Opposing roles for DNA replication initiator proteins ORC1 and CDC6 in control of Cyclin E gene transcription
Figures
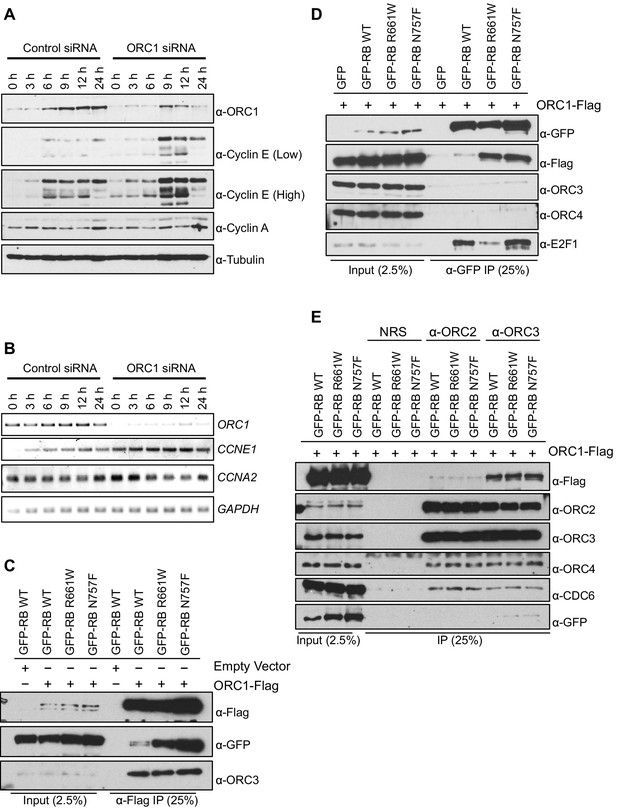
ORC1 represses Cyclin E gene expression and interacts with RB.
(A–B) Nocodazole arrested U2OS cells were transfected with control or ORC1 siRNA then released into the next cycle. (A) protein levels were estimated by immunoblotting with antibodies against ORC1, Cyclin E, Cyclin A and α-Tubulin. Low and high indicates different exposures. (B) mRNA levels of ORC1, Cyclin E (CCNE1), Cyclin A (CCNA2) and GAPDH. Quantitation of mRNA levels from multiple experiments is shown in Figure 1—figure supplement 1. (C–E) Interaction between ORC1 and RB or its pocket mutants. GFP, GFP-tagged wild-type or mutant RB were co-transfected in HEK293 cells with either ORC1-Flag or empty vector. Immunoprecipitation with anti-Flag antibody (C) or GFP antibody (D) from cell lysates followed by immunoblotting with the indicated antibodies. (E) Cell lysate from HEK293 cells overexpressing GFP-tagged wild type or mutant RB and ORC1-Flag were immunoprecipitated with normal rabbit serum (NRS) or ORC2 or ORC3 antibodies, immunoblotted with the indicated antibodies. Binding of ORC1 to wild-type and RB mutants and the effect of Cyclin E-CDK2 is shown in Figure 1—figure supplement 2. GFP, Green fluorescent protein; ORC, Origin Recognition Complex; RB, Retinoblastoma.
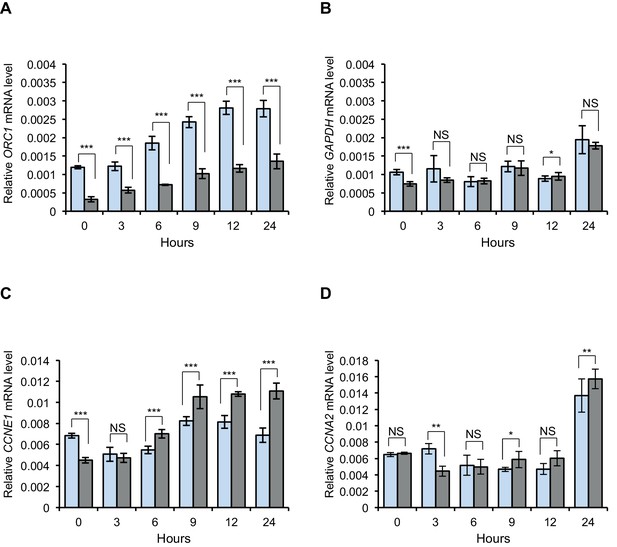
ORC1 represses Cyclin E gene expression and interacts with RB.
CCNE1 (Cyclin E) gene transcription is up-regulated upon ORC1 depletion. (A–D) Quantitative PCR analysis of ORC1, CCNE1 (Cyclin E), CCNA2 (Cyclin A) and GAPDH transcript levels in U2OS cells transfected with either control siRNA targeting GFP (blue bar) or ORC1 siRNA (grey bar). The siRNA treated cells were released after nocodazole arrest and mRNA levels estimated at different time points as indicated in hours. The values shown are average fold change (mean±SEM) from three independent experiment normalized to β-actin transcripts. Statistical analysis was performed using the Student’s t test. *p<0.01; **p<0.001; ***p<0.0001; NS, not significant. ORC, Origin Recognition Complex; RB, Retinoblastoma; GST, Glutathione S transferase.
