Regulation by the quorum sensor from Vibrio indicates a receptor function for the membrane anchors of adenylate cyclases
Figures
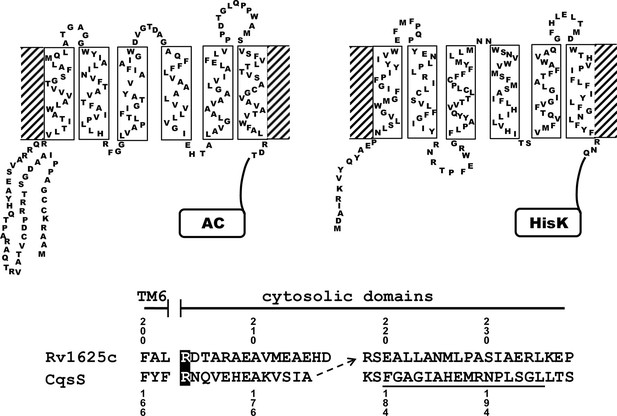
Two-dimensional models of the canonical class IIIa adenylate cyclase Rv1625c from M. tuberculosis (left) and the quorum-sensing receptor from V. harveyi (right) in the membrane.
Both proteins require dimerization to be catalytically active. The alignment below covers the amino acid sequences at the exit of TM6 of both proteins. The most efficient functional linkage of the CqsS receptor to the catalytic domain of Rv1625c is indicated by an arrow. The H-Box of the histidine-kinase domain is underlined. The numbering for CqsS and Rv1625c is indicated above and below the respective sequences.
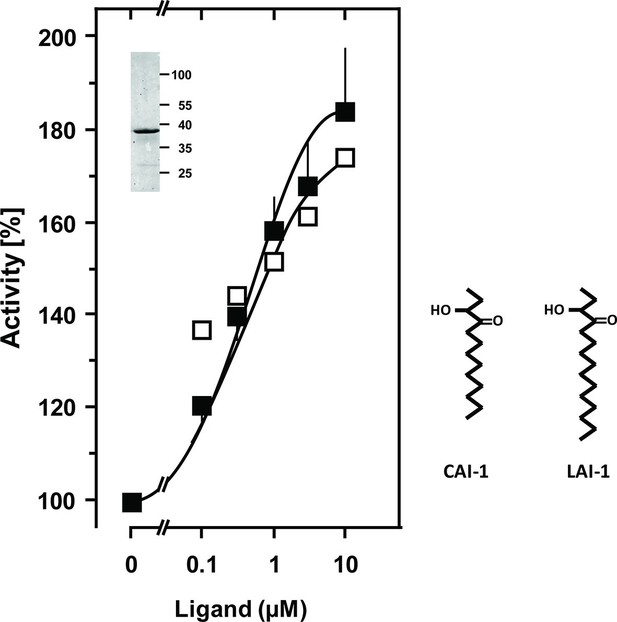
Stimulation of the chimera CqsS1-181-Rv1625c218-443 by the QS-ligands CAI-1 or LAI-1.
Basal activity was 5.5 nmol cAMP·mg-1·min-1. The EC50 concentrations were 400 nM. Filled squares, CAI-1 (n = 5–12; ± S.E.M.); open squares, LAI-1 (n= 1–2). CAI-1 stimulations were significant starting at 100 nM ligand. The insert shows a Western blot of the expression product with MW standards indicated at the side. The structure of the ligands is depicted at right. The catalytic domain of Rv1625c alone was not affected by CAI-1 or LAI-1 (not shown).
