-protein mixture diluted with increasing amounts of non-cross-linked E. coli lysates. Dark blue, inter-links; light blue, mono-links; green, loop-links; grey, regular peptides not modified by Leiker. (B) Number of cross-link identifications from E. coli lysates treated with Leiker or BS3. Shown in the left and right panels are the identified spectra and peptides, respectively.
-
Figure 3—source data 1
Ten standard proteins used to evaluate Leiker, mixed at equal amounts by mass.
- https://doi.org/10.7554/eLife.12509.012
-
Figure 3—source data 2
Summary of identified spectra from the ten-protein mixture.
- https://doi.org/10.7554/eLife.12509.013

Distance distributions of cross-linked lysine pairs in the undiluted ten-protein mixture.
https://doi.org/10.7554/eLife.12509.014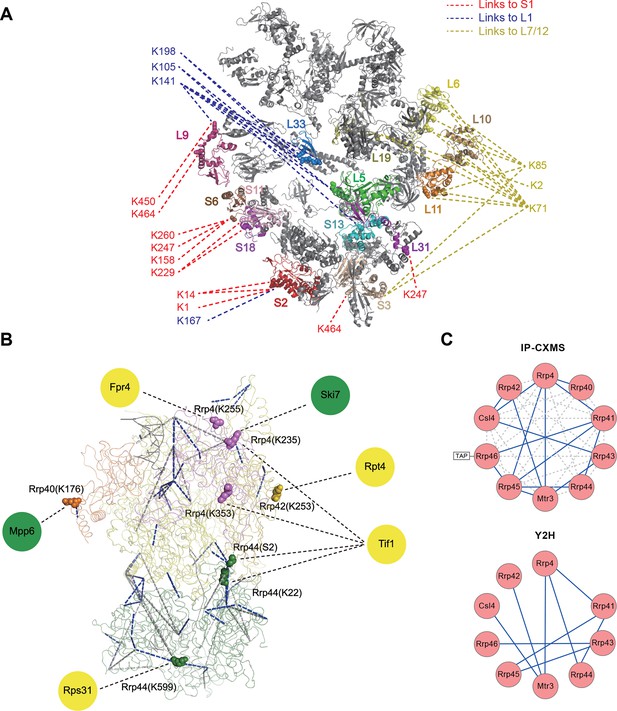
Leiker-based CXMS analyses of large protein assemblies.
(A) Analysis of a purified E. coli 70S ribosome revealed the locations of highly dynamic periphery ribosomal proteins S1, L1, and L7/12 that were refractory to crystallography and cryo-EM analysis. Cross-links to S1, L1, and L7/12 are colored red, blue, and yellow, respectively, and the cross-linked residues on these three proteins are numbered according to the Uniprot sequences. (B) Analysis of a crude immunoprecipitate of the yeast exosome complex. Dashed blue and grey lines denote 50 compatible and 22 incompatible cross-links, respectively, according to the structure of the RNA-bound 11-subunit exosome complex (PDB code: 4IFD). Rrp44, green; Rrp40, orange; Rrp4, violet; Rrp42, gold; other exosome subunits, yellow; RNA, black. Known and candidate exosome regulators revealed by Leiker-cross-links are shown along the periphery and highlighted in green and yellow circles, respectively. (C) Connectivity maps of the ten-subunit exosome core complex based on the inter-molecular cross-links identified in the current IP-CXMS experiments or on previous yeast two-hybrid (Y2H) studies (Stark et al., 2006; Uetz et al., 2000; Oliveira et al., 2002; Luz et al., 2007; Yu et al., 2008). Blue solid lines: experimentally identified putative direct protein-protein interactions; grey dashed lines: theoretical cross-links according to the crystal structure; Cα-Cα distance cutoff ≤30 Å.
-
Figure 4—source data 1
CXMS analysis of E. coli 70S ribosomes.
- https://doi.org/10.7554/eLife.12509.016
-
Figure 4—source data 2
Number of cross-linked lysine pairs classified by ribosomal proteins.
- https://doi.org/10.7554/eLife.12509.017
-
Figure 4—source data 3
Identified cross-linked lysine pairs involving L1.
- https://doi.org/10.7554/eLife.12509.018
-
Figure 4—source data 4
CXMS analysis of the Saccharomyces cerevisiae exosome complex.
- https://doi.org/10.7554/eLife.12509.019
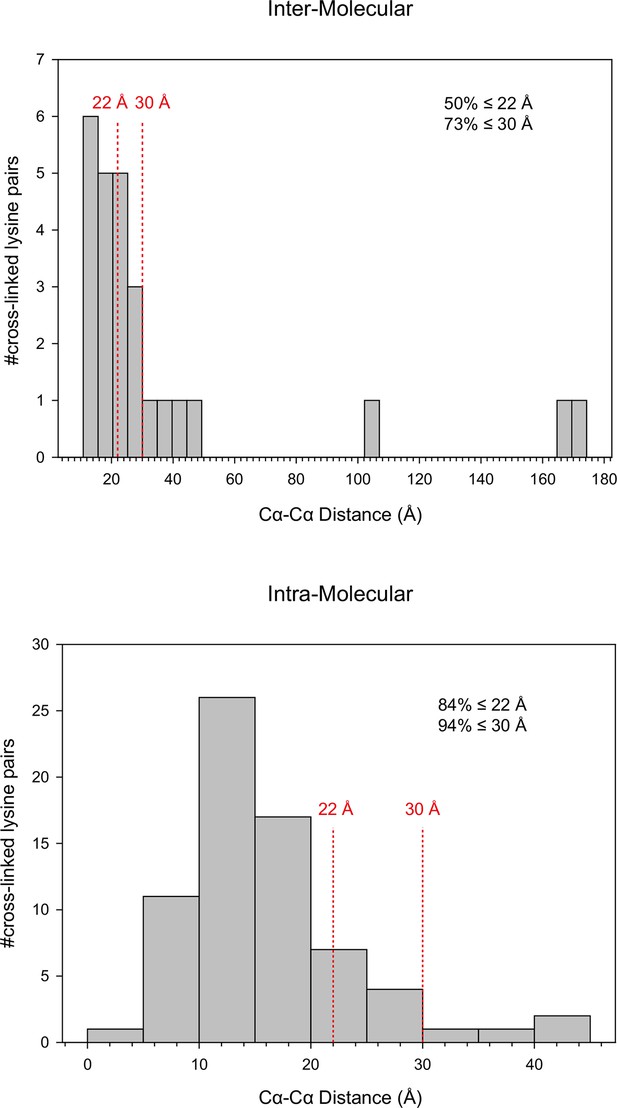
Distance distribution of the inter-molecular and intra-molecular cross-links identified in 70S ribosomes.
https://doi.org/10.7554/eLife.12509.020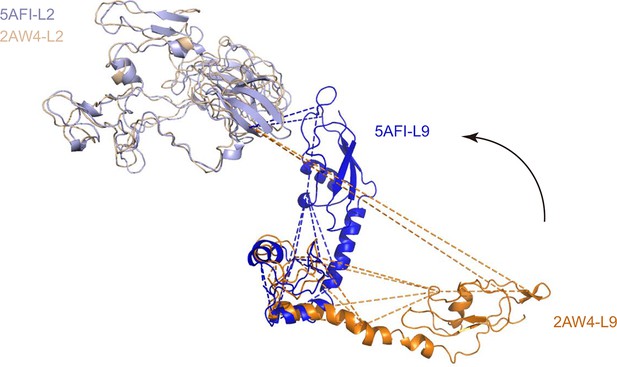
Alignment of L9 and L2 from the crystal structure (L9, orange; L2, wheat; PDB code: 2AW4) and their counterparts from the cryo-EM reconstruction (L9, blue; L2, lightblue; PDB code: 5AFI).
https://doi.org/10.7554/eLife.12509.021
Negative staining of non-cross-linked E. coli 70S ribosome.
https://doi.org/10.7554/eLife.12509.022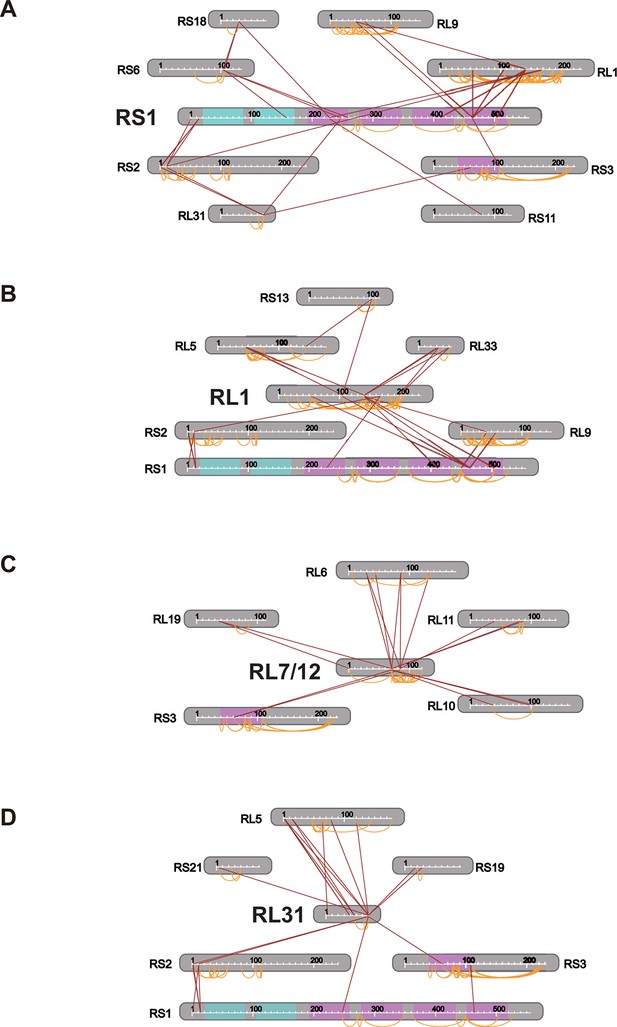
Connectivity maps of cross-links involving (A) S1, (B) L1, (C) L7/12, and (D) L31.