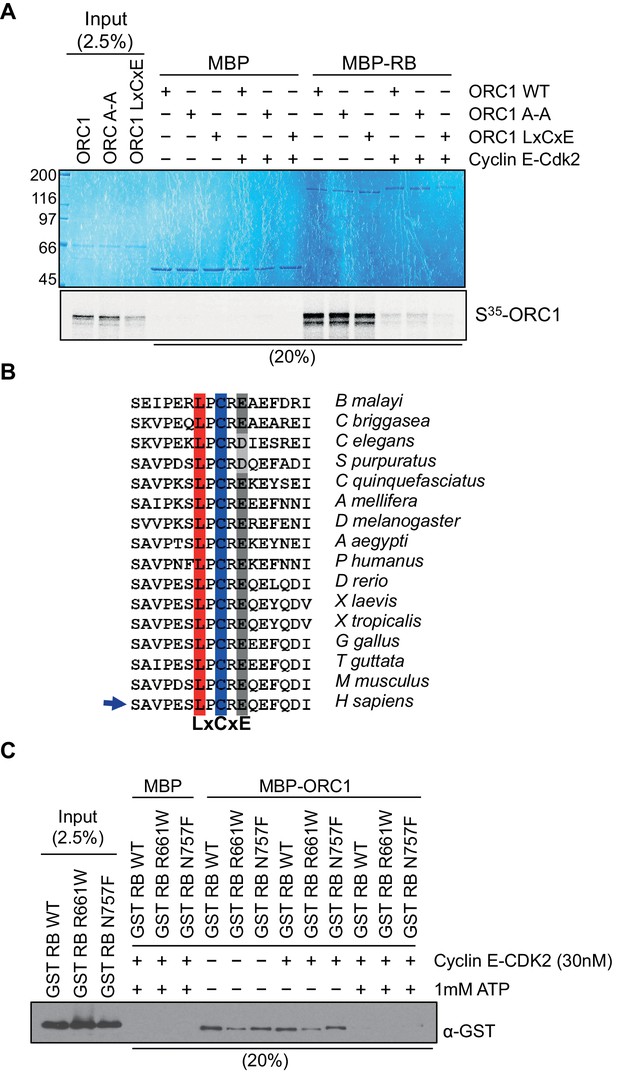
ORC1 represses Cyclin E gene expression and interacts with RB.
Cyclin E-CDK2 phosphorylation controls ORC1 and RB interaction. (A) Interaction between purified RB and ORC1 in a MBP pull down assay. MBP-fused wild-type RB was bound to amylose resin and further incubated with in vitro translated, S35-labeled wild type ORC1 or its mutants in the presence or absence of Cyclin E-CDK2 and 1 mM ATP. Beads were isolated and bound proteins were separated by gel electrophoresis. MBP was used as a control in the assay. (B) Alignment of ORC1 sequences is shown with conserved LxCxE motif. The conserved residues of LxCxE motif are indicated with different colors. The alignment shows conserved residues in ORC1 from different species in vertebrate and invertebrate classes (Invertebrates: Brugia malayi, Caenorhabditis briggsae, Caenorhabditis elegans, Strongylocentrotus purpuratus, Culex quinquefasciatus, Apis mellifera, Drosophila melanogaster, Aedes aegypti, and Pediculus humanus; Vertebrates: Danio rerio, Xenopus laevis, Xenopus tropicalis, Gallus gallus, Taeniopygia guttata, Mus musculus and Homo sapiens). (C) Interaction between ORC1 and RB in a MBP pull-down assay. GST-fused wild-type RB or its mutant proteins were incubated with wild type MBP-ORC1 in the presence or absence of Cyclin E-CDK2 and/or 1 mM ATP. Amylose-bead-bound proteins were isolated and bound proteins were separated by gel electrophoresis followed by immunoblotting with anti-GST antibody. Recombinant MBP was used as a control in the assay. ORC, Origin Recognition Complex; RB, Retinoblastoma; GST, Glutathione S transferase; MBP, Maltose binding protein.
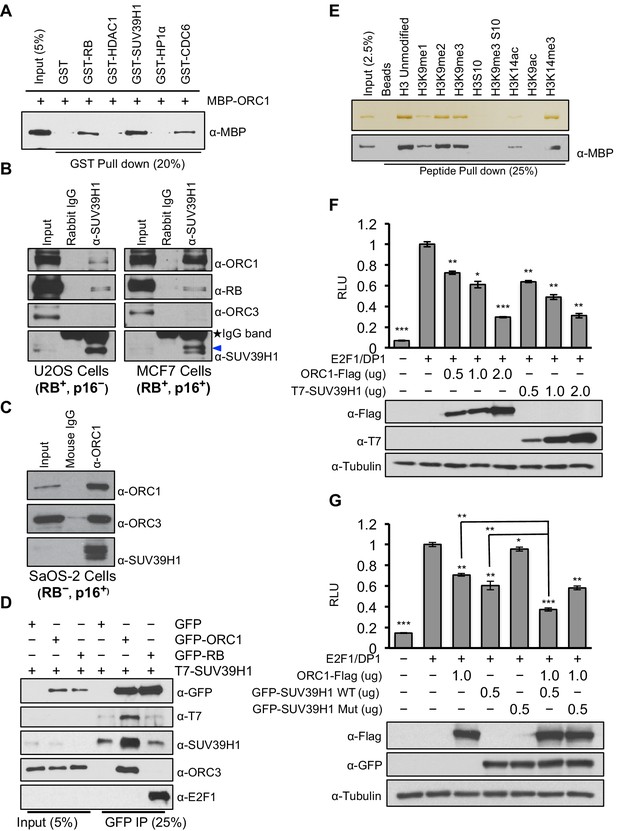
ORC1 binds SUV39H1 to control Cyclin E gene transcription.
