Zeb1 controls neuron differentiation and germinal zone exit by a mesenchymal-epithelial-like transition
Figures
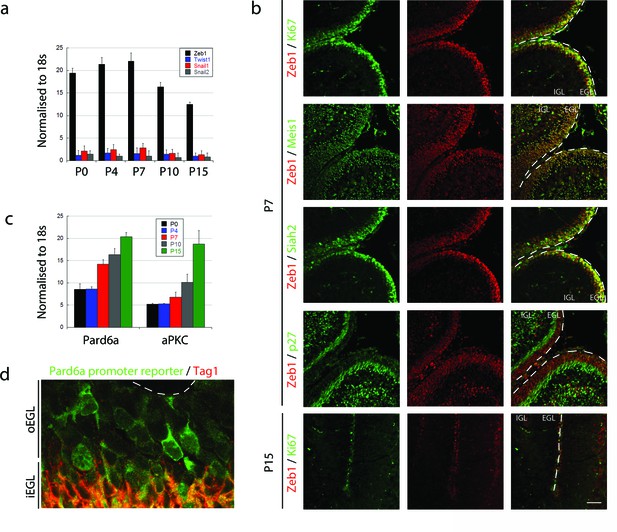
Zeb1 is the primary EMT regulator expressed in the developing cerebellum.
(a) qRT-PCR shows that Zeb1 mRNA is more abundant than other EMT factors (Twist, Snail1, Snail2) in GNPs. Zeb1 mRNA diminishes in GNPs at P10 and P15 (Zeb mRNA was significantly different at all times, t-test p<0.01). (b) Immunohistochemistry in P7 and P15 cerebellum shows Zeb1 (red) GNP expression at P7 coincident with that of Ki67, Meis1/2 and Siah2 (green) but complementary to the p27Kip marker (green). Zeb1 protein diminishes at P15. (c) qRT-PCR shows increasing Pard6a and Prkcz mRNA as GNPs at P10 and P15. (d) Immunohistochemistry in the P7 cerebellum of Pard6a-EGFP BAC transgenic mice shows little Pard6a promoter activity (green) in the outer EGL but elevated activity in the inner EGL with TAG1-positive CGNs (red).
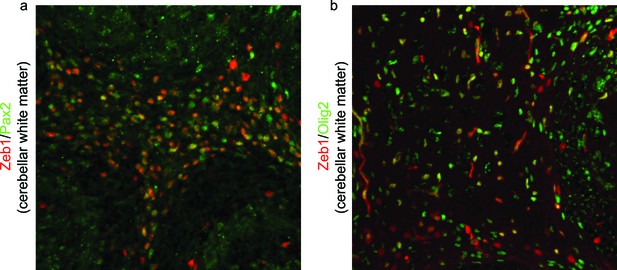
Zeb1 is expressed in Pax2 and Olig2 positive progenitors in the developing cerebellar white matter.
Immunohistochemistry in P7 cerebellum shows Zeb1 (green) expression at P7 partially overlaps with (a) Pax2 and (b) Olig2 in cerebellar white matter. This indicates that Zeb1 positive cells located in deeper cerebellar layers are interneuronal- or oligodendrocyte/glial-progenitors, not IGL resident CGNs.
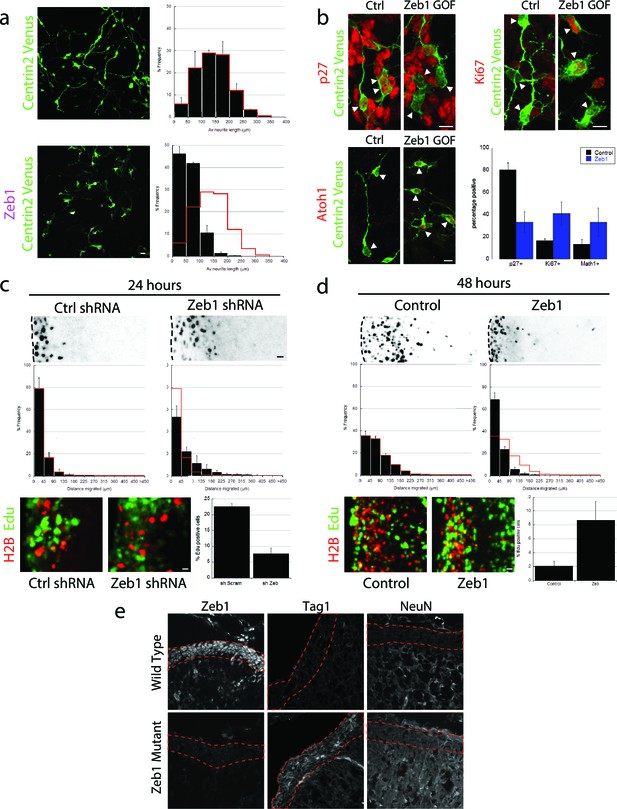
Zeb1 gain- or loss-of-function determines GNP differentiation.
(a) Micrographs of purified CGNs nucleofected with Centrin2-Venus alone (green) or Myc-Zeb1 (magenta). After 24 hr in culture, control cells extend long neurites ( = 139.8 ± 13.3 μm. n = 1045 cells), while Zeb1-expressing cells have short neurites ( = 59.6 ± 3.0 μm, n = 1164 cells, χ2 test, p<0.01). (b) Micrographs of purified CGNs nucleofected with Centrin2-Venus (green) cytoplasmic marker and Myc-Zeb1. After 24 hr, levels of p27 labeling decreased, while that of Ki67 and Atoh1 increased (t-test all conditions p<0.05). C,D. P7 EGL was co-electroporated with indicated vector and H2B-mCherry. After 24 (C) or 48 (D) hr of ex vivo culture, the migration distance of labeled CGN from the pial layer (dashed line) was analyzed in 3 experiments. Histograms show migration distributions. Zeb1-silenced cells incorporated EdU at lower rates than control cells. (c) Most control shRNA-expressing cells (black) remain within the EGL (dashed lines, = 34.2 ± 10.5 µm) at 24 hr, while Zeb1-silenced cells pre-maturely enter the ML and IGL ( = 67.5 ± 18.1 µm). (d) Control cells (black) entered the ML and IGL by 48h ( = 75.2 ± 3.5 µm), while Zeb1-expressing cells remain within the EGL ( = 40.2 ± 6.0 µm). T-tests and χ2 test showed significant differences in both conditions (p<0.01, n = 4500 to 9700 cells). (e) Immunohistochemistry in E18.5 cerebellum of wild type and Zeb1 mutant embryos shows the expected absence of Zeb1 expression in mutant embryos. Moreover, increased expression of Tag1 and NeuN differentiation markers is observed in the absence of Zeb1.