https://doi.org/10.7554/eLife.12509.023
Silver-stained SDS-PAGE gel of the crude immunoprecipitate of TAP-tagged Rrp46.
https://doi.org/10.7554/eLife.12509.024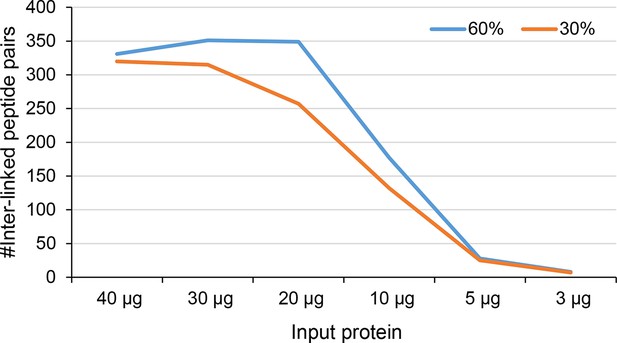
Number of identified inter-linked peptide pairs from decreasing amount of Leiker-cross-linked exosome immunoprecipitate (FDR < 0.05, E-value < 0.01).
After enrichment, 30% (orange) or 60% (blue) of each sample was analyzed by LC-MS/MS.
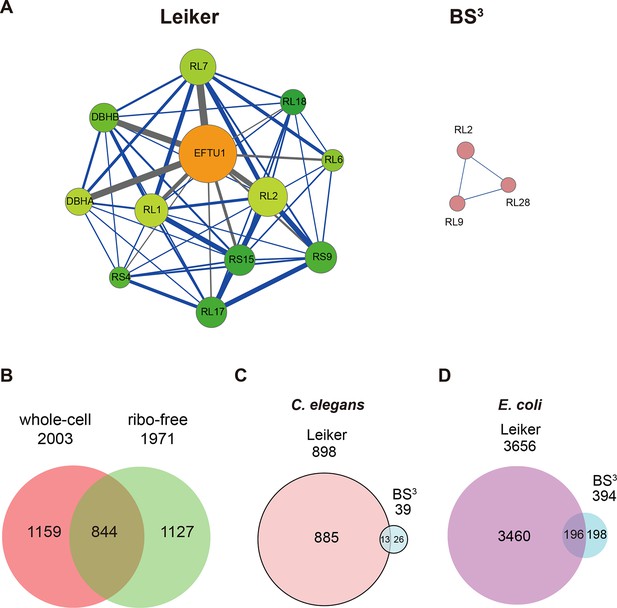
CXMS analyses of E. coli and C. elegans lysates.
(A) The best protein-protein interaction cluster extracted from the Leiker-identified or BS3-identified (Yang et al., 2012) inter-links from E. coli whole-cell lysates. Node size represents the degree of connectivity of the indicated protein in the network. Line width represents the spectral counts of every inter-molecular cross-link. The line color is set to blue when the two peptides of an inter-link are both attributed to unique proteins, to grey if either could be assigned to multiple proteins. All the lines connected to EF-Tu1 are grey because EF-Tu1 differs from EF-Tu2 by only one amino acid. (B) Comparison of the identified inter-links in E. coli whole-cell lysates and ribo-free lysates (5% FDR, E-value < 0.01, spectral count ≥ 3). (C and D) Comparison of the number of Leiker-identified inter-links and that of BS3-identified inter-links (Yang et al., 2012) from C. elegans (C) and E. coli (D) whole-cell lysates (5% FDR, E-value < 0.01, spectral count ≥ 1).
-
Figure 5—source data 1
CXMS analysis of E. coli whole-cell lysates.
- https://doi.org/10.7554/eLife.12509.027
-
Figure 5—source data 2
CXMS analysis of E. coli ribo-free lysates.
- https://doi.org/10.7554/eLife.12509.028
-
Figure 5—source data 3
CXMS analysis of C. elegans whole-cell lysates.
- https://doi.org/10.7554/eLife.12509.029
-
Figure 5—source data 4
CXMS analysis of C. elegans mitochondrial proteins.
- https://doi.org/10.7554/eLife.12509.030

Fractionation of digested, Leiker-treated E. coli lysates.
https://doi.org/10.7554/eLife.12509.031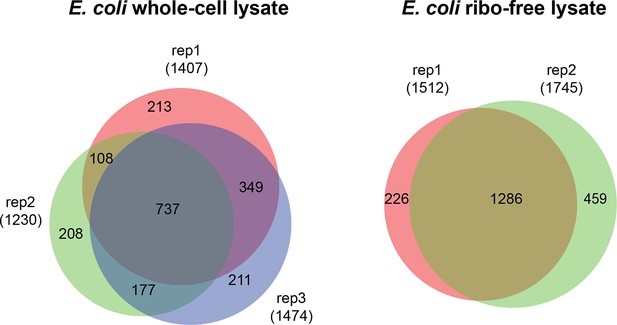
Overlap of cross-linked lysine pairs between biological replicates of E. coli lysates (FDR < 0.05, E-value < 0.01, and spectral count ≥ 3).
https://doi.org/10.7554/eLife.12509.032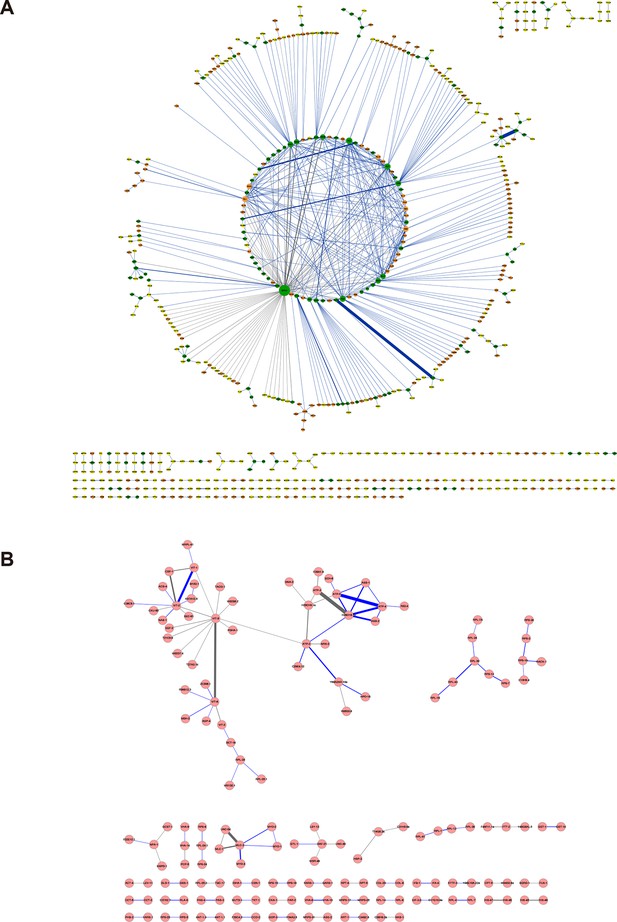
Protein-protein interaction networks constructed from the cross-links identified in (A) E. coli and (B) C. elegans.
The labeling scheme is the same as described in Figure 5A except for the node color. For E. coli, node color is set to orange if the protein was only identified in the whole-cell lysates, to yellow only identified in the ribo-free lysates, or to green if identified in both. There are 626 proteins in the E. coli network and 155 proteins in the C. elegans network.
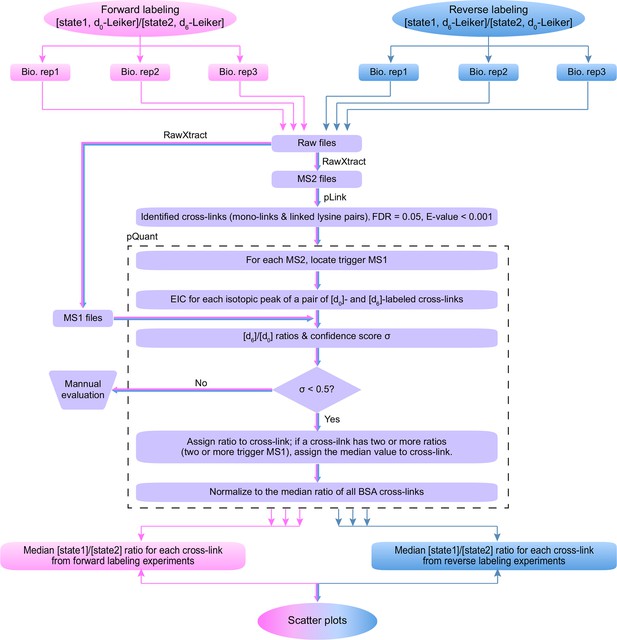
Workflow for quantification of cross-linked peptides using pQuant.
