Nucleosome breathing and remodeling constrain CRISPR-Cas9 function
Figures
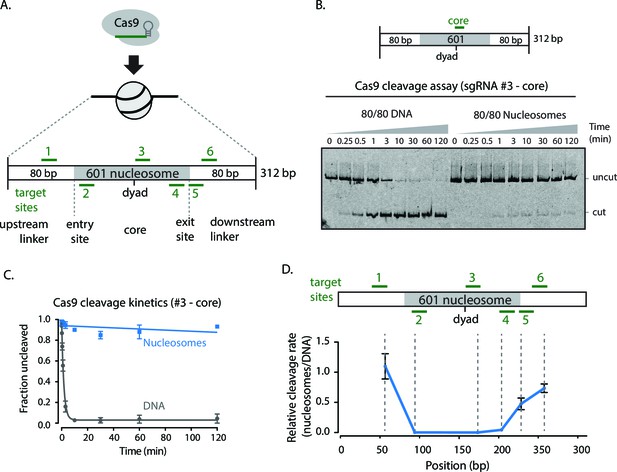
Cas9 DNA nuclease activity is hindered by nucleosomes.
(A) Schematic of sgRNAs designed against the assembled 601 80/80 nucleosome substrates targeting the flanking regions, entry/exit sites, and near the nucleosomal dyad. (B) Cleavage assay comparing Cas9 cleavage on 80/80 DNA and 80/80 nucleosomes when loaded with sgRNA #3. (C) Kinetics of cleavage with sgRNA #3. (D) Comparison of the relative rates of cleavage on nucleosomes to DNA at various positions along the 80/80 nucleosome construct. The position reported is the site of cleavage by Cas9. Represented values are mean ± SEM from three replicates.
-
Figure 1—source data 1
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNAs #2 and #6.
- https://doi.org/10.7554/eLife.13450.004
-
Figure 1—source data 2
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNAs #2 and #6.
- https://doi.org/10.7554/eLife.13450.005
-
Figure 1—source data 3
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNAs #2 and #6.
- https://doi.org/10.7554/eLife.13450.006
-
Figure 1—source data 4
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNA #5.
- https://doi.org/10.7554/eLife.13450.007
-
Figure 1—source data 5
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNA #5.
- https://doi.org/10.7554/eLife.13450.008
-
Figure 1—source data 6
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNA #1.
- https://doi.org/10.7554/eLife.13450.009
-
Figure 1—source data 7
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNA #1.
- https://doi.org/10.7554/eLife.13450.010
-
Figure 1—source data 8
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNA #3.
- https://doi.org/10.7554/eLife.13450.011
-
Figure 1—source data 9
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNA #3.
- https://doi.org/10.7554/eLife.13450.012
-
Figure 1—source data 10
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNA #4.
- https://doi.org/10.7554/eLife.13450.013
-
Figure 1—source data 11
Replicate gels of Cas9 cleavage of 80/80 601 DNA and nucleosomes with sgRNA #4.
- https://doi.org/10.7554/eLife.13450.014
-
Figure 1—source data 12
Quantification of Figure 1 Cas9 cleavage gels.
- https://doi.org/10.7554/eLife.13450.015
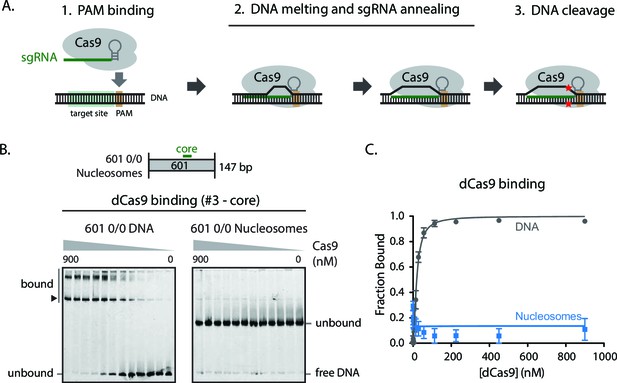
Nucleosome positioning blocks Cas9 from binding PAM sites on DNA.
(A) Schematic illustrating the stepwise mechanism of Cas9 binding to DNA targets and subsequent nucleolytic cleavage. (B) Gel shift assay comparing dCas9 binding to 0/0 DNA and nucleosomes while loaded with sgRNA #3 targeting the nucleosome dyad. Band shift pattern appears as discrete lower band (dCas9 bound to sgRNA-specified target, arrowhead) and higher, super shift bands (additional dCas9 PAM-only binding events). (C) Quantification of (B). Represented values are mean ± SEM from three replicates.