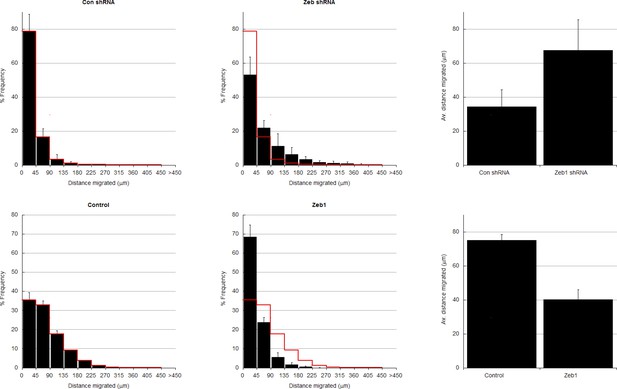
In depth quantitation of slice migration assays from Figure 2.
P7 EGL was co-electroporated with the indicated expression constructs and H2B-mCherry. After 24 (a–d) or 48 (e–f) hours of ex vivo culture, CGN migration distance was analyzed in 3 imaging experiments. Red overlay indicates the average migration distribution of control cells (error bar, SD). (a, b) Most control (n = 7,358 cells) migrated 34.2 ± 10.1 µm [ ± sd] at 24 hr, while Zeb1-silenced (n = 4,693 cells) migrated an Av. distance of 67.5 ± 18.1 µm. χ2 analysis showed distribution of data to be significantly different (p<0.01). (c) Binning distribution of the second mir30 based shRNA 846 used to confirm the precocious migration associated with Zeb1 silencing displayed in Figure 2C (n = 4879 cells). (d) Representative image of the mir30 based shRNA 846 illustrating Zeb1 silencing with a mirRNA based shRNA also spurs GZ exit. (e,f) While control (n = 9,744 cells) cells entered the ML and IGL after 48 hr (av distance = 75.2 ± 3.5 µm), However, Zeb1 (n = 5,359 cells) over-expressing cells remained in the EGL (Av. distance migrated = 40.2 ± 6.0 µm), which was shown to be significantly different by both χ2 analysis and t-test (p<0.03). (g) Summary of average distance migrated for the Zeb1-silencing and over-expression.
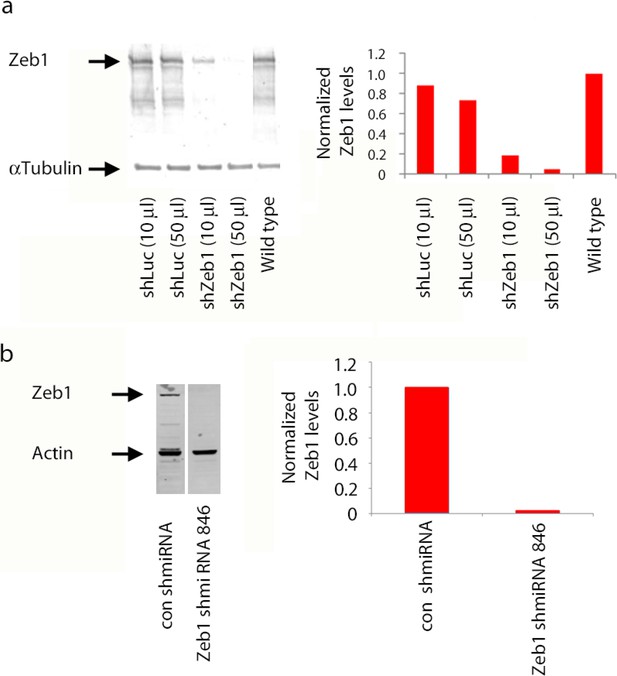
shRNA knockdown of Zeb1.
(a) Immunoblots of lysates of NS5 cells transduced with shLuc or shZeb1. Zeb1 levels are lower than control NS5 cell or cells expression a luciferase control shRNA. shZeb1 was used in Figure 2. (b) Immunoblots of lysates of HEK293 cells expressing Myc-Zeb1 with or without the corresponding shmiRNA constructs. After 48 hr Myc-Zeb1 protein levels are substantially less than those in controls, actin was loading control. The Zeb1 shmiRNA was used in Figure 7.
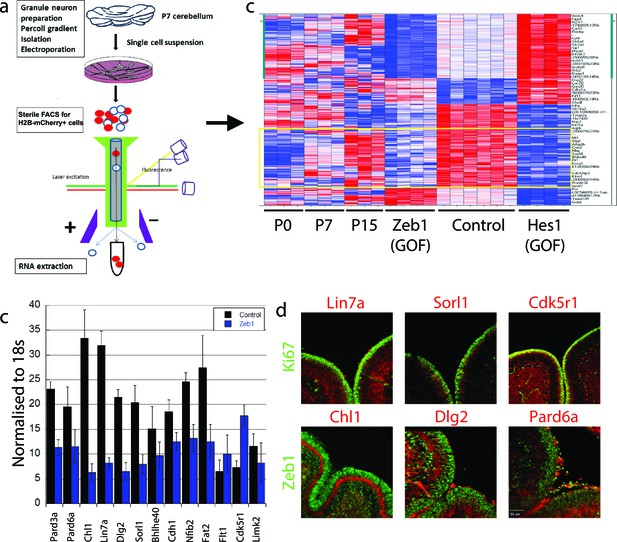
Zeb1 transcriptionally represses neuronal differentiation, cell polarity, and cell adhesion genes.
(a) Schematic of procedure used to produce pure populations of CGNs for array studies. (b) Heat map of the transcriptomes of GNPs and CGNs purified from P0, P7, and P15 compared to pure populations of control (e.g. H2B-mCherry vector alone), Zeb1-expressing (e.g. H2B-mCherry and Zeb1 vector) and HES1-expressing (e.g. H2B-mCherry and HES1 vector) GNPs cultured for 24 hr in vitro. Yellow rectangle highlights genes whose expression increases with development and are repressed by Zeb1. (c) qRT-PCR shows that ectopic Zeb1 expression inhibits transcription of most of the panel of CGN differentiation markers examined. (d) Immunohistochemistry in P7 cerebellum shows Zeb1 (red) and Ki67 (green) expression complementary with expression of the Lin7a, Sorl1, Cdk5r1, Chl1, Dlg2 and Pard6a (red) CGN markers.