(A) Purified MBP-ORC1 and various GST-fused proteins were mixed and proteins bound in a GST-pull down were detected by immunoblotting with anti-MBP antibodies. The purified proteins are shown in Figure 2—figure supplement 1. (B) U2OS and MCF7 cell lysates were immunoprecipitated with SUV39H1 antibody and immunoblotted with the indicated antibodies. Rabbit IgG served as control antibody. Asterisk indicates the cross-reacting antibody band; arrow indicates the SUV39H1 protein. (C) Immunoprecipitation from RB-negative SaOS-2 cell lysates with ORC1 antibody or IgG and immunoblotted with antibodies against ORC1, SUV39H1 or ORC3. (D) HEK293 cells were transiently co-transfected with GFP, GFP-ORC1 or GFP-RB plus T7-SUV39H1 plasmids (2.5 μg each). GFP antibody immunoprecipitates were immunoblotted with the indicated antibodies. The interaction between ORC1 and SUV39H1 and between RB and SUV39H1 is shown with purified proteins and quantitated in Figure 2—figure supplement 2. Higher levels of RB are required to demonstrate an interaction with SUV39H1 in vivo and ORC1 interacts with the SET domain of SUV39H1, Figure 2—figure supplement 3. (E) MBP-ORC1 was incubated with bead-bound histone peptides with or without the indicated modifications and bound MBP-ORC1 was observed by immunoblotting with anti-MBP antibody (lower box) or silver staining (upper box). (F–G) Wild-type CCNE1-luciferase reporter assay in U2OS cells. U2OS cells were transiently co-transfected with 500 ng of 10–4 CCNE1 promoter, 50 ng E2F1, 50 ng DP1 and 20 ng pCMV-LacZ plasmids along with the indicated amounts ORC1 and/or SUV39H1 plasmids. (F) Increasing amounts of ORC1-Flag or T7-SUV39H1 repress Cyclin E gene promoter. Experiments were carried out in triplicate. Expression of proteins was confirmed by Immunoblot; α-Tubulin as loading control. Statistical analysis was performed using the Student’s t test. *p<0.05; **p<0.005; ***p<0.001. (G) ORC1-Flag cooperates with wild type but not mutant SUV39H1 to repress CCNE1 gene expression. The experiments were carried out in triplicate. Expression of proteins was confirmed by Western blots. α-Tubulin served as a control for equal loading of each sample. Statistical analysis was performed using the Student’s t test. *p<0.01; **p<0.005; ***p<0.0001. Repression of transcription by ORC1 and SUV39H1 was also demonstrated using an artificial promoter and tethering the proteins via the GAL4 DNA binding domain in Figure 2—figure supplement 4. ORC, Origin Recognition Complex; MBP, Maltose binding protein; GST, Glutathione S transferase.
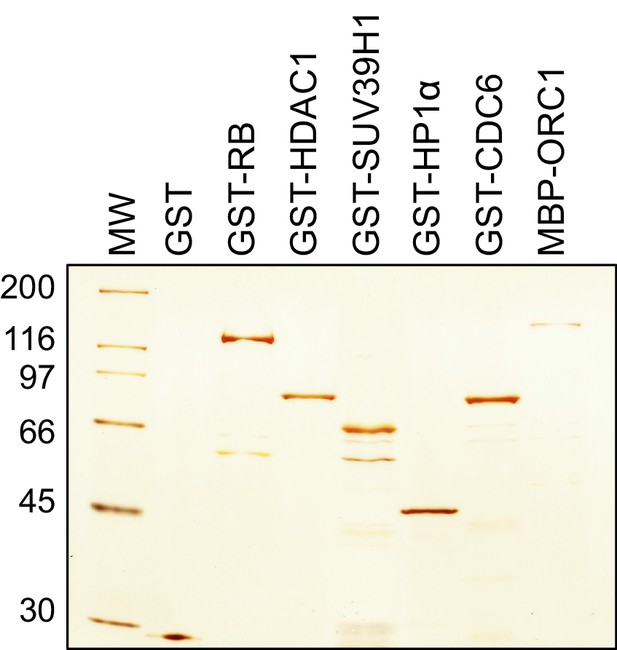
ORC1 binds SUV39H1 to control Cyclin E gene transcription.
Bacteria expressed and purified recombinant proteins. Silver stain of purified GST, GST-RB, GST-HDAC1, GST-SUV39H1, GST-HP1α, GST-CDC6 and MBP-ORC1 proteins. MW stands for protein molecular weight marker in kilodalton. ORC, Origin recogntion complex; MBP, Maltose binding protein; RB, Retinoblastoma; GST, Glutathione S transferase.