-
Figure 1—figure supplement 1—source data 1
-3Replicate gels of dCas9 binding to 0/0 601 DNA and nucleosomes with sgRNA #3.
- https://doi.org/10.7554/eLife.13450.017
-
Figure 1—figure supplement 1—source data 2
-3Replicate gels of dCas9 binding to 0/0 601 DNA and nucleosomes with sgRNA #3.
- https://doi.org/10.7554/eLife.13450.018
-
Figure 1—figure supplement 1—source data 3
-3Replicate gels of dCas9 binding to 0/0 601 DNA and nucleosomes with sgRNA #3.
- https://doi.org/10.7554/eLife.13450.019
-
Figure 1—figure supplement 1—source data 4
Quantification of Figure 1—figure supplement 1 gel shifts.
- https://doi.org/10.7554/eLife.13450.020
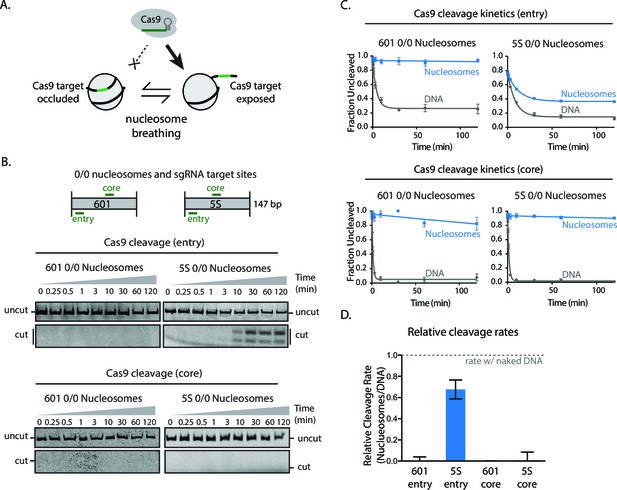
Higher nucleosomal breathing dynamics enhance Cas9 cleavage.
(A) Schematic illustrating nucleosome breathing and how it can enable Cas9 binding to a target in the nucleosome. (B) Cleavage assay comparing Cas9 cleavage of 601 and 5S 0/0 nucleosomes when loaded with sgRNAs targeting comparable positions at core and entry sites. (C) Quantification of (B). (D) Cas9 cleavage rates on 601 and 5S nucleosomes when targeted to core and entry sites. Values were normalized against naked DNA control rates. Represented values are mean ± SEM from three replicates. Additional gel panels shown in Figure 2—figure supplement 1.
-
Figure 2—source data 1
Replicate gels of cleavage of 0/0 5S DNA and nucleosomes with sgRNA core.
- https://doi.org/10.7554/eLife.13450.022
-
Figure 2—source data 2
Replicate gels of cleavage of 0/0 5S DNA and nucleosomes with sgRNA core.
- https://doi.org/10.7554/eLife.13450.023
-
Figure 2—source data 3
Replicate gels of cleavage of 0/0 5S DNA and nucleosomes with sgRNA entry.
- https://doi.org/10.7554/eLife.13450.024
-
Figure 2—source data 4
Replicate gels of cleavage of 0/0 5S DNA and nucleosomes with sgRNA entry.
- https://doi.org/10.7554/eLife.13450.025
-
Figure 2—source data 5
Replicate gels of cleavage of 0/0 601 DNA and nucleosomes with sgRNA entry.
- https://doi.org/10.7554/eLife.13450.026
-
Figure 2—source data 6
Replicate gels of cleavage of 0/0 601 DNA and nucleosomes with sgRNA entry.
- https://doi.org/10.7554/eLife.13450.027
-
Figure 2—source data 7
Quantification of Figure 2 Cas9 cleavage gels.
- https://doi.org/10.7554/eLife.13450.028
-
Figure 2—source data 8
Quantification of Figure 2 Cas9 cleavage gels.