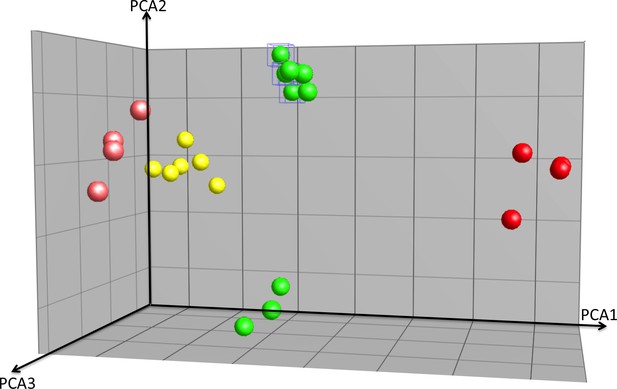
PCA analysis of array experiments shown in Figure 3.
Principal component analysis (PCA) of Zeb1, Hes1-overexpression GNPs, controls and cerebellum granule neuron (CGN) cells purified at P0, P7 and P15. Total of 40.9% of variation among these samples can be explained by the first three component (PCA1 = 20.2%, PCA2 = 10.6%, PCA3 = 10.1%). Hes1 samples are well separated from the rest along the first component and Zeb1 are separated from the controls along PCA3. Comparing with the Hes/Zeb controls, if the distance between the centroids of P7 CGNs and the control samples is 1, the distance for P0 CGNs, P15 CGNs, Zeb1 samples and Hes1 samples are 1.3, 1.2, 1.2 and 2.4 indicating that the P7 CGNs are most similar to the control samples.
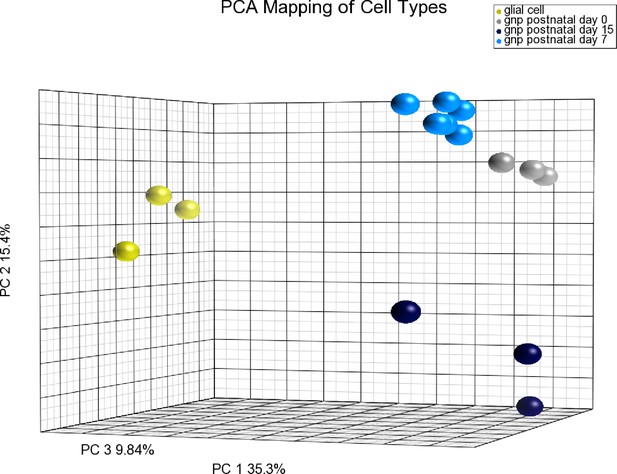
PCA demonstrating purity of GNPs/CGNs prepared at different developmental stages from Figure 3.
Principal component analysis (PCA) of GNP/CGN cells purified at P0, P7 and P15, compared to purified cerebellar glial cells. Variation among these samples can be explained by the first three components (PCA1 = 35.3%, PCA2 = 15.4%, PCA3 = 9.84%). The purified glial cells are well separated indicating low levels of this most common contaminating cell population. The purified cells from each developmental stage are well clustered statically verifying the consistency of the purification procedure at the level of the whole transcriptome. P0 and P7 are most similar. The separation of P15 cells from the earlier developmental stages is on a principle component axis that unique from the purified glial cell population, indicating the differences are related to developmental changes in the cell population than contamination with a non-GNP/CGN cell population.
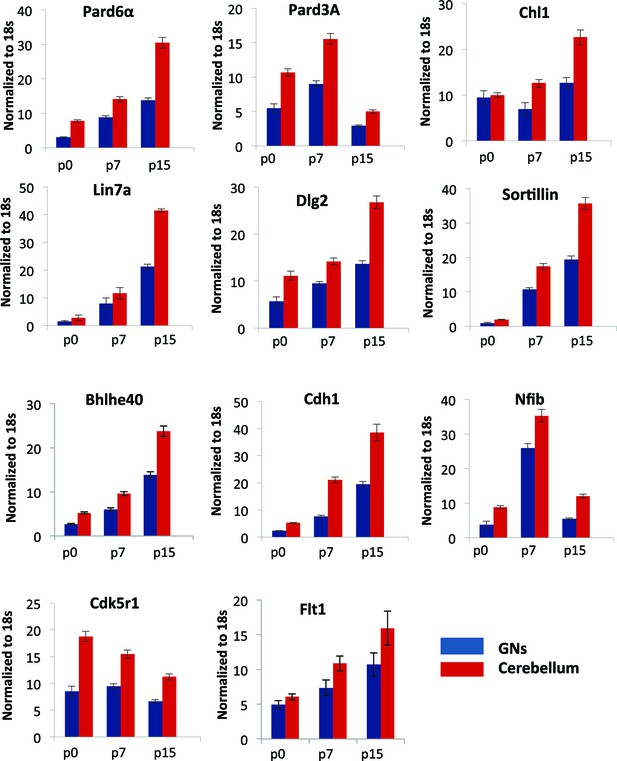
qRT-PCR analysis of Zeb1 target mRNA expression in GNPs or whole cerebellum.
RNA from purified GNPs (blue bars) or whole cerebellum (red bars) was extracted at p0, p7, and p15. qRT-PCR shows validate Zeb1 target mRNA expression increases as GNPs mature. Cdk5r1 expression declines as GNPs mature.
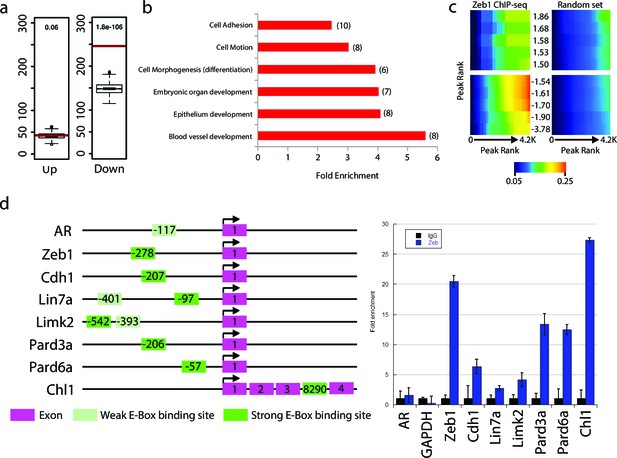
Zeb1 binds to the genomic loci of target genes identified in the expression screen.
(a) Zeb1 binding events are significantly associated with down-regulated genes (right) but not with up-regulated genes (left) between the NS5 CHIP-Seq and CGN expression array data. Red bars: total number of binding events associated with each group of genes; boxplots: distribution of binding events associations with 1000 random sets of genes. Test data are represented as a boxplot showing the test median and 1st and 3rd quartiles; whiskers are ± 1.5 the interquartile range. (b) Biological processes representing clusters of gene ontology terms enriched among genes directly targeted by Zeb1. Parentheses show number of genes associated with each term. (c) Heat-map displaying the cumulative fraction of deregulated genes that are directly regulated by Zeb1 (up-top left panel; down-bottom left panel). Transcripts are divided in equal bins of decreasing expression fold change and plotted against Zeb1 binding events with increasing p-value. Control: 100 sets of random binding events (right panels, the mean value shown). (d) CHIP PCR Validation of Zeb1 binding in P7 GNPs. The schematic on the left displays gene structure. Exons are pink rectangles, Zeb1 binding unoccupied motifs are colored light green and validated Zeb1 binding sites are colored dark green. The graph on the right shows fold enrichment at the listed genes.