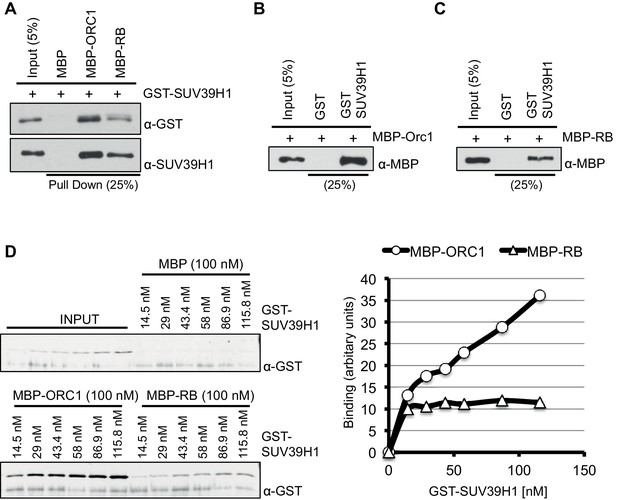
ORC1 binds SUV39H1 to control Cyclin E gene transcription.
SUV39H1 interaction with ORC1 and RB. (A) Interaction between purified MBP-ORC1 or MBP-RB with GST-SUV39H1 in a MBP pull down assay. MBP-fused proteins were bound to amylose resin and incubated with GST-SUV39H1. Bound proteins were separated by gel electrophoresis followed by immunoblotting with either anti-GST or anti-SUV39H1 antibodies. Recombinant MBP was used as a control. (B, C) GST-SUV39H1 protein was bound to the resin and incubated with either MBP-ORC1 or MBP-RB proteins. Bead-bound proteins were immunoblotted with anti-MBP antibody. Recombinant GST was used as a control. (D) Concentration-dependent interaction with increasing levels of GST-SUV39H1 and either 100 nM of MBP or MBP-ORC1 or MBP-RB followed by pull down with amylose beads and subsequently immunoblotted with anti-GST antibody. Bands were quantified and represented in a graph after background subtraction with MBP control protein. ORC, Origin recogntion complex; MBP, Maltose binding protein; GST, Glutathione S transferase.
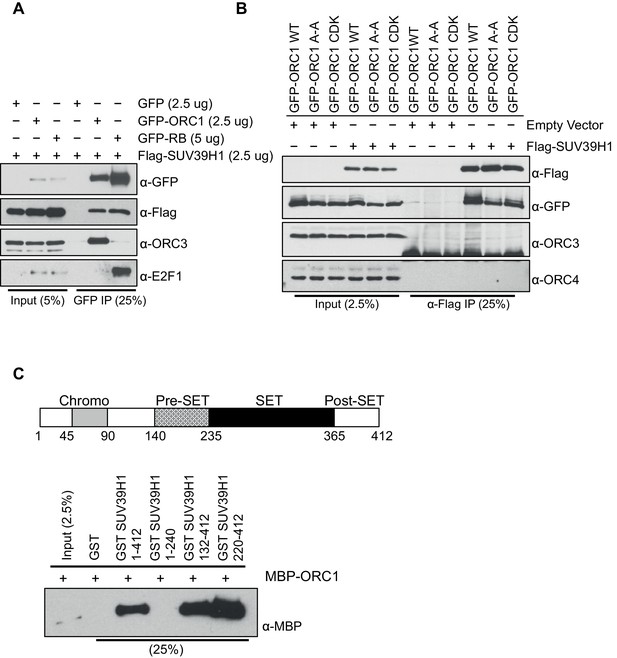
ORC1 binds SUV39H1 to control Cyclin E gene transcription.
ORC1 interaction with SET domain of SUV39H1 does not involve other ORC subunits. (A) HEK293 cells were transiently co-transfected with GFP, GFP-ORC1 or GFP-RB and T7-SUV39H1-expressing plasmids at the indicated amounts in micrograms. The whole cell lysate prepared from HEK293 cells expressing the indicated constructs were immunoprecipitated with GFP antibody followed by immunoblotting with specific antibodies. (B) GFP-tagged wild-type ORC1 or its mutants (A-A: [ORC235ARA237'Cy' motif mutant] or ORC1S258A,S273A,T375A [CDK: with mutants in CDK target sites]) were co-transfected in HEK293 cells with either Flag-SUV39H1 or its empty vector. Immunoprecipitation with anti-Flag antibody from cell lysates of HEK293 cells overexpressing the indicated constructs followed by immunoblotting with the indicated antibodies. (C) Schematic showing domains of human SUV39H1 protein. In the GST-pull down assay, GST-SUV39H1 or its truncation mutant proteins were incubated with MBP-ORC1 protein as indicated and immunoblotted with anti-MBP antibody. GST protein served as negative control. ORC, Origin recogntion complex; MBP, Maltose binding protein; GST, Glutathione S transferase.