- https://doi.org/10.7554/eLife.13450.029
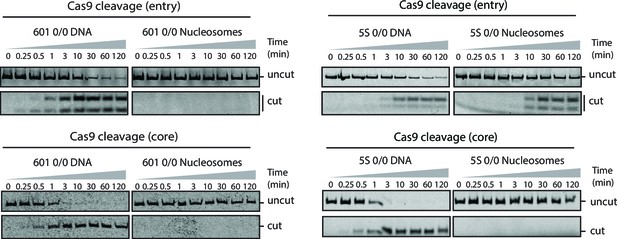
Cas9 cleavage assay with 601 and 5S 0/0 nucleosomes.
Representative gel images of Cas9 cleavage experiments with 601 (left) and 5S (right) 0/0 particles using sgRNAs targeting entry (top) or core (bottom) sites, including DNA control experiments. Samples were resolved on 12% (entry) or 8% (core) polyacrylamide gels. Cleavage with sgRNAs targeting the entry site on both 5S and 601 substrates generates a short cleavage product (13 bp) which partially denatures and runs as two bands (single and double stranded) on 12% polyacrylamide gels.
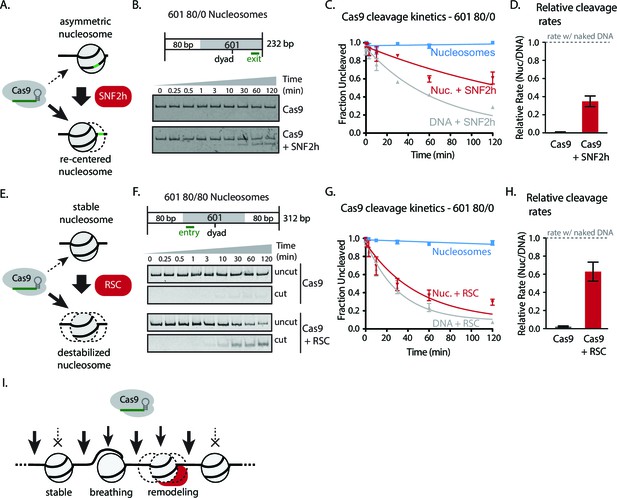
Chromatin remodeling improves Cas9 cleavage of nucleosomal substrates.
(A) Schematic of Cas9 cleavage assay with remodeling. Cas9 is presented with 601 nucleosomes either untreated or previously remodeled with SNF2h or RSC remodelers. (B) Assay comparing cleavage on untreated and remodeled 80/0 nucleosomes when Cas9 is targeted to exit site (depicted in green). These asymmetric nucleosomes are recentered by SNF2h, exposing the exit target site to Cas9 (C) Quantification of (B). (D) Cleavage rates of 80/0 nucleosomes by Cas9 relative to naked DNA, in the presence or absence of SNF2h. SNF2h improves Cas9 cleavage to ~35% of the naked DNA cleavage rate. (E) Assay comparing Cas9-mediated cleavage at entry site of 80/80 symmetric 601 nucleosomes, either untreated or previously treated with RSC remodeler. RSC can destabilize nucleosome structure and reposition nucleosomes towards the DNA ends. (F) Quantification of (E) (G) Comparison of the rates of cleavage of nucleosomes normalized to DNA control with and without the action of RSC chromatin remodeler. Mean enhancement rates of Cas9 activity by chromatin remodeling are shown. (H) Cleavage rates of 80/80 nucleosomes by Cas9 relative to naked DNA, in the presence or absence of RSC. Cas9 cleavage is substantially enhanced by RSC, attaining ~63% of the naked DNA cleavage rate. Represented values are mean ± SEM from three replicates. Additional gel panels shown in Figure 3—figure supplement 1. (I) Model of Cas9 activity in vivo in eukaryotes. Left, stable and strongly positioned nucleosomes impede Cas9 activity (downward arrows). However, nucleosomes in vivo are generally more dynamic (breathing), allowing Cas9 opportunities to target underlying DNA (center). Cas9 accessibility to nucleosomal DNA can be further enhanced by the activity of chromatin remodelers that destabilize and/or reposition nucleosomes (right).
-
Figure 3—source data 1
Replicate gels of cleavage of 80/0 DNA and nucleosomes using sgRNA #4 with or without prior remodeling by Snf2h.
- https://doi.org/10.7554/eLife.13450.032
-
Figure 3—source Data 2
Replicate gels of cleavage of 80/0 DNA and nucleosomes using sgRNA #4 with or without prior remodeling by Snf2h.