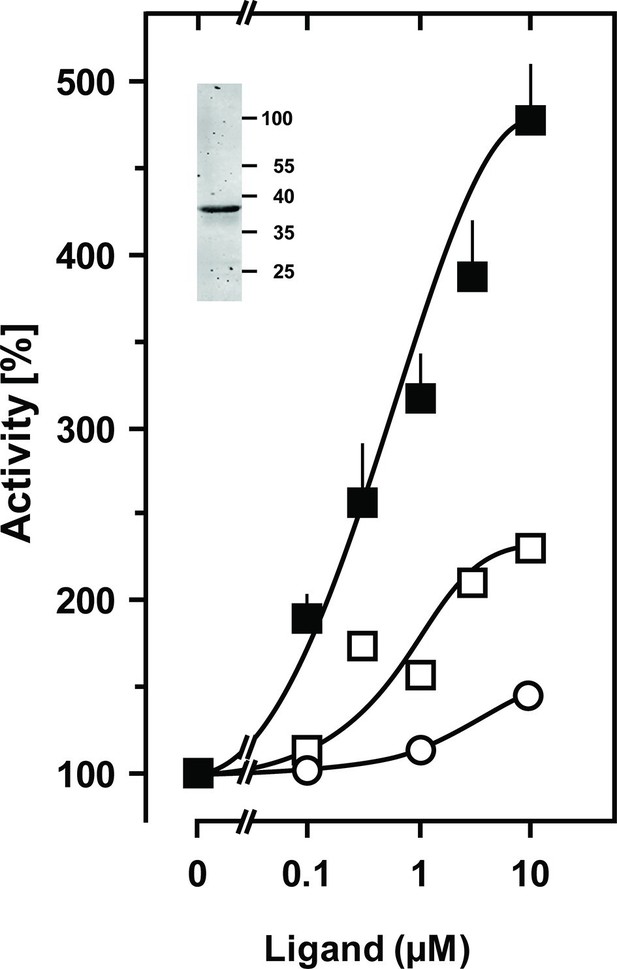
Stimulation of the chimera CqsS1-181F166L-Rv1625c218-443 by the QS-ligands CAI-1 or LAI-1.
Basal activity was 4 nmol cAMP·mg-1·min-1. Filled squares, CAI-1 (n = 5–11; ± S.E.M.); open squares, LAI-1 (n=2); open circles, 3,4-tridecanediol. The EC50 concentrations were 400 nM CAI-1, 900 nM LAI-1, and 2000 nM 3,4-tridecanediol. CAI-1 stimulations were significant starting at 100 nM ligand. Insert: Western blot of expression product.
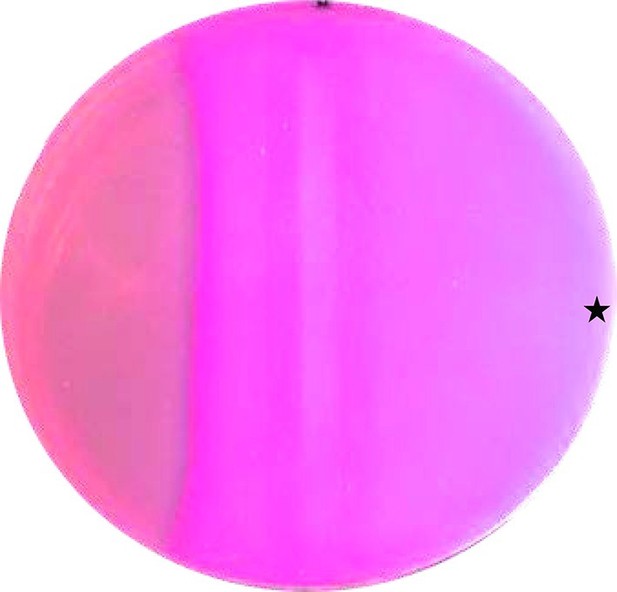
CAI-1 stimulates cAMP formation in vivo.
A MacConkey maltose agar plate with E. coli cya-99 crp*144 transformed with CqsS1-181F166L-Rv1625c218-443 was induced by a filter strip soaked with 1 mM IPTG (running from top to bottom in the middle). 10 µl of 100 µM CAI-1 in DMSO/water was spotted at the asterisk, the plate was tipped and the solution was allowed to move left. As a surface active compound it regularly spread over a large area. Note that the bacterial lawn at left was not induced. Picture was taken from the bottom of the Petri-dish (three independent experiments were carried out, each with at least three agar plates and different concentrations of CAI-1 and IPTG; controls with solvent were negative).