For each identified cross-link spectrum, an extracted ion chromatogram (EIC) is constructed for each isotopic peak of the [d0]- and [d6]-labeled precursor. The [d6]/[d0] ratios can be calculated based on the monoisotopic peak, the most intense peak, or the least interfered peak of each isotopic cluster as specified by users. The accuracy of the ratio calculation was evaluated with the confidence score σ (range: 0–1, from the most to the least reliable). If a cross-link have ratios with σ < 0.5, the median of these ratios is assigned to this cross-link. The cross-link ratios of the proteins of interest are normalized to the median ratio of all BSA cross-links. For each cross-link, the median [state1]/[state2] ratio of three independent forward labeling experiments is plotted against the median ratio of three independent reverse labeling experiments. Cross-links that are only present in state1 or state2 due to a dramatic conformational change cannot be quantified as described above because the ratios would be zero or infinite and their σ values would be 1. Therefore, if a cross-link does not have a valid ratio after automatic quantification, the EICs were manually inspected to determine if it was an all-or-none change.
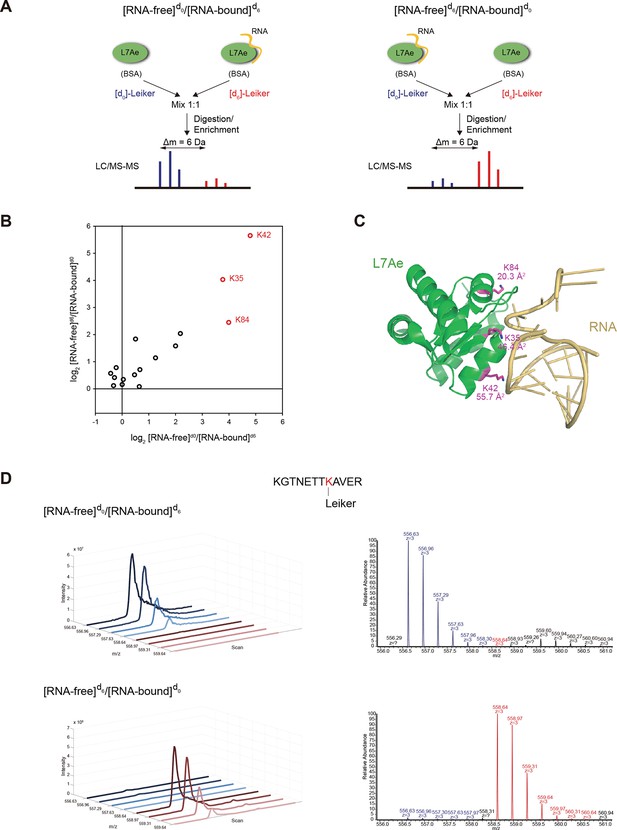
Quantitative CXMS analysis of the L7Ae-RNA complex.
(A) Reciprocal labeling of RNA-free (F) and RNA-bound (B) L7Ae with [d0]/[d6]-Leiker. (B) Abundance ratios of mono-links (F/B) in the forward (F[d0]/B[d6]) and the reverse labeling experiment (F[d6]/B[d0]). Each circle represents a mono-linked lysine residue and is colored red if it has a ratio greater than five in both labeling schemes. (C) The three lysine residues affected by RNA binding are highlighted in the structure model (PDB code: 2HVY). The number below each such lysine residue indicates the buried surface area (Å2) upon RNA binding. (D) Extracted ion chromatograms (left) and representative MS1 spectra (right) of a K42 mono-link.
-
Figure 7—source data 1
Quantitative CXMS analysis of L7Ae with or without the H/ACA RNA.
- https://doi.org/10.7554/eLife.12509.036

Extracted ion chromatograms (left) and representative MS1 spectra (right) of a mono-linked peptide corresponding to (A) K35 and (B) K84.
https://doi.org/10.7554/eLife.12509.037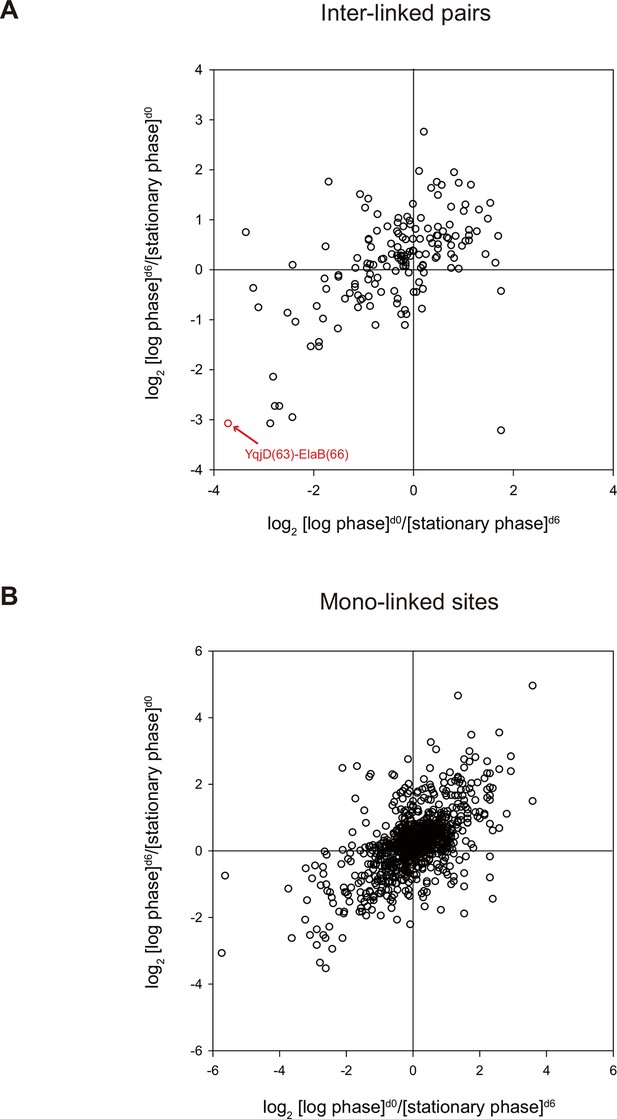
Quantitative CXMS analysis of E. coli lysates.
Abundance ratios of (A) inter-linked lysine pairs and (B) mono-linked sites in the forward ([log phase]d0/[stationary phase]d6) and the reverse labeling experiment ([log phase]d6/[stationary phase]d0).
-
Figure 8—source data 1
Quantitative CXMS analysis of E. coli lysates.
- https://doi.org/10.7554/eLife.12509.039
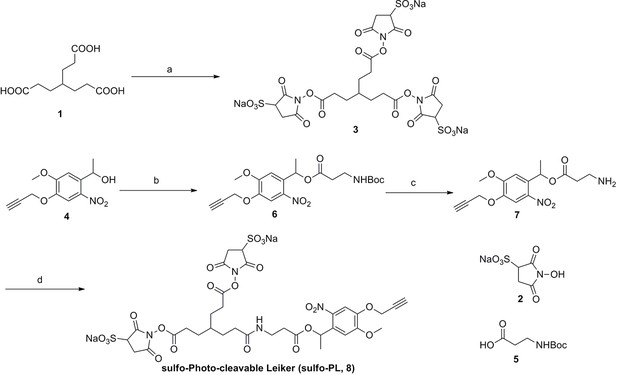
Synthesis of sulfo-Photo-cleavable Leiker (sulfo-PL, 8)
https://doi.org/10.7554/eLife.12509.040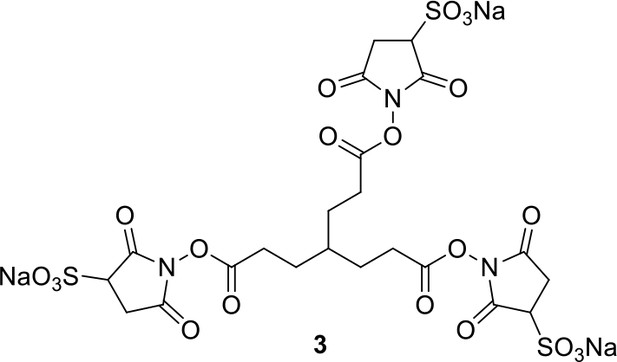
Compound 3.
https://doi.org/10.7554/eLife.12509.041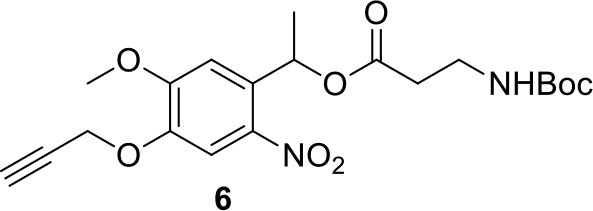
Compound 6.
https://doi.org/10.7554/eLife.12509.042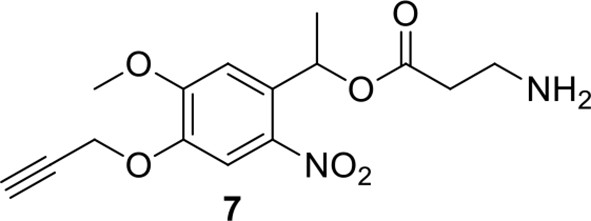
Compound 7.