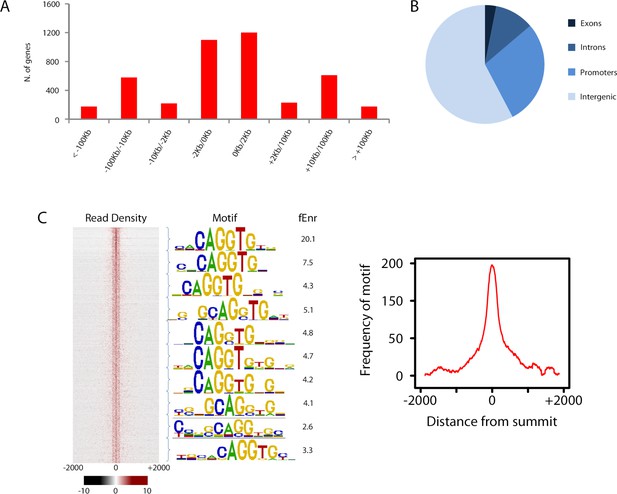
Overview of Zeb1 ChIP-Seq dataset in NS5 neural stem cells.
(a) Location of Zeb1 binding events respective to the closest annotated TSS. (b) Locations of Zeb1 binding events respective to various genomic features. (c) Density plot of Zeb1 ChIP-seq reads mapping to the 4 Kb genomic regions surrounding peak summits. Signal intensity represents the ChIP-seq normalized tag count (left). Total bound sites were divided in 10 bins and the top motif found enriched at vicinity of summits is shown for each bin, with respective fold enrichment over genomic background (middle). The frequency of E-box motif is shown, centered on peak summits (right).
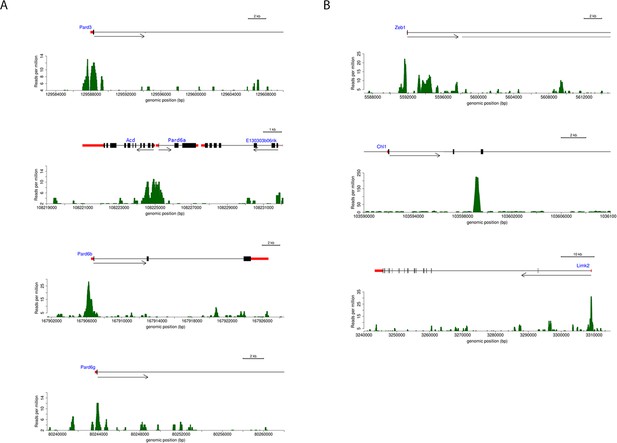
Annotated ChIP peaks in polarity genes and putative Zeb1 targets identified in NS5 data set.
Visual representation of Zeb1 ChIP-seq enrichment in the vicinity of various putative Zeb1 targets. UTRs are represented as red rectangles, translated exons as black rectangles and the direction of transcription of a locus is represented by an arrow. The graph below each gene indicate the relative Zeb1 ChIP-Seq reads per million at each genomic position near the displayed genes. (a) Shows Zeb1 binding enrichment at core PAR complex genes. Each core member of the PAR complex contains a region of enriched Zeb1 binding near Exon 1. (b) Shows Zeb1, Chl1 and Limk2 genes. A scale bar indicates that size of each locus.
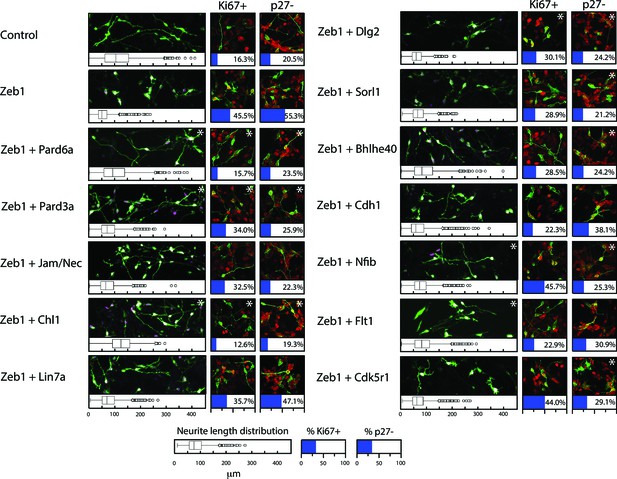
Restored expression of Zeb1-Target genes rescues neurite extension and CGN differentiation status in vitro.
The rectangular images show representative morphological information and myc-Zeb1 expression; box plot below each quantifies neurite lengths in each experimental condition. On average control cells extended neurites 115.4 ± 17.7 µm [ ± sd] compared to 55.2 ± 2.6 µm. Asterisks indicate conditions significantly different to the Zeb1 data as determined by t-test (p<0.01). Images on right show representative Ki67 or p27 labeling, quantified below. Asterisks indicate statistically significant rescue of the Zeb1 phenotype by target expression determined by t-test (p<0.01).
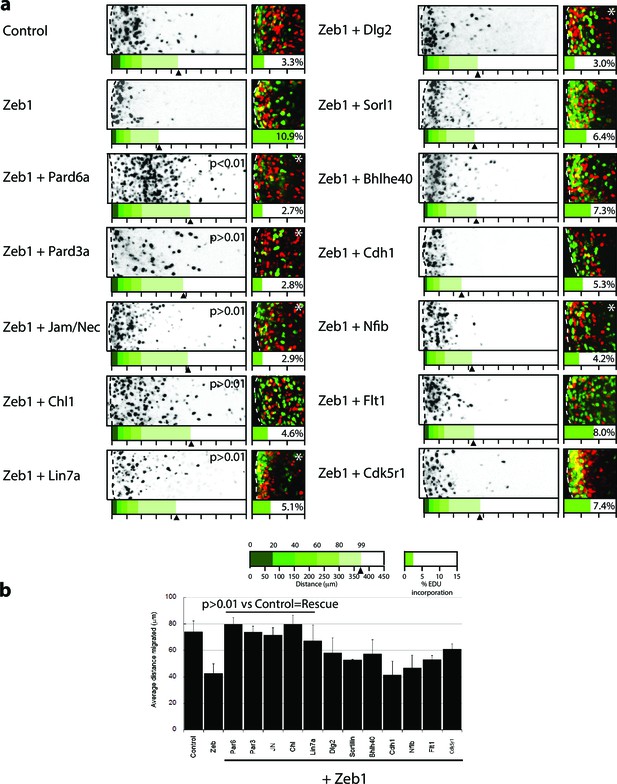
Restored expression of Zeb1 target genes rescues GNP proliferation, GZ exit and IGL-directed migration in ex vivo cerebellar slices.