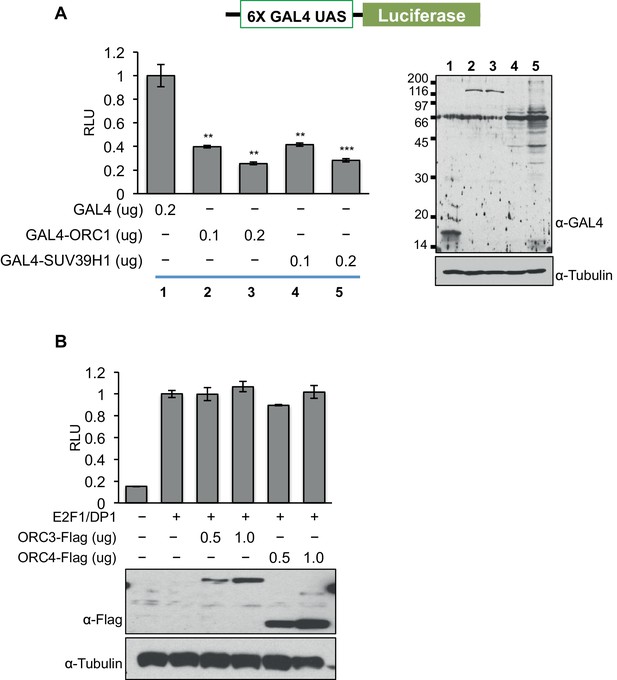
ORC1 binds SUV39H1 to control Cyclin E gene transcription.
ORC1, but not ORC3 or ORC4 can repress gene transcription. (A) The U2OS cells were transfected with a Gal4-driven luciferase reporter as shown in the schematic with increasing amounts of Gal4DBD-ORC1 or Gal4DBD-SUV39H1 together with pCMV-LacZ plasmids. Relative luciferase activity was determined and normalized to lacZ activity. Experiments were carried out in triplicate. The whole cell extract was immunoblotted with anti-Gal4 antibody for expression of Gal4DBD fusion plasmids. α-Tubulin served as a loading control. Statistical analysis was performed using the Student’s t test. **p<0.01; ***p<0.005. (B) Wild-type CCNE1 promoter-luciferase reporter assay in U2OS cells. The U2OS cells were transiently co-transfected with 500ng of the 10–4 CCNE1 promoter, 50 ng E2F1, 50 ng DP1 and 20 ng pCMV-LacZ plasmids along with the indicated amounts ORC3-Flag or ORC4-Flag plasmids. The increasing amounts of ORC3-Flag or ORC4-Flag do not repress the CCNE1 gene promoter as indicated by relative light units (RLU) normalized to β-galactosidase activity. Experiments were carried out in triplicate. Expression of proteins was confirmed by Western blot. α-Tubulin served as a loading control. ORC, Origin Recognition Complex.
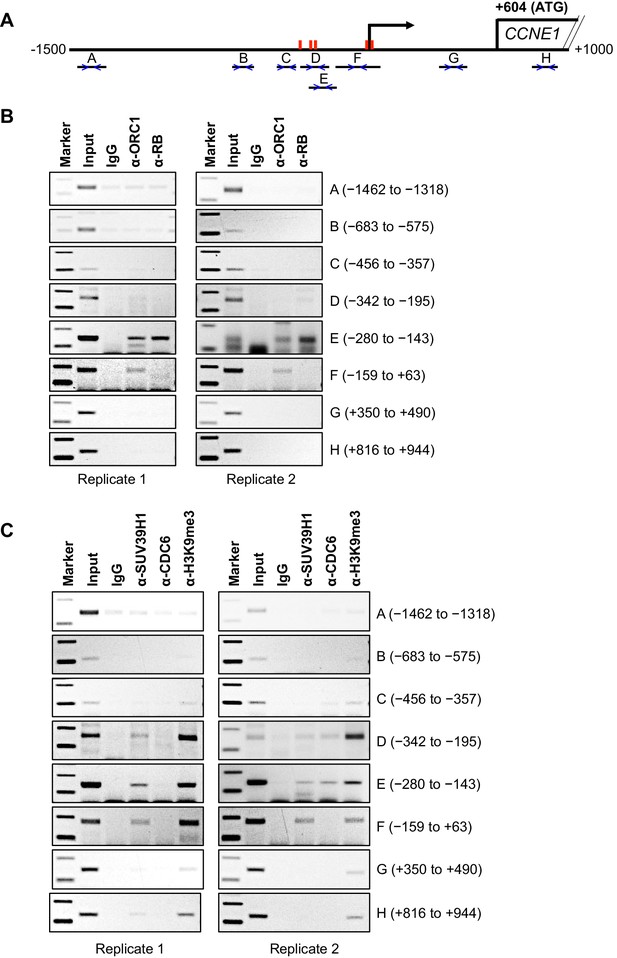
Binding of ORC1, RB, SUV39H1 and CDC6 proteins to the CCNE1 promoter.
(A) Schematic of the CCNE1 promoter and the regions amplified with different primer pairs used for ChIP assay were indicated as follows: A (−1462 to −1318); B (−683 to −575); C (−456 to −357); D (−342 to −195); E (−280 to −143); F (−159 to +63); G (+350 to +490); H (+816 to +944). The red bars indicate five E2F1 consensus sites. The truncated box indicates the first exon of the CCNE1 gene. (B–C) The occupancy of ORC1, RB, SUV39H1 and CDC6 proteins was analyzed by chromatin immunoprecipitation at the CCNE1 promoter in asynchronous growing MCF7 cells. ORC1 and RB are mouse antibodies, while SUV39H1 and CDC6 are rabbit antibodies. In the marker lanes, the two bands are 100 and 200 base pairs. The experiments were done in triplicate and two of these experiments are shown.