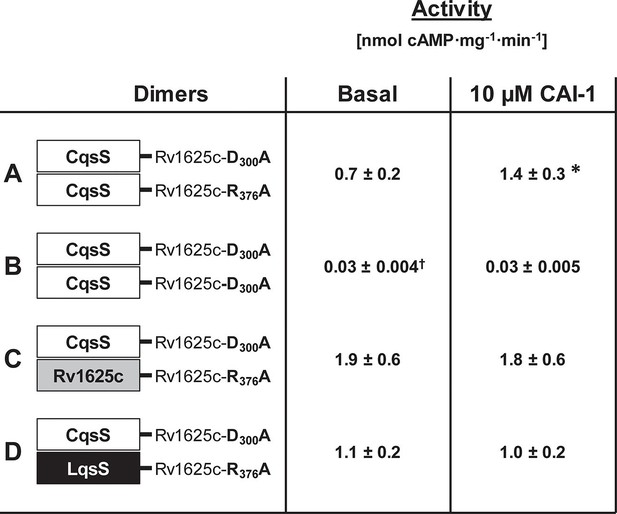
Homodimerization of the CqsS receptor is required for signaling.
(A) with complementary Rv1625c point mutations Rv1626cD300A and Rv1625cR376A a regulated dimeric chimera was generated (*p<0.05 compared to respective basal activity). (B) as a control the construct CqsS-Rv1625cD300A was expressed alone. It was inactive. (C, D) complementing mutants with differing membrane domains were active, yet unregulated. Basal activity of construct B significantly differed from those in constructs A, C, and D (†p<0.001).
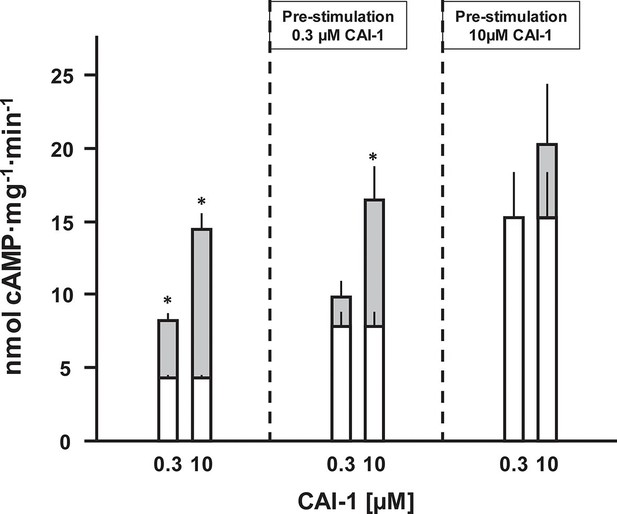
CAI-1 ligand binding to the CqsS QS-receptor is irreversible.
Membranes containing CqsS1-181F166L-Rv1625c218-443 were stimulated with 0.3 or 10 µM CAI-1 (left), re-isolated and re-stimulated with 0.3 and 10 µM CAI-1. Only the stimulations marked with an asterisk differed significantly from the respective unstimulated controls. White bars represent basal AC activities, gray bars on top represent additive CAI-1-stimulated activities (S.E.M., n = 6).