(a) Rectangles show representative P7 EGL slice images assessing GZ exit and IGL-directed migration. Labeled cell (black) migrate from the lateral surface (dashed line) to the IGL (to the right). Below each image is a cumulative distribution plot of all cells relative to a 450 μm scale. Arrowhead indicates the 99th percentile of the total population. Control cells migrated 74.0 ± 8.3 μm ( ± sd) while Zeb1 migrated 42.4 ± 7.6 μm. Images at right show representative EdU labeling with% labeling index. A statistically significant rescue of a Zeb1 phenotype in the slice migration assay is indicated by the presence of p>0.01 (t-test mean migration distance vs. control). Zeb1 and additional target expression conditions had a p-value < 0.01 vs. control indicating GZ was not rescued. Asterix indicates a statistical difference of EdU incorporation between Zeb1 and target expression condition by t-test [both p<0.01]). Reduced EdU labeling indicates a rescue of elevated proliferation in the Zeb1 gain-of-function condition. b Average migration distance shown in accompanying graph, a Student’s t-test shows rescue conditions (Pard6a, Pard3a, Chl1, Jam/Nec, and Lin7a) with a p value>0.01 indicating no statistical difference from the control. Zeb1 alone and Zeb1 plus Dlg2, Sorl1, Bhlhe40, Cdh1, Nfib, Flt1 or Cdk5r1 migration differences were statistically lower than the control (t-test p<0.01), indicating GZ was not rescued with these targets.
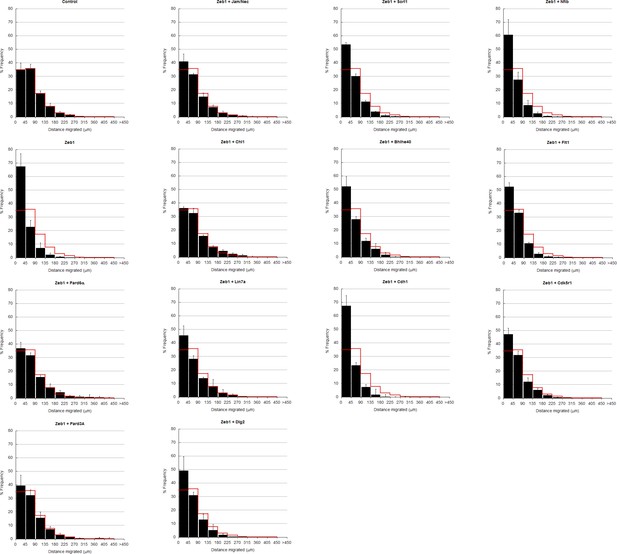
In depth quantitation of slice migration assays from Figure 6.
P7 EGL was co-electroporated with the indicated expression constructs and H2B-mCherry. After 48 hr of ex vivo culture, CGN migration distance was analyzed in 3 imaging experiments. Red overlay indicates the average migration distribution of control cells (error bar, SD). While control (13,064 cells, 74.0 ± 8.3 µm [n, ± sd]) cells entered the ML and IGL after 48 hr and Zeb1 over-expressing cells (13,424 cells, 42.4 ± 7.6 µm) remained in the EGL, Addition of Pard6a (3,886 cells, 79.8 ± 5.2 µm), Pard3a (8,622 cells, 73.9 ± 4.5 µm), Jam/Nectin (11,333 cells, 71.7 ± 5.5 µm)), Chl1 (3.006 calls, 79.4 ± 6.9 µm) to Zeb1-expressing CGNs restored migration, suggesting that the Zeb1 migration phenotype are dependent on key polarity or cell adhesion molecule repression. Note: the criteria for rescue was set if the condition resulted in cell distribution that was 80% similar to the Control distribution (χ2-test p>0.8) and the average migration distance was less than 3% similar than the Zeb1 condition. Control vs Zeb1, [χ2 test] p(χ2) = 1.9 x 10–7, [t-test] p(t) = 6.44 x 10–5. Control vs Pard6a p(χ2) = 0.96, p(t) = 3.68 x 10–6. Control vs Pard3a p(χ2) = 0.99, p(t) = 5.67 x 10–5. Control vs Jam/Nec p(χ2) = 0.98, p(t) = 3.67 x 10–4. Control vs Chl1 p(χ2) = 0.98, p(t) = 3.68 x 10–6. Control vs Lin7a p(χ2) = 0.84, p(t) = 0.02. Control vs Dlg1 p(χ2) = 0.25, p(t) = 0.06. Control vs Sorl1 p(χ2) = 0.04, p(t) = 0.01. Control vs Bhlhe40 p(χ2) = 0.13, p(t) = 0.06. Control vs Cdh1 p(χ2) = 5.57 x 10–13, p(t) = 0.44. Control vs Flt1 p(χ2) = 1.70 x 10–4, p(t) = 0.27. Control vs Cdk5r1 p(χ2) = 0.31, p(t) = 0.01.
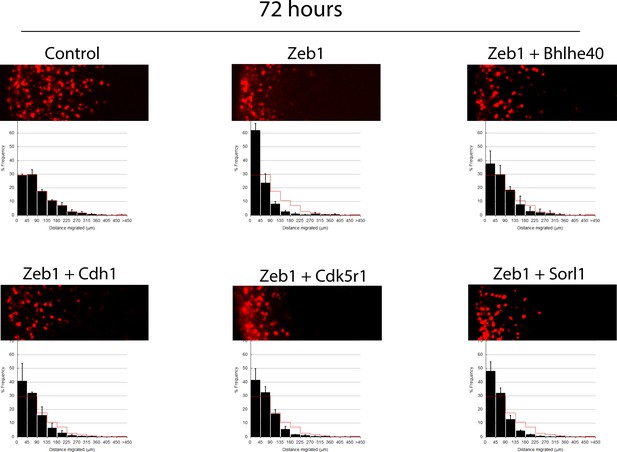
Longer term ex vivo epistasis analysis.