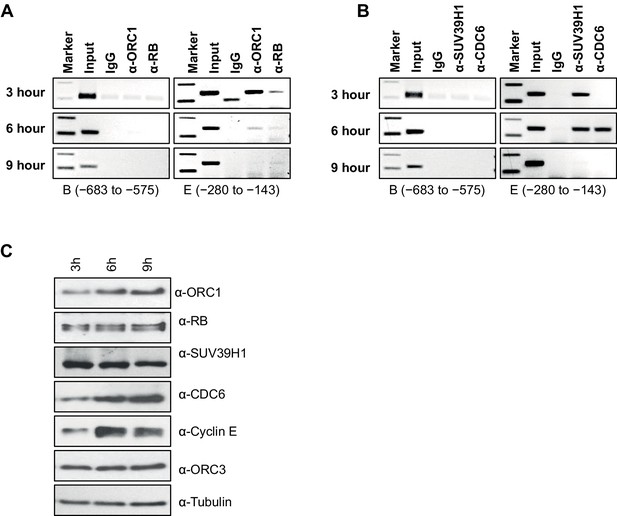
Dynamic association of ORC1, RB, SUV39H1 and CDC6 proteins to the CCNE1 promoter during the cell cycle.
(A–B) Nocodazole arrested U2OS cells were released for different times (3, 6 and 9 hr) and analyzed for occupancy of ORC1, RB, SUV39H1 and CDC6 proteins at the CCNE1 promoter by ChIP assay. The primer pairs used to analyze two different regions of the CCNE1 promoter are indicated. The experiments were done in triplicate with results similar to those shown. (C) Whole cell protein levels of nocodazole arrested and released U2OS cells at different time points (as indicated in hours) by immunoblotting with antibodies against ORC1, RB, SUV39H1, CDC6, Cyclin E, ORC3 and α-Tubulin. ORC, Origin Recognition Complex; RB, Retinoblastoma.
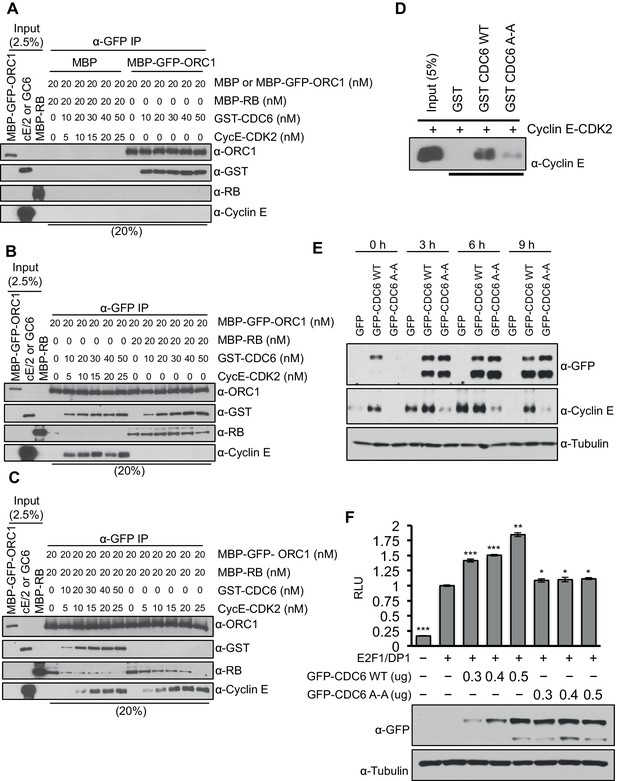
CDC6 co-operates with Cyclin E-CDK2 to activate E2F1-dependent CCNE1 gene transcription.
(A–C) Equimolar amounts of MBP-GFP-ORC1 and MBP-RB proteins were incubated with increasing amounts of GST-CDC6 and/or Cyclin E-CDK2. MBP-GFP-ORC1 protein was immunoprecipitated with GFP antibody, then immunoblotted with the indicated antibodies. The purified proteins used in these experiments are shown in Figure 5—figure supplement 1 (A) MBP-GFP-ORC1 binds GST-CDC6. MBP protein served as control. (B) The binding of MBP-GFP-ORC1 protein to either GST-CCD6 in the presence of Cyclin E-CDK2 (left section) or MBP-RB (right section). (C) The binding of MBP-GFP-ORC1 to MBP-RB in the presence of increasing molar amounts of Cyclin E-CDK2 (right section) or both GST-CDC6 and Cyclin E-CDK2 (left section). (D) GST-pull down assay using GST-CDC6 wild type or CDC694ARA96 mutant (CDC6A-A) with purified Cyclin E-CDK2 protein followed by Immunoblotting with Cyclin E antibody. GST protein served as control. (E) Nocodazole arrested U2OS cells were transfected with 500 ng of GFP, GFP-CDC6 wild type or CDC694ARA96 mutant (CDC6A-A) plasmids, then released into the next cell cycle. At indicated times, whole cell extracts were immunoblotted with specific antibodies against GFP and Cyclin E. α-Tubulin served as loading control. (F), CCNE1 promoter-luciferase reporter assay in U2OS cells. Cells transiently co-transfected with 500 ng of 10–4 CCNE1 promoter, 50 ng E2F1, 50 ng DP1 and 20 ng pCMV-LacZ plasmids together with increasing amounts GFP-CDC6 WT or CDC694ARA96 plasmids for 24 hr. Relative luciferase activity was normalized to co-transfected LacZ control. Experiments in triplicate. Protein expression determined by immunoblot; α-Tubulin as loading control. Statistical analysis was performed using the Student’s t test. *p<0.05; **p<0.001; ***p<0.0005. GFP, Green fluorescent protein; MBP, Maltose binding protein; GST, Glutathione S transferase.