- https://doi.org/10.7554/eLife.13450.033
-
Figure 3—source data 3
Replicate gels of cleavage of 80/0 DNA and nucleosomes using sgRNA #4 with or without prior remodeling by Snf2h.
- https://doi.org/10.7554/eLife.13450.034
-
Figure 3—source data 4
Quantification of Cas9 cleavage gels from Figure 3—source data 1–3.
- https://doi.org/10.7554/eLife.13450.035
-
Figure 3—source data 5
Replicate gels of cleavage of 80/80 DNA and nucleosomes using sgRNA 601_2 with or without prior remodeling by RSC.
- https://doi.org/10.7554/eLife.13450.036
-
Figure 3—source data 6
Replicate gels of cleavage of 80/80 DNA and nucleosomes using sgRNA 601_2 with or without prior remodeling by RSC.
- https://doi.org/10.7554/eLife.13450.037
-
Figure 3—source data 7
Replicate gels of cleavage of 80/80 DNA and nucleosomes using sgRNA 601_2 with or without prior remodeling by RSC.
- https://doi.org/10.7554/eLife.13450.038
-
Figure 3—source data 8
Quantification of Cas9 cleavage gels from Figure 3—source data 5–7.
- https://doi.org/10.7554/eLife.13450.039
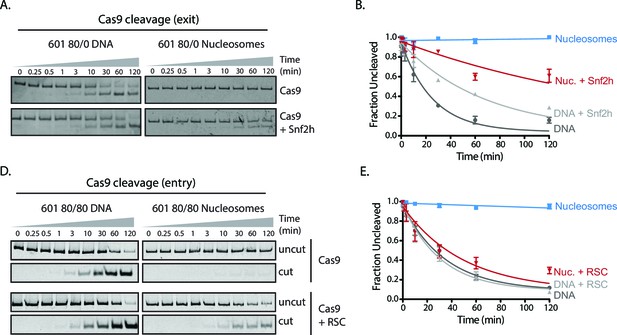
Cas9 cleavage assays with SNF2h and RSC chromatin remodelers.
(A) Representative gel images of Cas9 cleavage experiments with 601 80/0 asymmetric particles and SNF2h chromatin remodeler, including DNA control experiments. Cas9 was loaded with sgRNA targeting the exit site of the nucleosome. SNF2h re-centers asymmetric nucleosomes such as 80/0 (Figure 3—figure supplement 3), exposing the Cas9 target site. (B) Quantification of (A). (C) Representative gel images of Cas9 cleavage experiments with 601 80/80 symmetric nucleosomes and RSC chromatin remodeler, including DNA control experiments. Cas9 was targeted to the entry site of the nucleosome. RSC destabilizes nucleosomal structure and repositions nucleosomes to the ends of the DNA molecule. (D) Quantification of (C). Represented values are mean ± SEM from three replicates.
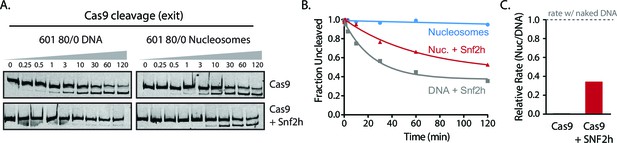
Simultaneous chromatin remodeling and Cas9 cleavage of nucleosomal substrates.
(A) Assay comparing Cas9 cleavage of 601 80/0 nucleosomes simultaneously with chromatin remodeling byr SNF2h. The 601 80/0 asymmetric nucleosomes are recentered by SNF2h, exposing the exit target site to Cas9 (B) Quantification of (A). (C) Cleavage rates of 80/0 nucleosomes by Cas9 relative to naked DNA, in the presence or absence of SNF2h. SNF2h improves Cas9 cleavage to ~40% of the naked DNA cleavage rate, similarly to sequential remodeling and cleavage assays shown in Figure 3A–D. Results shown for single experiment performed.
-
Figure 3—figure supplement 2—source data 1
Gel of cleavage of 80/0 DNA and nucleosomes using sgRNA #4 with or without simultaneous remodeling by Snf2h.