https://doi.org/10.7554/eLife.12509.043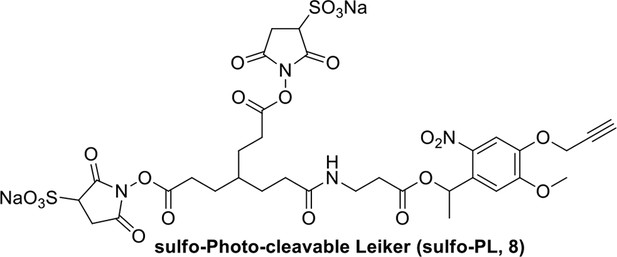
Compound 8.
https://doi.org/10.7554/eLife.12509.044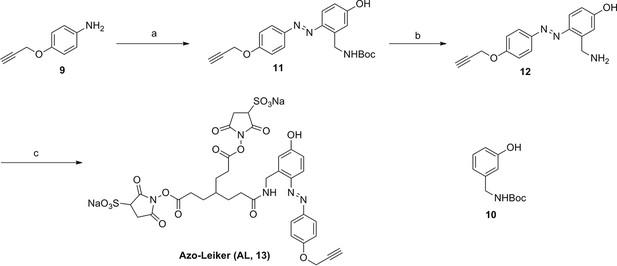
Synthesis of Azo-Leiker (AL, 13).
https://doi.org/10.7554/eLife.12509.045
Compound 11.
https://doi.org/10.7554/eLife.12509.046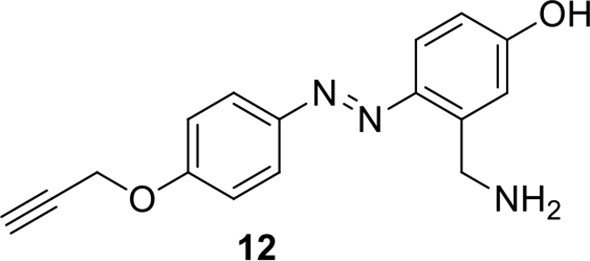
Compound 12.
https://doi.org/10.7554/eLife.12509.047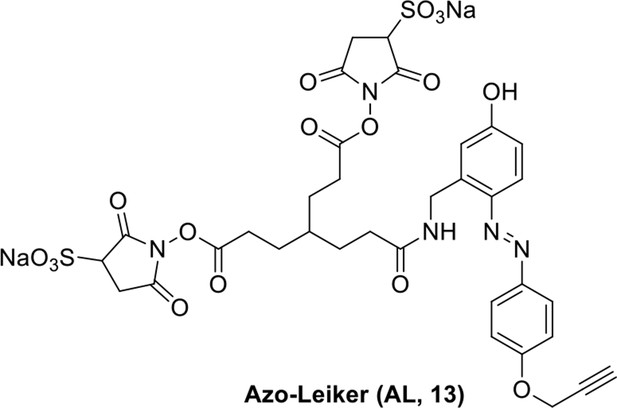
Compound 13.
https://doi.org/10.7554/eLife.12509.048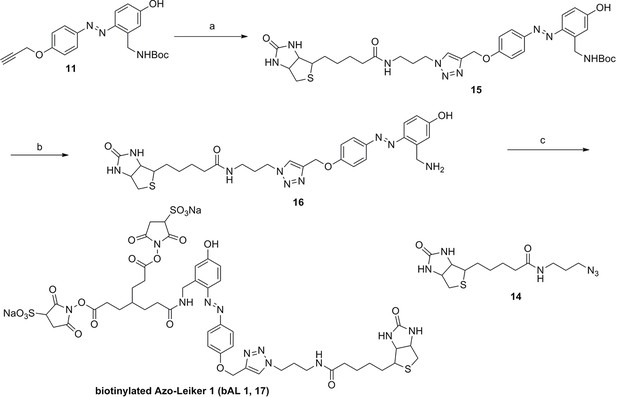
Synthesis of biotinylated Azo-Leiker 1 (bAL 1, 17).
https://doi.org/10.7554/eLife.12509.049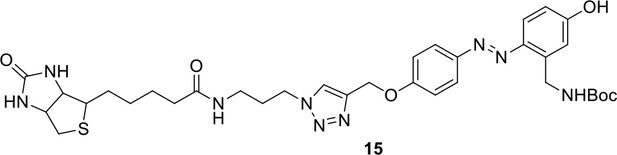
Compound 15.
https://doi.org/10.7554/eLife.12509.050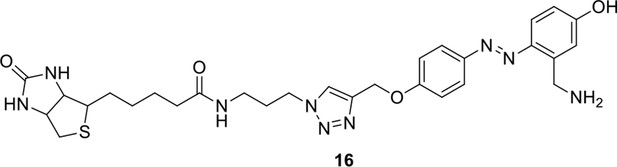
Compound 16.
https://doi.org/10.7554/eLife.12509.051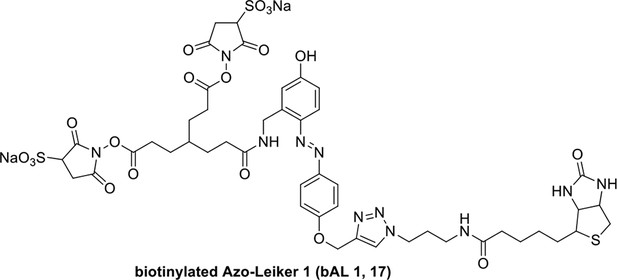
Compound 17.
https://doi.org/10.7554/eLife.12509.052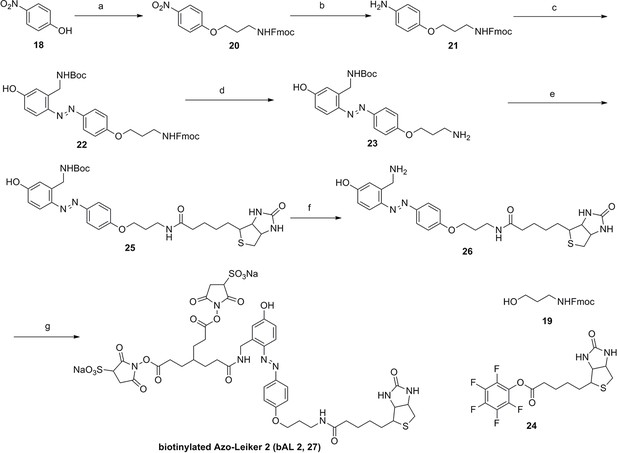
Synthesis of biotinylated Azo-Leiker 2 (bAL 2, 27).
https://doi.org/10.7554/eLife.12509.053
Compound 20.
https://doi.org/10.7554/eLife.12509.054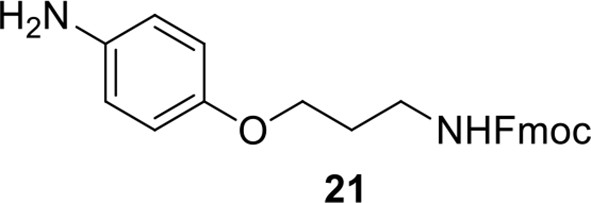
Compound 21.
https://doi.org/10.7554/eLife.12509.055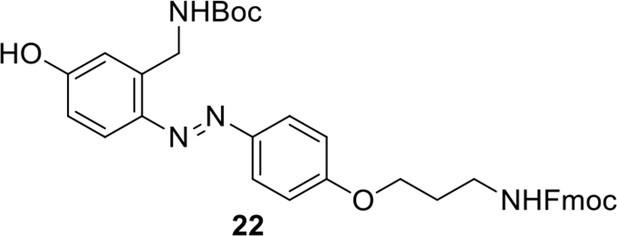
Compound 22.
https://doi.org/10.7554/eLife.12509.056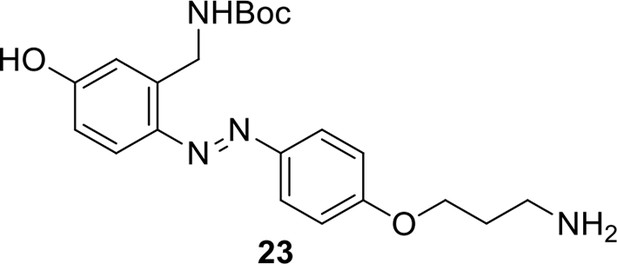
Compound 23.
https://doi.org/10.7554/eLife.12509.057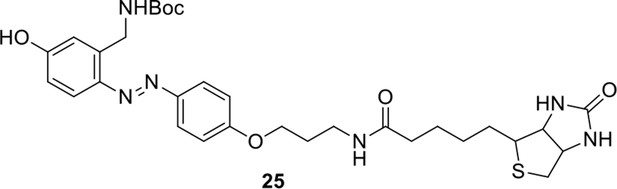
Compound 25.
https://doi.org/10.7554/eLife.12509.058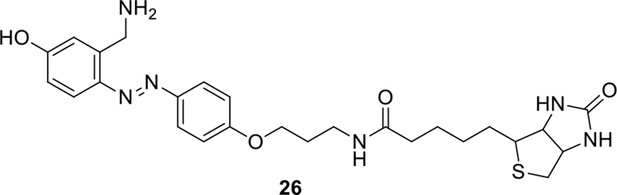
Compound 26.