CDC6 co-operates with Cyclin E-CDK2 to activate E2F1-dependent CCNE1 gene transcription.
Purified Proteins. Coomassie Brilliant Blue stained gel of purified MBP, MBP-GFP-ORC1, MBP-RB and GST-CDC6 proteins. MW stands for protein molecular weight marker in kilodalton. MBP, Maltose binding protein; RB, Retinoblastoma; GST, Glutathione S transferase.
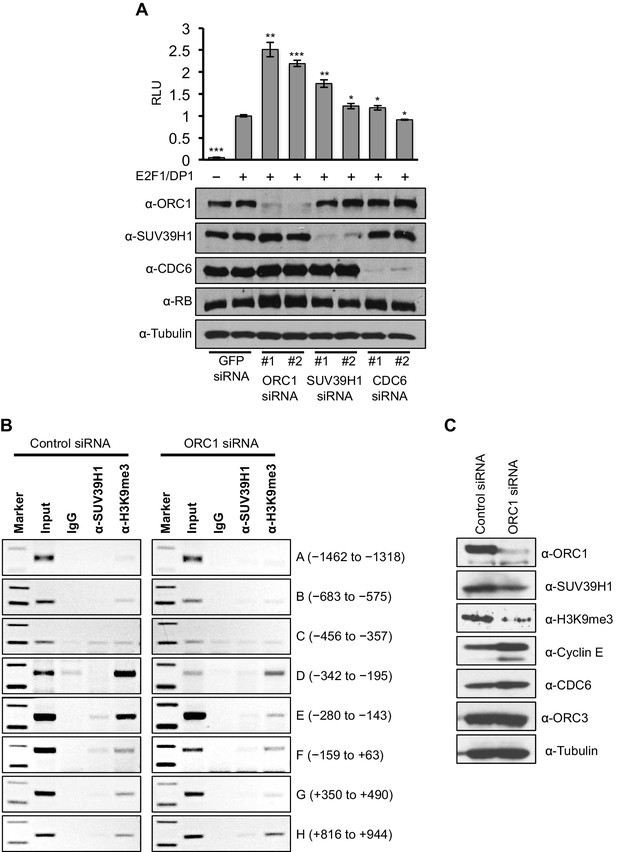
ORC1 depletion decreases association of SUV39H1 and H3K9me3 with the CCNE1 promoter and increases CCNE1 gene transcription.
(A) CCNE1-luciferase reporter assay in U2OS cells. U2OS cells transiently co-transfected with 500 ng of 10–4 CCNE1 promoter, 50 ng E2F1, 50 ng DP1 and 20 ng pCMV-LacZ. U2OS cells were also transiently transfected for 24 hr with two different siRNAs targeting either ORC1, SUV39H1 or CDC6. GFP siRNA was used as a control. Relative luciferase activity was normalized to co-transfected LacZ control. Depletion of proteins was confirmed by Immunoblot; α-Tubulin as loading control. Statistical analysis was performed using the Student’s t test. *p<0.005; **p<0.001; ***p<0.0005. (B) The CCNE1 promoter was analyzed for SUV39H1 binding and the presence of the H3K9me3 mark by ChIP assay in U2OS cells treated with either control siRNA or ORC1 siRNA for 48 hr. The experiments were done in triplicate and one experiment is shown. (C) Immunoblot of protein levels following control and ORC1 siRNA treatment at different times post nocodazole release. α-Tubulin was used as loading control.
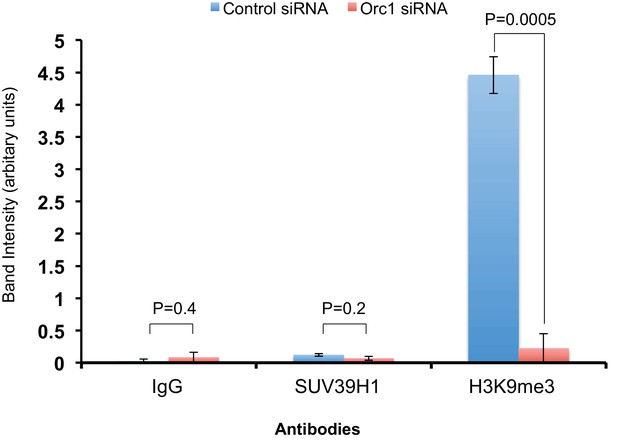
The ChIP qPCR bands were quantified using ImageJ software to analyze the extent of binding of SUV39H1 and histone H3K9me3 to the CCNE1 promoter region (-280 to -143 bp) in ORC1 siRNA treated U2OS cells compared to control siRNA-treated cells.