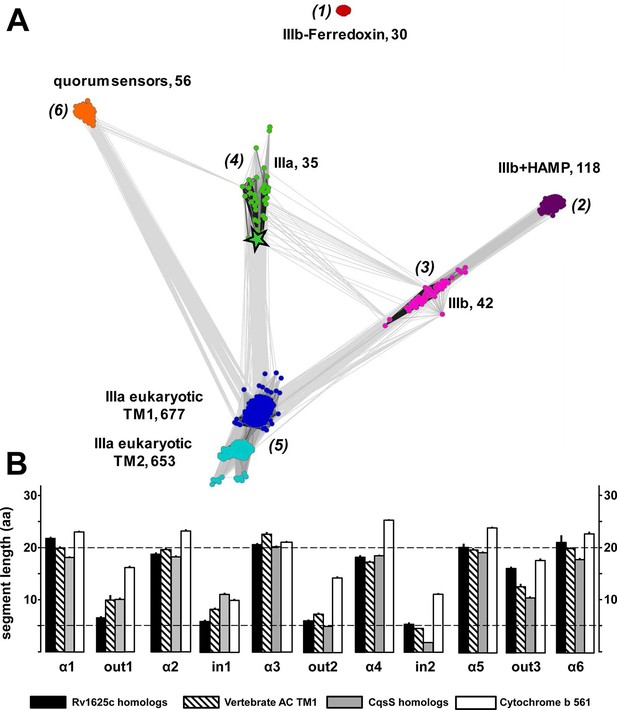
(A) Cluster map of 6TM domains of adenylate cyclases and CqsS-like sensory receptors.
A comprehensive set of 6TM AC anchors was extracted from 408 eukaryotic and 1456 bacterial proteomes and clustered in CLANS (HHsearch p-value cutoff 5E-4, attraction value 10, repulsion value 5); outliers were removed. Each dot represents a single 6TM domain. Above threshold HHsearch hits are shown as connecting lines between AC pairs in different clusters; the darker line color, the more similar the protein sequences. 6TM anchors of ACs form five clusters of high pairwise sequence similarity: cluster (1), anchors of bacterial class IIIb ACs characterized by the presence of a cytosolic ferredoxin domain (30 sequences from the α and β branches of proteobacteria). Cluster (2), anchors of bacterial class IIIb ACs characterized by a signal-transducing HAMP domain (118 sequences mainly from Actinobacteria, but also from α-proteobacteria, δ-proteobacteria, Chlorobia and Thermoleophilia). Cluster (3), anchors of bacterial class IIIb ACs similar to HAMP-associated anchor domains but which lack a HAMP domain (42 sequences mainly from α-proteobacteria). Cluster (4), anchors from bacterial class IIIa ACs prototypically represented by the mycobacterial AC Rv1625c (35 from many different phyla of bacteria, including Actinobacteria, Proteobacteria, Chlorophyta, Spirochaetes, and Bacteriodetes). The enlarged asterisk denotes the position of the mycobacterial AC Rv1625c. Cluster (5), anchors of the pseudoheterodimeric eukaryotic class IIIa ACs (TM1 677 sequences, TM2 653 sequences). CqsS Cluster (6), 6TM domains of sensory His-Kinases similar to CqsS (from Bacteriodetes, Chlorobia, α-, β-, and γ-proteobacteria). (B) Length comparisons of the transmembrane helices and loops of 6TM membrane anchors/receptors/sensors. The data sets from clusters 4, 5 and 6 from Figure 7A were used supplemented with 250 cytochrome b561 proteins. In case no S.E.M. is visible the size of the vertical bar is within the line thickness of the respective bar. ‘α’-Numbering denotes the consecutive TM helices starting from the N-terminus, 'out' and 'in’ denote sequential extra- and intracellular loop sequences. The two horizontal lines are at the 5 and 20 aa level.
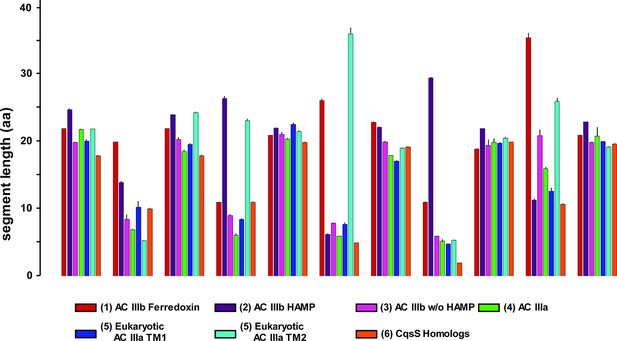
Length comparisons of α-helices and loops of 6TM modules from adenylate cyclases and quorum sensors.