- https://doi.org/10.7554/eLife.13450.042
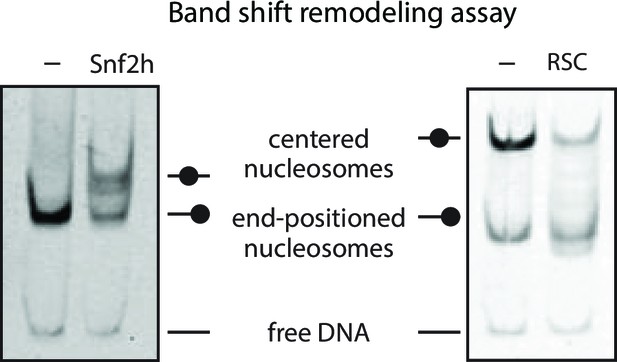
SNF2h and RSC remodel nucleosomes prior to Cas9 cleavage.
Gel‐shift nucleosome remodeling assay comparing positioned and SNF2h‐remodeled 80/0 nucleosomes (left) or RSC-remodeled 80/80 nucleosomes (right). Migration pattern for all three forms (centered, end-positioned nucleosomes and free DNA) is illustrated (center). Remodeling reactions were carried out 1 hr before being quenched and run on a 5% Acrylamide 0.5x TBE gel. SNF2h remodels 80/0 end‐positioned nucleosomes in a range of nucleosome positions biased towards the center of the DNA molecule, whereas RSC has the opposite bias of repositioned centered nucleosomes towards the ends of the DNA molecule. Images shown for single experiments performed, as previously described (Yang et al., 2006; Rowe and Narlikar, 2010).
-
Figure 3—figure supplement 3—source data 1
Test remodeling gel of 80/0 nucleosomes with Snf2h.
- https://doi.org/10.7554/eLife.13450.044
-
Figure 3—figure supplement 3—source data 2
Test remodeling gel of 80/80 nucleosomes with RSC.
- https://doi.org/10.7554/eLife.13450.045
Tables
Spacer sequences for sgRNAs used in biochemistry experiments.
| sgRNA # | Guide sequence | PAM | Target strand | Figures where used |
|---|---|---|---|---|
| 601_1 | CGAGTTCATCCCTTATGTGA | TGG | Antisense | Figure 1D |
| 601_2 (entry) | AATTGAGCGGCCTCGGCACC | GGG | Sense | Figure 1D, Figure 2B–D, Figure 2—figure supplement 1, Figure 3E–H, Figure 3—figure supplement 1D–E |
| 601_3 (core) | CCCCCGCGTTTTAACCGCCA | AGG | Antisense | Figure 1B–D, Figure 1—figure supplement 1B–C, Figure 2B–D, Figure 2—figure supplement 1 |
| 601_4 | GTATATATCTGACACGTGCC | TGG | Sense | Figure 1D |
| 601_5 | TCGCTGTTCAATACATGCAC | AGG | Sense | Figure 1D |
| 601_6 | GCGACCTTGCCGGTGCCAGT | CGG | Antisense | Figure 1D |
| 5S_1 (entry) | TCTGATCTCTGCAGCCAAGC | AGG | Sense | Figure 2B–E, Figure 2—figure supplement 1 |
| 5S_2 (core) | TATGGCCGTAGGCGAGCACA | AGG | Antisense | Figure 2B–E, Figure 2—figure supplement 1 |
Sequences for DNA molecules used for biochemical assays (Positioning sequence highlighted in grey).
| Name | Sequence |
|---|---|
| 601 80/80 | CGGGATCCTAATGACCAAGGAAAGCATGATTCTTCACACCGAGTTCATCCCTTATGTGATGGACCCTATACGCGGCCGCCCTGGAGAATCCCGGTGCCGagGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGTGCATGTATTGAACAGCGACCTTGCCGGTGCCAGTCGGATAGTGTTCCGAGCTCCCACTCTAGAGGATCCCCGGGTACCGA |
| 601 0/0 | CTGGAGAATCCCGGTGCCGagGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGT |
| 601 80/0 | CGGGATCCTAATGACCAAGGAAAGCATGATTCTTCACACCGAGTTCATCCCTTATGTGATGGACCCTATACGCGGCCGCCCTGGAGAATCCCGGTGCCGagGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGT |
| 5S 0/0 | GGCCCGACCCTGCTTGGCTGCAGAGATCAGACGATATCGGGCACTTTCAGGGTGGTATGGCCGTAGGCGAGCACAAGGCTGACTTTTCCTCCCCTTGTGCTGCCTTCTGGGGGGGGCCCAGCCGGATCCCCGGGCGAGCTCGAATT |