https://doi.org/10.7554/eLife.12509.059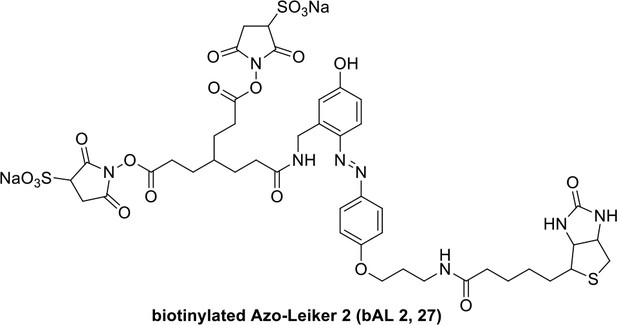
Compound 27.
https://doi.org/10.7554/eLife.12509.060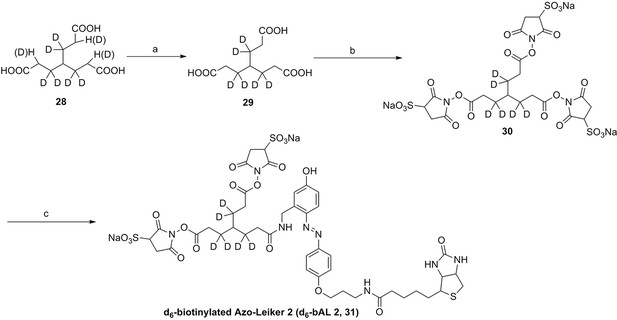
Synthesis of d6-biotinylated Azo-Leiker 2 (bAL 2, 31).
https://doi.org/10.7554/eLife.12509.061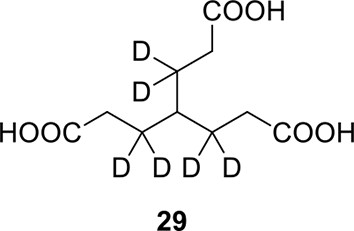
Compound 29.
https://doi.org/10.7554/eLife.12509.062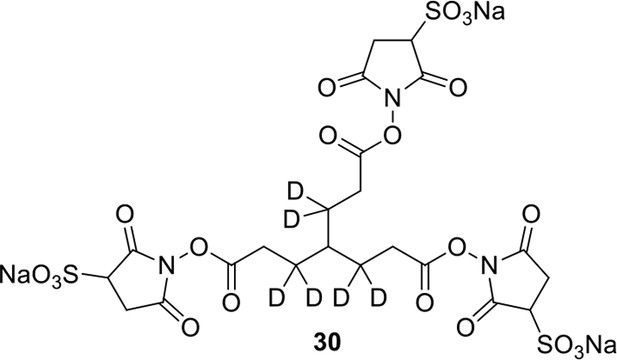
Compound 30.
https://doi.org/10.7554/eLife.12509.063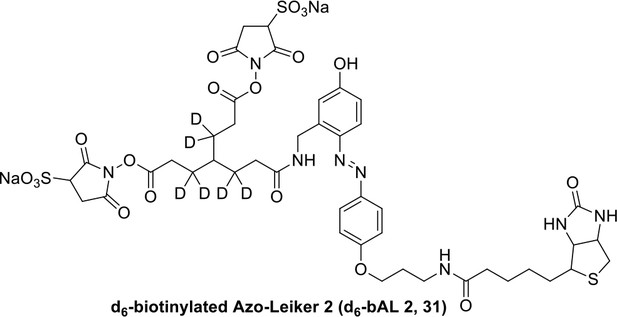
Compound 31.
https://doi.org/10.7554/eLife.12509.064
1H NMR spectra of compound 6.
https://doi.org/10.7554/eLife.12509.065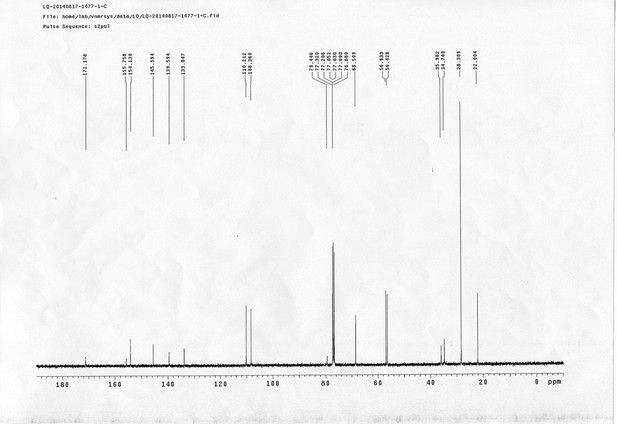
13C NMR spectra of compound 6.
https://doi.org/10.7554/eLife.12509.066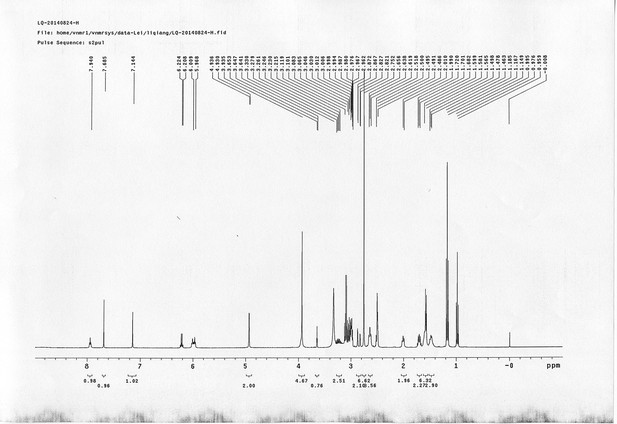
1H NMR spectra of sulfo-PL.
https://doi.org/10.7554/eLife.12509.067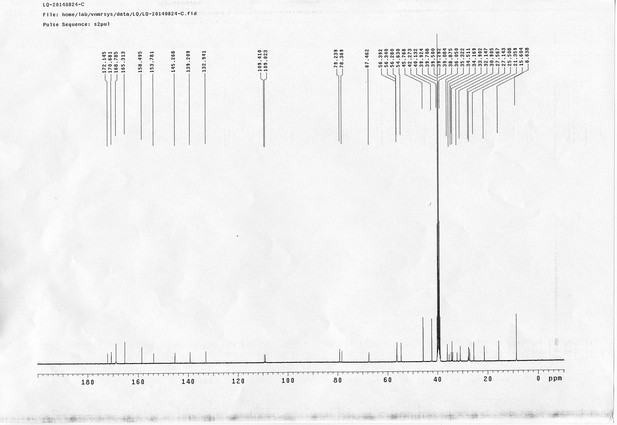
13C NMR spectra of sulfo-PL.
https://doi.org/10.7554/eLife.12509.068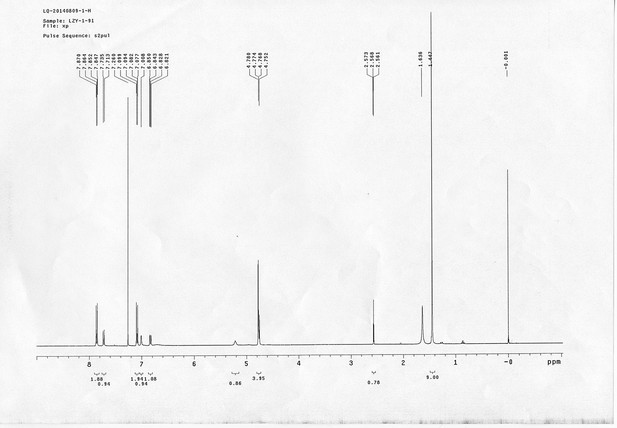
1H NMR spectra of compound 11.
https://doi.org/10.7554/eLife.12509.069
13C NMR spectra of compound 11.
https://doi.org/10.7554/eLife.12509.070
1H NMR spectra of AL.
https://doi.org/10.7554/eLife.12509.071
13C NMR spectra of AL.
https://doi.org/10.7554/eLife.12509.072
1H NMR spectra of compound 15.
https://doi.org/10.7554/eLife.12509.073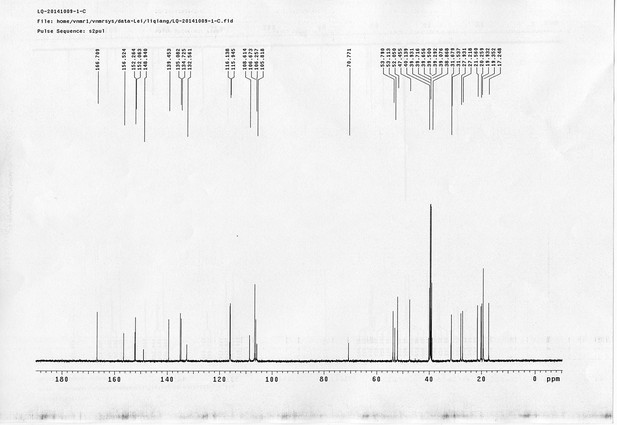
13C NMR spectra of compound 15.