All values were normalized to the corresponding input DNA. Statistical analysis was performed using the Student’s t test. The p-values are indicated above the bars. The reduction in SUV39H1 was not significant due to one out of the three replicates showing little reduction in ORC1 siRNA-treated U2OS cells compared to the control. Binding of SUV39H1 to the CCNE1 promoter was also low compared to histone H3K9me3 in asynchronous U2OS cells treated with siRNAs.
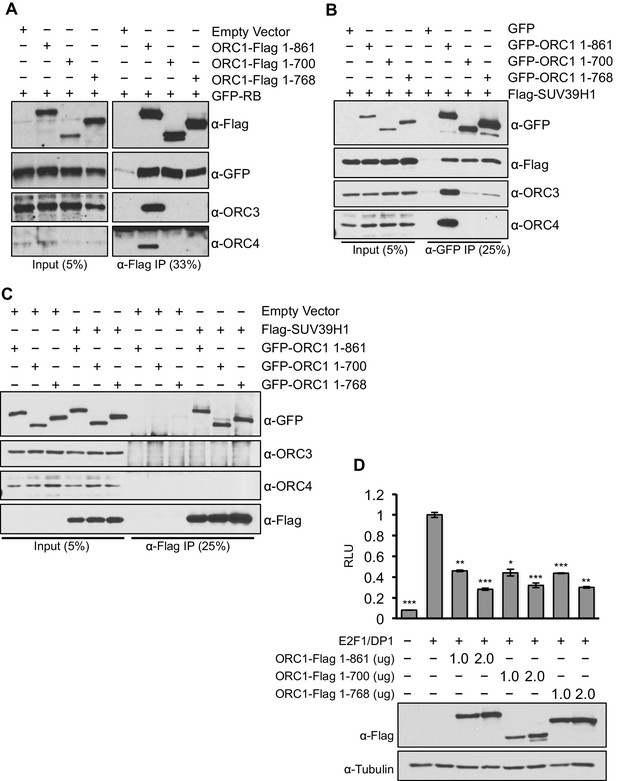
ORC1 mutants separate its role as a transcription co-repressor from its role in DNA replication.
(A) HEK293 cells were transfected with ORC1-Flag or its truncation mutants and GFP-RB as indicated. Whole cell extracts were immunoprecipitated with anti-Flag antibody and immunoprecipitates were analyzed by immuoblot with the indicated antibodies. (B and C) In vivo interaction between SUV39H1 and ORC1 or its truncation mutants. GFP-tagged wild-type ORC1 (1–861) or its truncation mutants (1–700 and 1–768) plasmids were co-transfected into HEK293 cells with Flag-SUV39H1 plasmid and either GFP-vector or empty vector as a control plasmids. Immunoprecipitation with anti-GFP antibody (B) or anti-Flag antibody (C) from cell lysates of HEK293 cells expressing the indicated constructs, followed by immunoblotting with the indicated antibodies. (D). U2OS cells transiently transfected for 24 hr with increasing amounts of wild type ORC1-Flag (1–861) or truncation mutants (1–700 and 1–768). Relative luciferase activity normalized to co-transfected lacZ control. Experiments were in triplicate. Expression of proteins determined by Immunoblot; α-Tubulin as loading control. Statistical analysis was performed using the Student’s t test. *p<0.005; **p<0.001; ***p<0.0005. ORC, Origin Recognition Complex.
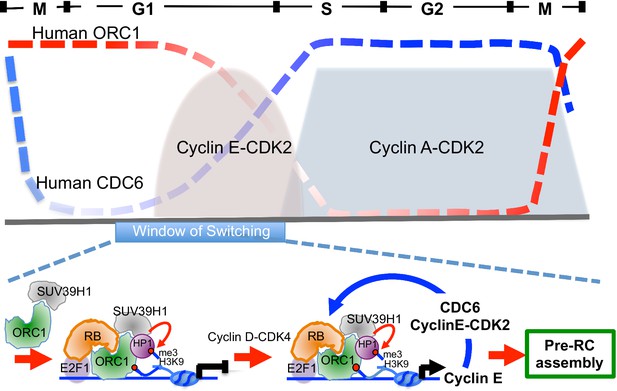
Model showing contrasting roles of DNA replication proteins ORC1 and CDC6 in the regulation of CCNE1 transcription and commitment to pre-RC assembly and cell division.
Top panel summarizes the cycle levels of ORC1 and CDC6. The bottom panel shown the ORC1 associated complexes at different stages of the cell division cycle. Blue arrow, positive feedback inhibition of ORC1-RB interaction.

Protein levels in U2OS cells and SaOS-2 cells detected by immunoblot.
https://doi.org/10.7554/eLife.12785.021Additional files
-
Supplementary file 1
Oligonucleotides employed for this research.
- https://doi.org/10.7554/eLife.12785.019
-
Supplementary file 2
DNA polymerases used for each primer pair in ChIP analysis.
- https://doi.org/10.7554/eLife.12785.020