Data are means ± S.E.M. of the sequence groups of all clusters shown in Figure 7A as indicated below. The color-coding used in Figure 7A has been conserved. The designations ‘in’ and ‘out’ indicate intra- and extracellular loops.
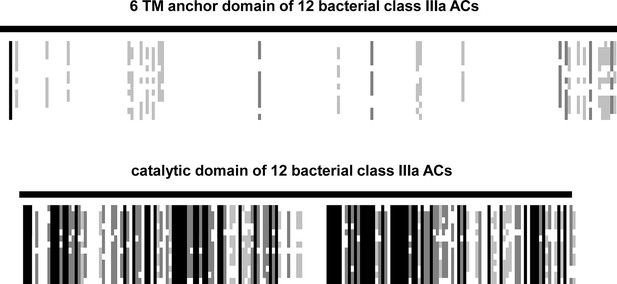
The 12 sequences used for the alignment were from Agrobacterium albertimagni, WP_006725538; Arthrospira maxima, B5VUZ0; Arthrospira platensis, D5A5G2; Beggiatoa, A7BXS6; Dechloromonas, Q47AI8; Hyphomicrobium, C6QBG1; Lyngbya, A0YQ82; Mesorhizobium, LSHC420B00; Microcoleus sp., PCC_7113; Oscillatoria acuminata, PCC_6304; Nostoc, YP_001866931; Mycobacterium tuberculosis, ALB18789 (Rv1625c).
The alignment was made by ClustalW and adjusted and converted into a ‚bargraph ‘style alignment in Genedoc. Black lines reflect sequence identity, shades of grey different degrees of similarity, and white patches denote sequence diversity. Note the striking dissimilarity of the TM domains and the conservative nature of the catalytic domains.

AC_1 Sequences used for the alignment: Macaca fasci, XP_005551359.1; Bos taurus, NP_776654.1; Anas platyrhynchos, XP_005014606.1; Danio rerio, NP_001161822.1; Mesocricetus auratus, XP_005083033.1; Pseudopodoces humilis, XP_005525280.1; Gallus gallus, XP_418883.4; Homo sapiens, NP_066939.1; Mouse, GI:62512159; Heterocephalus glaber, XP_004840600.1; Orcinus orca, XP_004283063.1; Pan troglodytes, XP_519081.3; Ficedula albicollis, XP_005041477.1; Sarcophilus harrisii, XP_003762599.1; Odobenus rosmarus divergens, XP_004409496.1; Melopsittacus undulatus, XP_005148091.1.
AC_2 sequences used for the alignment: Jaculus jaculus, XP_004670691.1; Ictalurus punctatus, AHH38946.1; Myotis brandtii, EPQ02509.1; Anas platyrhynchos, XP_005024930.1; Monodelphis domestica, XP_001363692.1; Ornithorhynchus anatinus, XP_001519046.2; Macaca mulatta, NP_001252581.1; Rattus norwegicus, NP_112269.1; Rabbit, XP_002721866.1; Latimeria chalumnae, XP_006007585.1; Mouse, Q80TL1; Callithrix jacchus, XP_002745178.1; Homo sapiens, Q08462.5; Ficedula albicollis, XP_005041747.1; Columba livia, XP_005508897.1; Dog, XP_535798.3; Bos taurus, XP_587884.4; Myotis brandtii, EPQ02509.1; Alligator mississippiensis, XP_006274555.1. On top are the joint alignments of TM1 and TM2 of AC1 and AC2, respectively. Similarities are limited. Deconstruction of the AC1 and AC2 alignment results in individual AC1 and AC2 alignments. The similarities within one isoform throughout evolution are striking.
Additional files
-
Supplementary file 1
List of sense (s) and antisense (as) primers used for generating the diversity of chimeras used in this study.
- https://doi.org/10.7554/eLife.13098.013