https://doi.org/10.7554/eLife.12509.074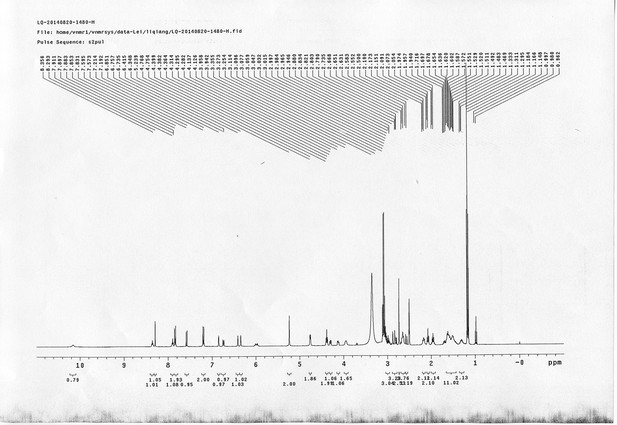
1H NMR spectra of bAL 1.
https://doi.org/10.7554/eLife.12509.075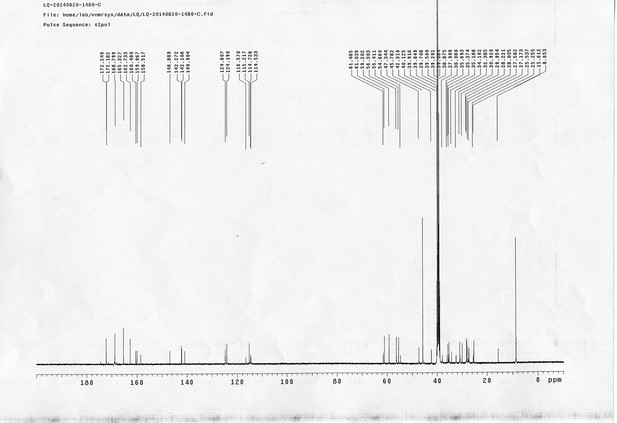
13C NMR spectra of bAL 1.
https://doi.org/10.7554/eLife.12509.076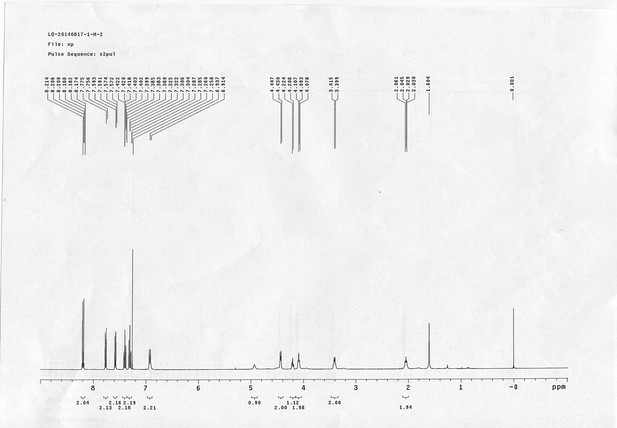
1H NMR spectra of compound 20.
https://doi.org/10.7554/eLife.12509.077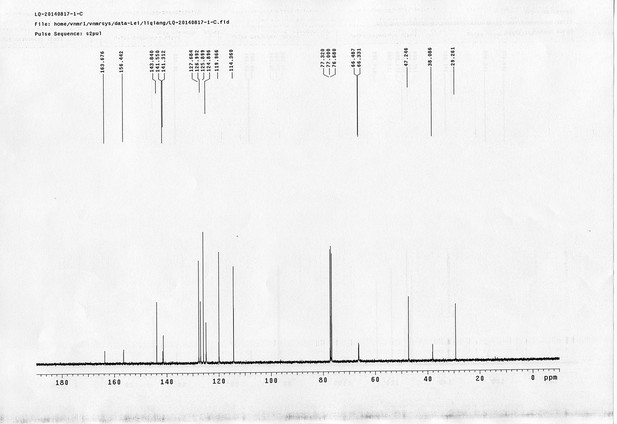
13C NMR spectra of compound 20.
https://doi.org/10.7554/eLife.12509.078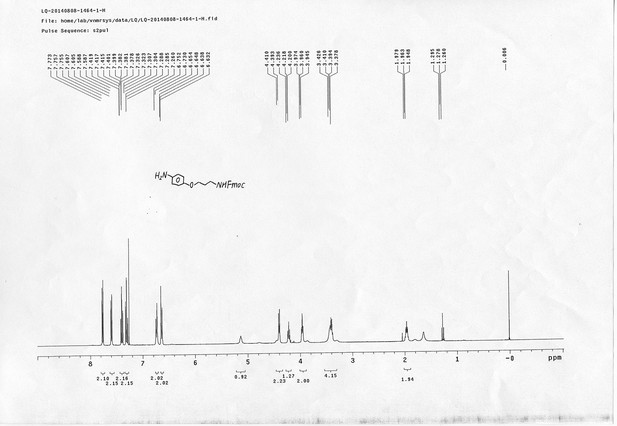
1H NMR spectra of compound 21.
https://doi.org/10.7554/eLife.12509.079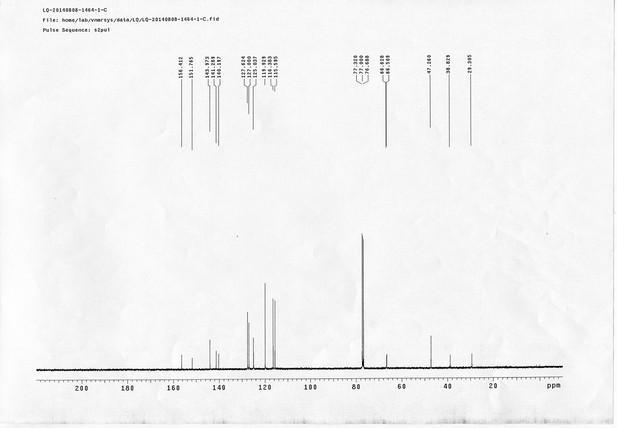
13C NMR spectra of compound 21.
https://doi.org/10.7554/eLife.12509.080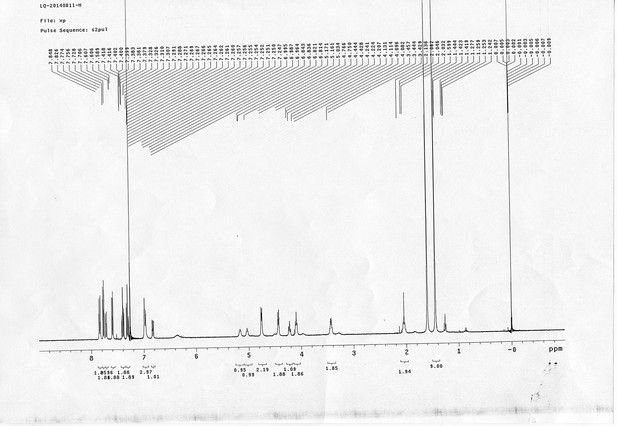
1H NMR spectra of compound 22.
https://doi.org/10.7554/eLife.12509.081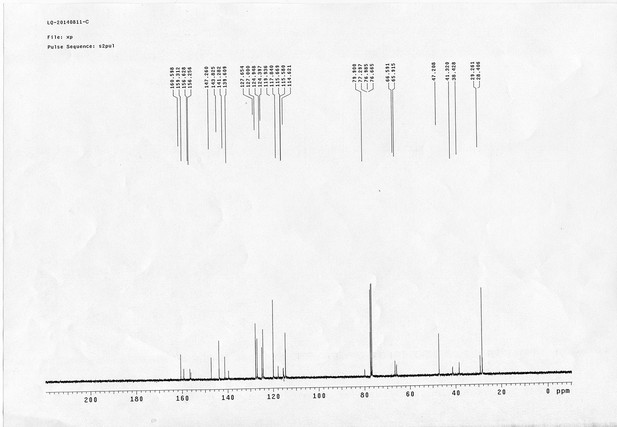
13C NMR spectra of compound 22.
https://doi.org/10.7554/eLife.12509.082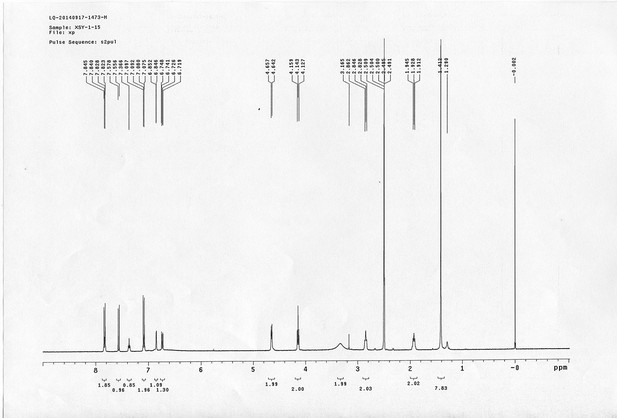
1H NMR spectra of compound 23.