P7 EGL was co-electroporated with the indicated expression constructs and H2B-mCherry. After 72 hr of ex vivo culture, CGN migration distance was analyzed for a minimum of 4000 nucleofected cells in each experimental condition. Red overlay indicates the average migration distribution of control cells (error bar, SD). While control cells entered the ML and IGL after 72 hr, Zeb1 over-expressing cells remained in the EGL even with longer-term incubation. Bhlhe40 expression, but not Cdh1, Cdk5r1 and Sorl1, significantly restores IGL-directed migration of the context of Zeb1 gain-of-function (determined by Student t-test).
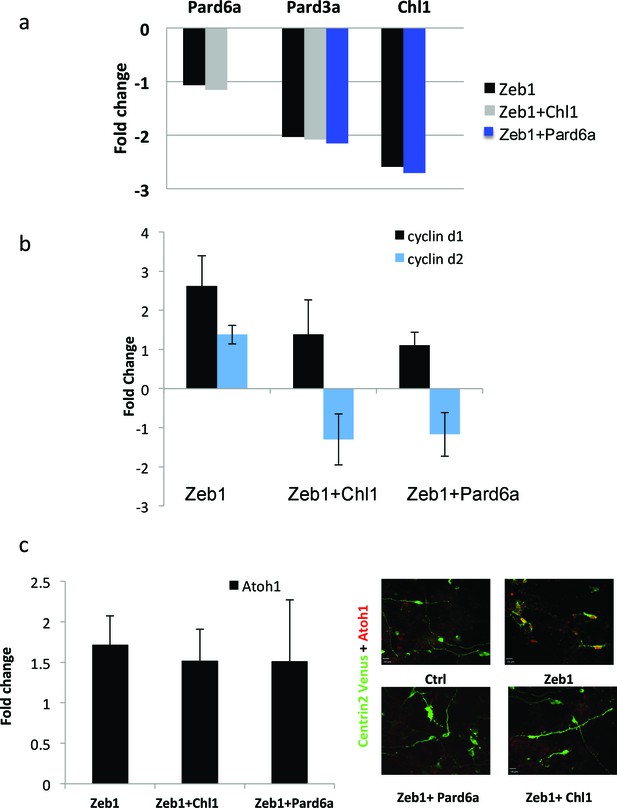
Pard6a and Chl1 rescue neuronal differentiation in the Zeb1 gain-of-function context.
Cultured CGNs were nucleofected with a marker plasmid encoding H2B mCherry (or Centrin2-Venus in Panel c) alone or in combination with plasmids encoding Myc-Zeb1 plus single plasmids encoding Pard6a or Chl1 in our array expression screen. After 24 hr in culture, nucleofected cells were FACS sorted to isolate mRNA (a, b, c) or stained with antibodies to highlight morphology/Atoh1 expression (c). a. qRT-PCR analyses shows that: 1) Pard6a and Pard3a expression continues to be suppressed in Chl1 rescued GNPs and 2) Pard3 and Chl1 expression continues to be suppressed by Zeb1 Pard6a rescued GNPs. NS = not shown. (b) qRT-PCR analyses shows that Zeb1 gain-of-function induced CyclinD1 and CyclinD2 mRNA expression and that both restored expression of Chl1 and Pard6a reduces D-type cyclin expression. (c) qRT-PCR analyses shows that Zeb1 gain-of-function mildly induced Atoh1 mRNA expression. While Chl1 and Pard6a rescue have little affect on Atoh1 mRNA expression, restored expression of both these genes strongly reduce Atoh1 protein expression detected by immunocytochemistry.
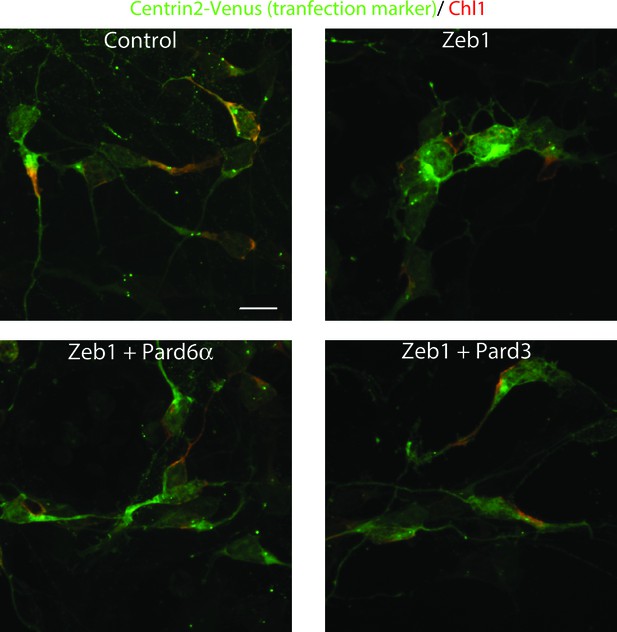
Immunocytochemical analysis of Chl1 expression in Control, Zeb1-expressing or Pard6a and Pard3 rescued CGNs.
Dissociated CGNs were prepared and nucleofected with the indicated expression constructs. 18 hr post-nucleofection, cultures were fixed and stained with antibodies recognizing EGFP and Chl1. Control neurons express robust levels of Chl1 protein in their somas and proximal leading process. In contrast, Zeb1 expressing as well as Pard6a or Pard3 rescued cells expressed lower amounts of Chl1 immunoreactivity.
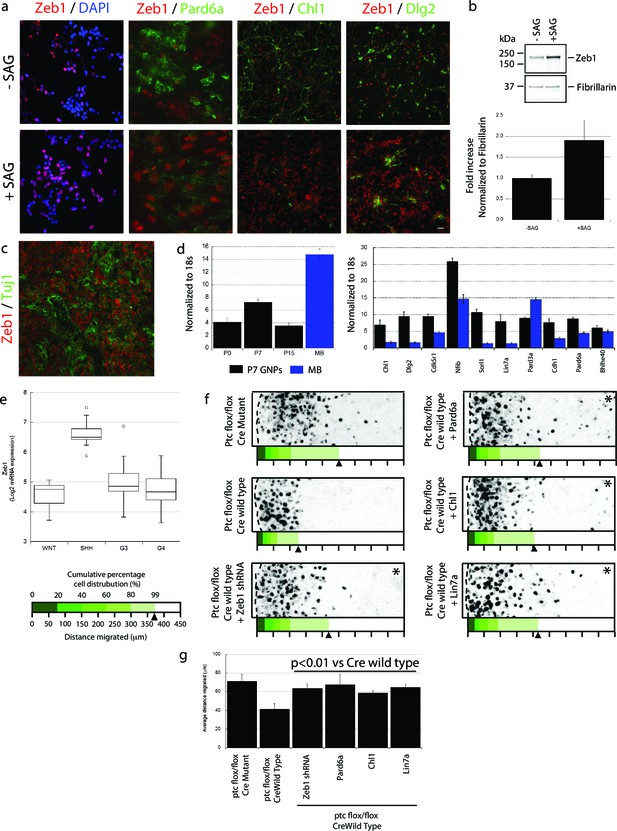
Zeb1 expression is linked to SHH signaling, and restoring polarity of Ptch1-deficient GNPs rescues GZ exit.
(a) GNPs were cultured in the presence or absence of SAG, a small-molecule agonist of SHH, fixed and stained for DAPI (blue), Zeb1 (red) or the Zeb1 targets Pard6a, Chl1 and Dlg2. Zeb1 expression was maintained, but Zeb1 target expression diminished. (b) Western blotting with anti-Zeb1 confirmed that Zeb1 expression was maintained in the presence of SAG. Fibrillarin was loading control (t-test, p<0.01). (c) Immunohistochemistry shows maintained expression of Zeb1 (red) in a Ptch1+/-, Cdkn2c-/- SHH-type mouse MB; Zeb1 expression is complementary to Tuji1 staining (green). (d) qRT-PCR of mRNA from Ptch1+/-, Cdkn2c-/- mouse MBs shows much higher Zeb1 mRNA expression in MB cells than in P7 GNPs. Most of the targets in our screen are expressed at a lower level in SHH MB than in P7 GNPs. (e) Zeb1 mRNA expression in 4 MB subgroups. Data set includes 74 MBs (WNT n = 8; SHH n = 11; G3 n = 17; G4 n = 38) profiled on the Affymetrix U133plus2 array. (f) The migration distance of CGNs (black dots) from the pial layer (dashed line) was analyzed (n = 8,800 to 11,300 cells). Control cells expressing catalytically inactive Cre enter the ML and IGL (71.2 ± 7.8 µm [ ± sd]), while Ptch1-deficient GNPs expressing wild-type Cre remain within the EGL (41.4 ± 5.8 µm). Zeb1 silencing and restored expression of Pard6a, Chl1 and Lin7a rescued the defective GZ exit (Asterisks indicate conditions where rescue observed (χ2 test vs Cre mutant, p>0.8; t-test vs Cre WT, p<0.01). Below each image is a cumulative distribution plot showing the area relative to a 450 μm scale. Arrowhead indicates 99th population percentile. (g) Average migration distance shown in accompanying graph, a Student’s t-test shows rescue conditions with a p value <0.01 vs Cre wild type.
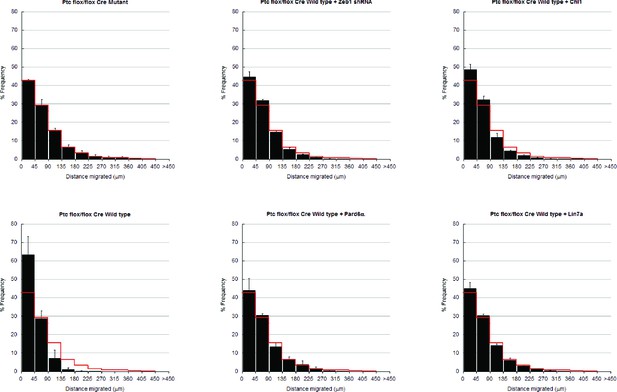
In depth quantitation of slice migration assays from Figure 8.
P7 EGL of Ptch1 flox/flox animals was co-electroporated with the indicated expression constructs and H2B-mCherry. After 48 hr of ex vivo culture, CGN migration distance was analyzed in 3 imaging experiments. Red overlay indicates the average migration distribution of control cells (error bar, SD). While Cre Mutant (n = 9,471 cells) cells entered the ML and IGL after 48 hr and Cre wild type (n = 9,872) over-expressing cells remained in the EGL, Zeb1 silencing (n = 11,383) or addition of Pard6a (n = 8,839), Lin7a (n = 10,543), or Chl1 (n = 11,348) to Zeb1-expressing CGNs restored migration, suggesting that the GZ exit phenotype Ptch1 deficient GNPs is dependent on Zeb1 repression of its targets. Average migration distance shown in accompanying graph. Note: the criteria for rescue was set if the condition resulted in cell distribution that was 80% similar to the Cre Mut distribution (χ2- test p>0.8) and the average migration distance was less than 3% similar than the Cre WT condition (t-test p<0.03). Cre Mut vs Cre WT, [χ2 test] p(χ2) = 2.86 x 10–3, [t-test] p(t) = 3.03 x 10–3. Controls vs Zeb1 shRNA p(χ2) = 0.48, p(t) = 0.03. Controls vs Pard6a p(χ2) = 1.00, p(t) = 0.01. Controls vs Chl1 p(χ2) = 0.86, p(t) = 4.78 x 10–3. Controls vs Lin7a p(χ2) = 1.00, p(t) = 1.99 x 10–3.
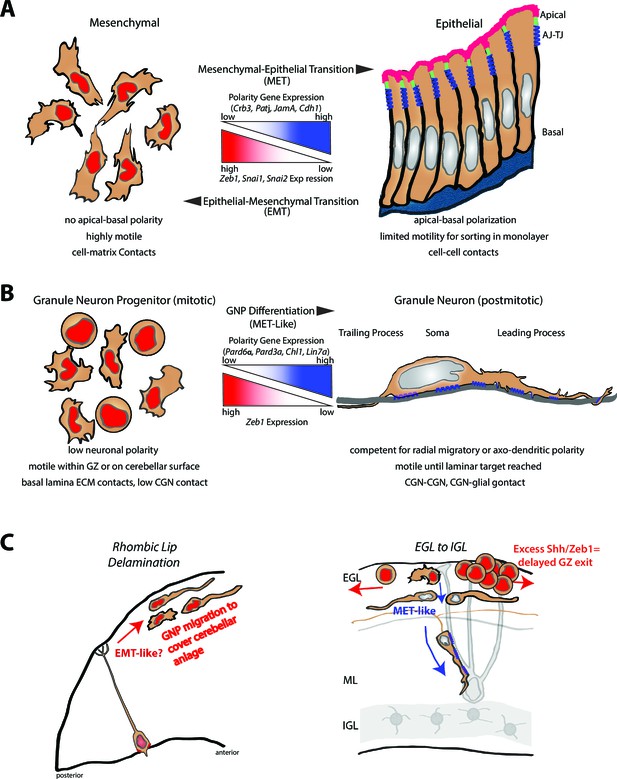
Model comparing MET to GNP differentiation.