https://doi.org/10.7554/eLife.12509.083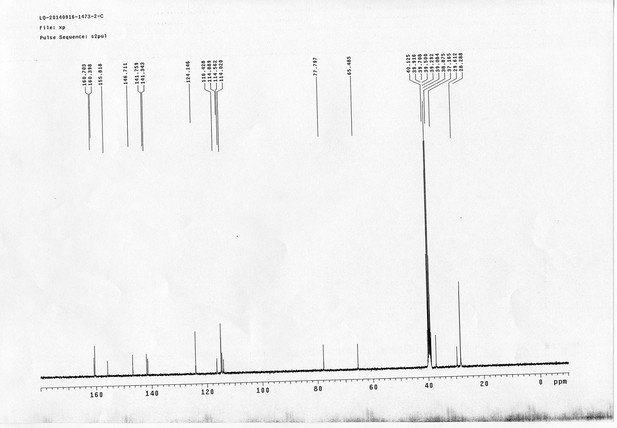
13C NMR spectra of compound 23.
https://doi.org/10.7554/eLife.12509.084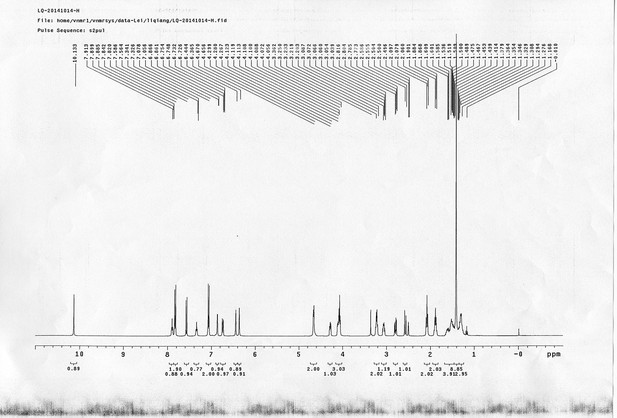
1H NMR spectra of compound 25.
https://doi.org/10.7554/eLife.12509.085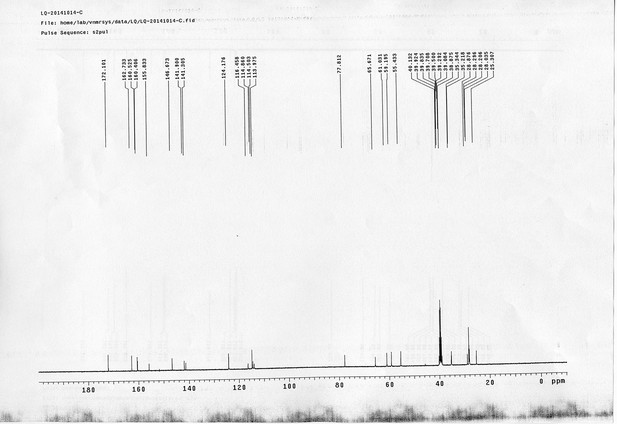
13C NMR spectra of compound 25.
https://doi.org/10.7554/eLife.12509.086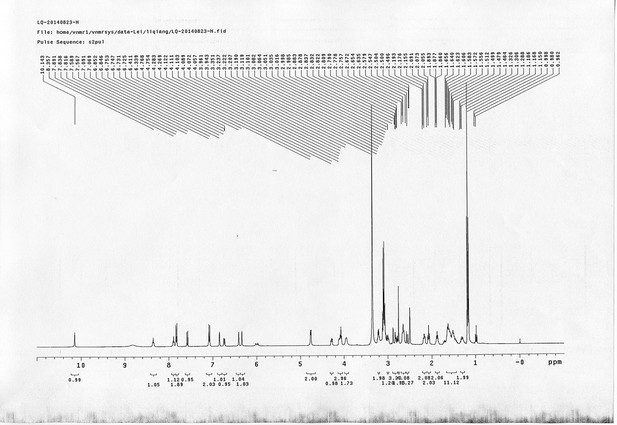
1H NMR spectra of bAL 2.
https://doi.org/10.7554/eLife.12509.087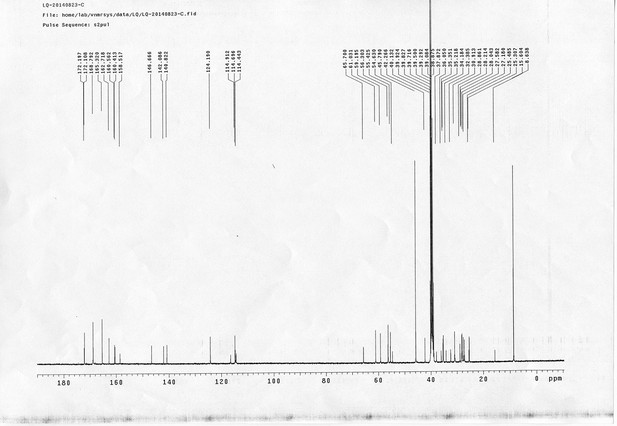
13C NMR spectra of bAL 2.
https://doi.org/10.7554/eLife.12509.088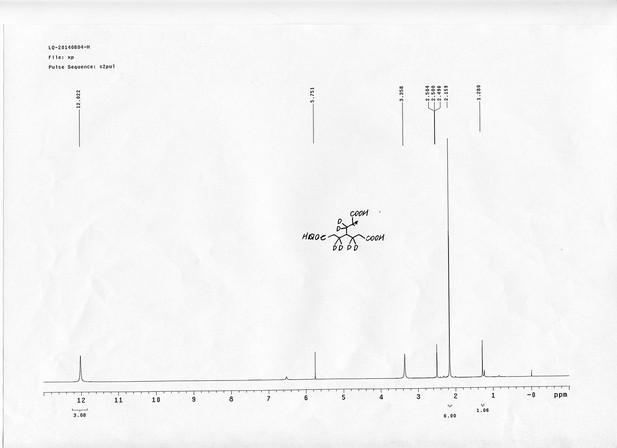
1H NMR spectra of compound 29.
https://doi.org/10.7554/eLife.12509.089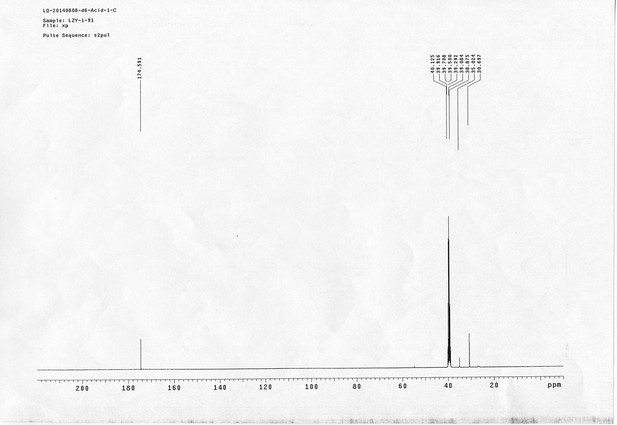
13C NMR spectra of compound 29.
https://doi.org/10.7554/eLife.12509.090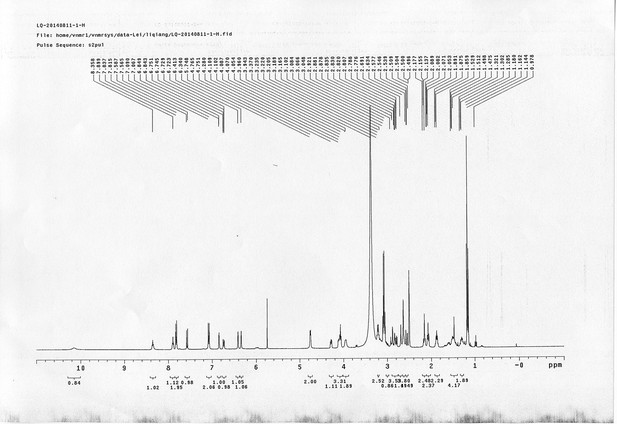
1H NMR spectra of d6-bAL 2.
https://doi.org/10.7554/eLife.12509.091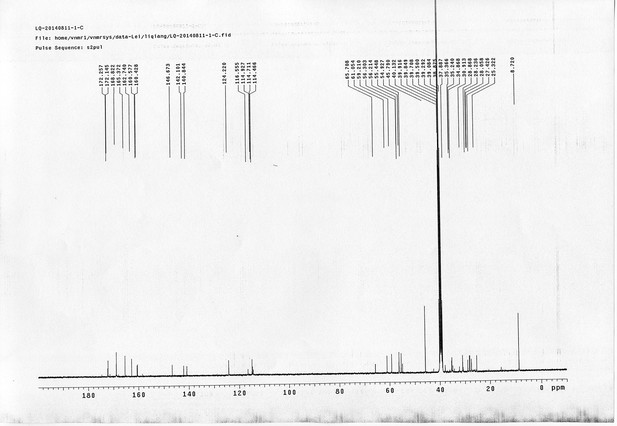
13C NMR spectra of d6-bAL 2.
https://doi.org/10.7554/eLife.12509.092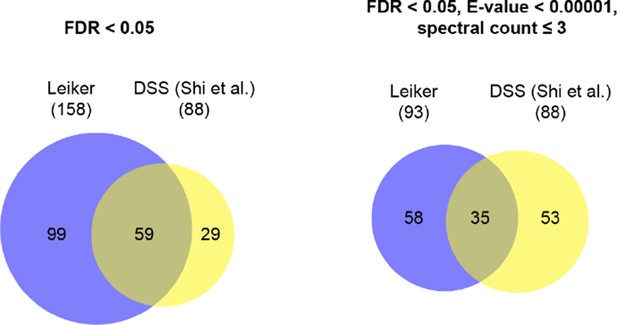
Overlap of inter-links between subunits of the exosome core complex.
https://doi.org/10.7554/eLife.12509.094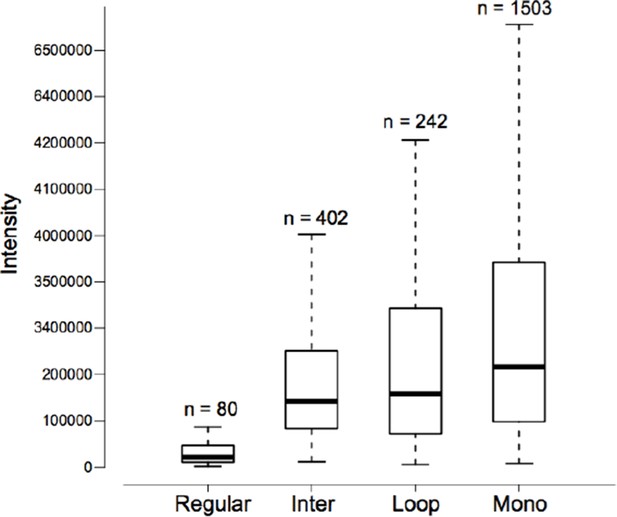
Intensity distributions of the reporter ion of m/z 122.0606 for different types of peptides.