(a) Mesenchymal-epithelial transition. Left: Mesenchymal cells are nonpolar, highly motile, with prominent cell-matrix contacts. Right: epithelial cells possess apical-basal polarity. Apical membrane (pink) is separated from basolateral and basal membranes by tight junctions (parallel blue rectangles) and adherens (blue springs) junctions. MET-EMT balance is controlled by antagonism between transcriptional regulators and polarity genes (center panel). (b) Left: GNP. As in MET, GNPs lose Zeb1 expression as they differentiate, relieving polarity gene repression. Center panel: Change in gene expression with GNP differentiation to CGNs. Right: CGNs morphologically mature, exit their GZ and make contacts with other CGNs or glia (blue springs depict adhesion to grey glial fiber). (c) Transition from tangential migration within the EGL by GNPs and nascent CGNs to radial migration (red arrows) by polarized T-shaped CGNs is MET-like, given falling Zeb1 expression (red to grey nuclei). Blue springs depict neuron-glial adhesions. Elevated SHH signaling drives Zeb1 expression to delay GZ exit at early stage of MB tumorigenesis.
Videos
Representative time lapse imaging sequence of a CGN migrating in a dissociated culture labeled with Centrin2-Venus (green, centrosome) and RFP-UTRCH ABD (f-actin).
The focused cell undergoes typical two-stroke nucleokinesis with f-actin accumulation in the leading process. Time stamp= hours: minutes: seconds. Scale bar = 10 μm.
Representative time lapse imaging sequence of Zeb1 over-expressing CGNs migrating in a dissociated culture labeled with Centrin2-Venus (green, centrosome) and RFP-UTRCH ABD (f-actin).
The featured cells undergo random amoeboid movements with isotropic f-actin decorating the cell periphery. Note the centrosome does not adopt a polarized configuration as in Video 1. Time stamp= hours: minutes: seconds. Scale bar =10 μm.
Representative time lapse imaging sequence of control CGNs labeled with Centrin2-Venus (green, centrosome) and H2B-mCherry (nucleus) in a dissociated culture.
The migrating cells in the field undergo typical two-stroke nucleokinesis with centrosome entering the leading process prior to somal translocation. Note: even stationary cells extend long neurites. Time stamp= hours: minutes: seconds. Scale bar= 10 μm.
Representative time lapse imaging sequence of Zeb1 over-expressing CGNs labeled with Centrin2-Venus (green, centrosome) and H2B-mCherry (nucleus) in a dissociated culture.
The migrating cells in the field undergo random amoeboid movements where the centrosome adopts an unpolarized position in the cell body. Note: even stationary cells extend do not extend long neurites. Time stamp = hours: minutes: seconds. Scale bar = 10 μm.
Representative time lapse imaging sequence of Zeb1 over-expressing CGNs with restored Pard6a expression labeled with Centrin2-Venus (green, centrosome) and H2B-mCherry (nucleus) in a dissociated culture.
The migrating cells in the field undergo typical two-stroke nucleokinesis with centrosome entering the leading process prior to somal translocation. Note: even stationary cell extend long neurites. Time stamp= hours: minutes: seconds. Scale bar = 10 μm.
Representative time lapse imaging sequence of Zeb1 over-expressing CGNs with restored Pard3a expression labeled with Centrin2-Venus (green, centrosome) and H2B-mCherry (nucleus) in a dissociated culture.
The migrating cells in the field undergo typical two-stroke nucleokinesis with centrosome entering the leading process prior to somal translocation. Note: even stationary cell extend long neurites. Time stamp = hours: minutes: seconds. Scale bar = 10 μm.
Representative time lapse imaging sequence of Zeb1 over-expressing CGNs with restored Chl1 expression labeled with Centrin2-Venus (green, centrosome) and H2B-mCherry (nucleus) in a dissociated culture.
The migrating cells in the field undergo typical two-stroke nucleokinesis with centrosome entering the leading process prior to somal translocation. Note: even stationary cell extend long neurites. Time stamp= hours: minutes: seconds. Scale bar = 10 μm.
Representative time lapse imaging sequence of control CGNs labeled with JAM-C-pHluorin (green, adhesions) and H2B-mCherry (nucleus) in a dissociated culture.
Note: exuberant cell contacts are observed among most cells. Time stamp= hours: minutes: seconds. Scale bar = 10 μm.
Representative time lapse imaging sequence of Zeb1 over-expressing CGNs labeled with JAM-C-pHluorin (green, adhesions) and H2B-mCherry (nucleus) in a dissociated culture.
Sparse cell contacts are observed among most cells. Time stamp = hours: minutes: seconds. Scale bar = 10 μm.
Representative time lapse imaging sequence of Zeb1 over-expressing CGNs with restored Pard6a expression labeled with JAM-C-pHluorin (green, adhesions) and H2B-mCherry (nucleus) in a dissociated culture.
Note: note cell contacts are observed among most cells. Time stamp = hours: minutes: seconds. Scale bar = 10 μm.
Representative time lapse imaging sequence of Zeb1 over-expressing CGNs with restored Pard3a expression labeled with JAM-C-pHluorin (green, adhesions) and H2B-mCherry (nucleus) in a dissociated culture.
Note: restored cell contacts are observed among most cells. Time stamp= hours: minutes: seconds. Scale bar = 10 μm.
Representative time lapse imaging sequence of Zeb1 over-expressing CGNs with restored Chl1 expression labeled with JAM-C-pHluorin (green, adhesions) and H2B-mCherry (nucleus) in a dissociated culture.
Note: restored cell contacts are observed among most cells. Time stamp= hours: minutes: seconds. Scale bar = 10 μm.
Additional files
-
Supplementary file 1
The results of mouse EMT pathway focused RT2 Profiler PCR array.
A fold change filtering was performed from three independent experiments using 2-ΔΔCt (where ΔΔCT=ΔCT of FACS sorted Zeb1 overexpressing GNPs–ΔCT of control GNPs) and is represented as tables. The threshold for cut off was a fold change ≥ +2.0 or ≤ -2.0. Supplementary Table 1A: Functional gene grouping shows an upregulation in expression of several genes such as Anhak, Col3A1, Gng11, MMP2 and3, Serpine (Pals-1) and Vim all of which are documented to be highly expressed during EMT. Supplementary Table 1B: Reciprocal expression of several key genes that are also downregulated during EMT included Dsp, Fgfbp1, Mst1r. Additionally Krt14, Nodal and Sox10 genes that are involved in differentiation showed a reduced expression when Zeb1 was overexpressed.
- https://doi.org/10.7554/eLife.12717.036
-
Supplementary file 2
- https://doi.org/10.7554/eLife.12717.037
-
Supplementary file 3
(A) List of primers used for RT-PCR in microarray validation and developmental profile expression analysis. (B) Primer Sequences used for validating the ChIP -PCR studies. (C) List of Antibodies.
- https://doi.org/10.7554/eLife.12717.038