In each category, the FDR of peptide-spectrum match (PSM) is 5%. The data came from the ten standard proteins.
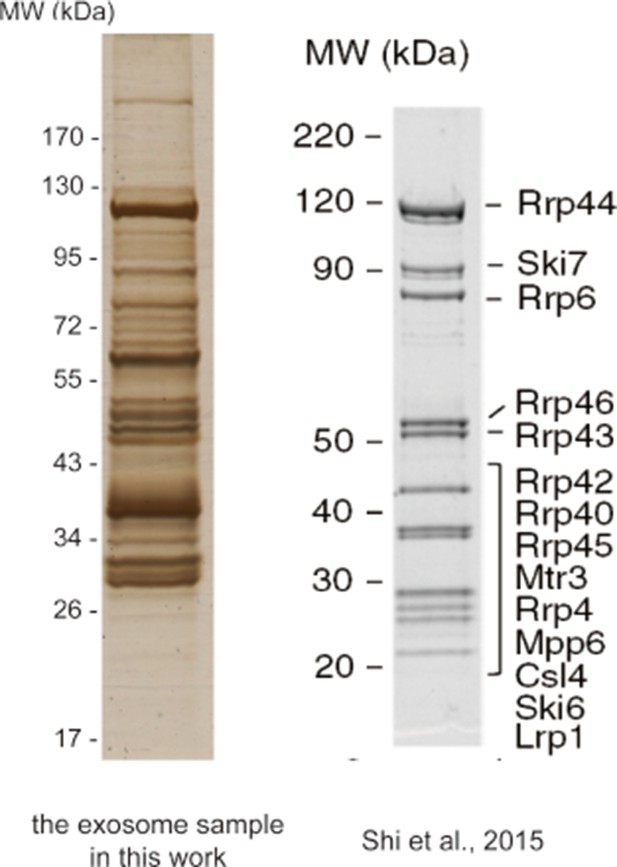
Silver-stained SDS-PAGE of the crude immunoprecipitate of TAP-tagged Rrp46 (left) and the SDS-PAGE of the purified exosome sample of Shi et al. (right).
https://doi.org/10.7554/eLife.12509.096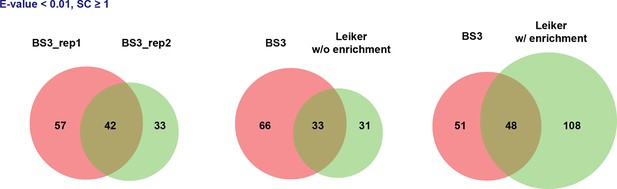
Overlap between two CXMS experiments using BS3 or Leiker on the same 10-protein mix.
https://doi.org/10.7554/eLife.12509.097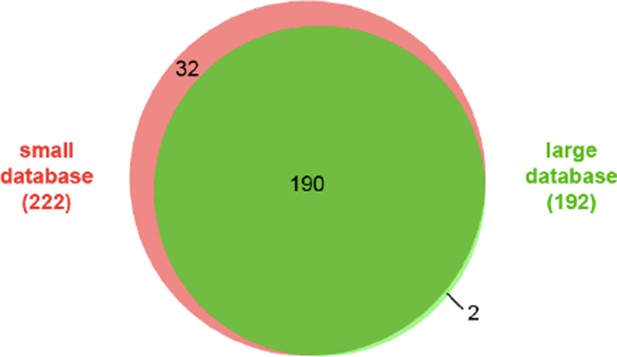
Comparison of identified cross-links using a small database (62 proteins) and a large database (562 proteins).
Filtering criteria: FDR < 5%, E-value < 0.00001, and spectral count ≥ 5.
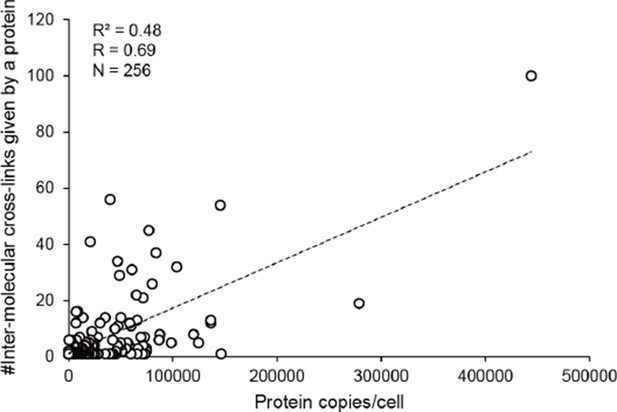
The number of inter-molecular cross-links observed for a protein in E. coli whole-cell lysates is positively correlated with the abundance of this protein.
R, Pearson correlation coefficients; N, number of values.
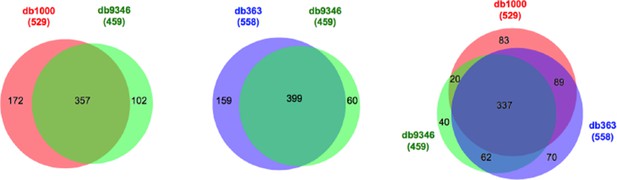
Comparison of identified cross-links using databases of different size.
db9346, containing all the proteins identified in the sample; db1000, containing the top 1000 abundant proteins; db363, containing all the proteins for which intra-molecular cross-links had been identified. Filtering criteria: FDR < 5%, E-value < 0.01, and spectral count ≥ 3.
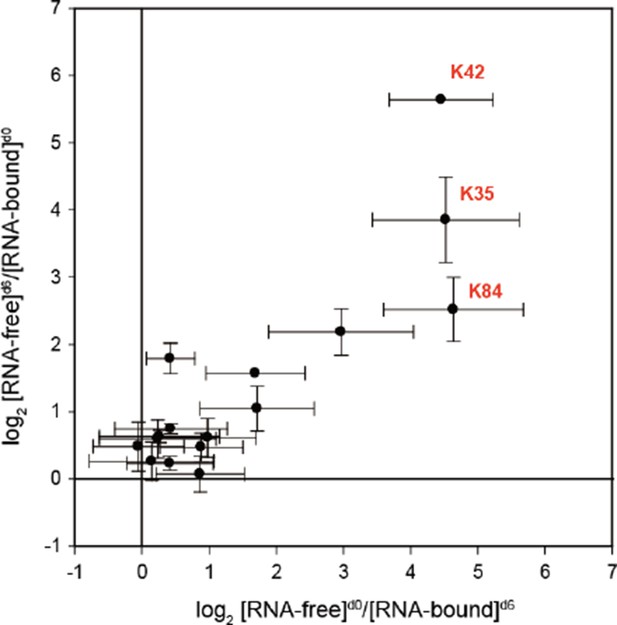
Abundance ratios of mono-links (F/B) in the forward (F[d0]/B[d6]) and the reverse labeling experiment (F[d6]/B[d0]).
Mean ± SEM.
Tables
Comparison of this study and Shi et al. study of the exosome complex.
Leiker | DSS (Shi et al.) | ||
filtering criteria | FDR < 0.05 | FDR < 0.05, E-value < 0.00001, spectral count ≥ 3 | FDR < 0.05, manual inspection |
#total cross-links | 625 | 195 | 211 |
#inter-subunit cross-links | 362 | 43 | 79 |
#intra-subunit cross-links | 263 | 152 | 132 |
amount of sample | 40 μg | 15 μg | |
#proteins in the sample | 740 (FDR 0.1%) | 44 (FDR ~1%) | |
sample purity | poor | good | |
type of exosome | cytosolic exosome (in the Rrp6 deletion background) | cytosolic exosome and nuclear exosome | |
database for pLink search | 740 proteins | 17 proteins | |
Replicates of the E. coli and C. elegans lysates.
Sample | Biological Replicate | Technical Replicate | Fractionation |
E. coli whole-cell lysates | 3 | 2 | 10-11 fractions |
E. coli ribo-free lysates | 2 | 3 | 10-12 fractions |
C. elegans whole-cell lysates | 1 | 2 | 8 fractions |
C. elegans mitochondrial proteins | 1 | 2 | 9 fractions |





