Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor
Figures
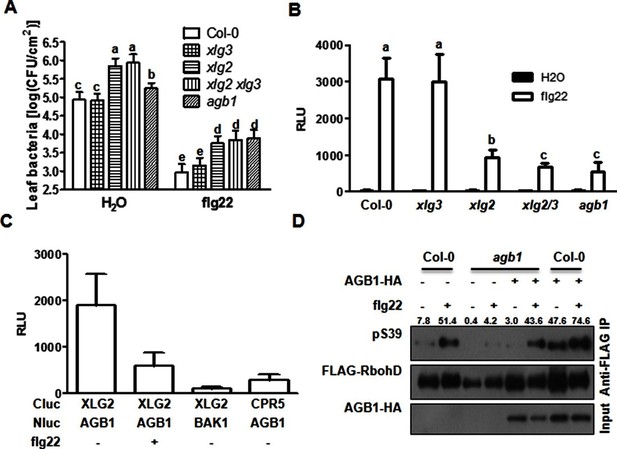
G proteins are required for FLS2-mediated immunity.
(A) XLG2/3 and AGB1 play overlapping but not identical roles in disease resistance to Pst. Plants of indicated genotypes were infiltrated with H2O and flg22 1 day before infiltration with P. syringae DC3000, and bacteria number was determined 2 days later (mean ± SD; n ≥ 6; p<0.05, Student’s t-test; different letters indicate significant difference). (B) xlg2/3 and agb1 are similarly compromised in flg22-induced ROS burst. Leaves of the indicated genotypes were examined for flg22-induced ROS production, and peak RLU values are shown (mean ± SD; n ≥ 6; p<0.05, Student’s t-test; different letters indicate significant difference). (C) Flg22 treatment disrupts XLG2-AGB1 interaction. Cluc-XLG2 and AGB1-HA-Nluc constructs are transiently expressed in Nb leaves, relative luminescence unit (RLU) was measured 2 days later. Cluc-CPR5 and BAK1-HA-Nluc were used as negative control (mean ± SD; n ≥ 6). (D) Flg22-induced RbohD phosphorylation is impaired in agb1. FLAG-RbohD and/or AGB1-HA constructs were expressed under control of the 35S promoter in WT or agb1 protoplasts. The FLAG-RbohD protein was affinity purified and subject to anti-FLAG and anti-pSer39 immuoblot analyses. Numbers indicate arbitrary units of RbohD pS39 phosphorylation calculated from densitometry measurements normalized to total FLAG-RbohD protein. Each experiment was repeated three times, and data of one representative experiment are shown.
-
Figure 1—source data 1
Raw data and exact p value of Figure 1A, B and Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.13568.004
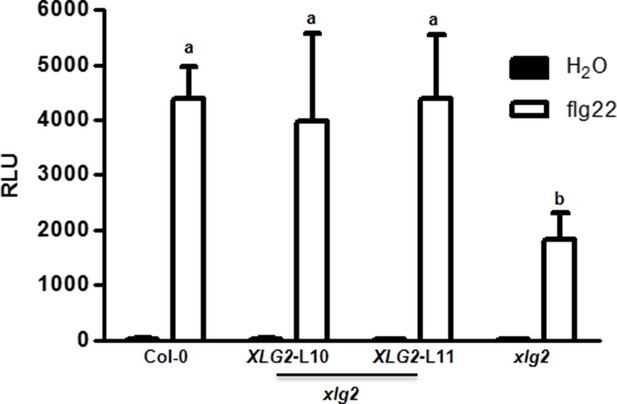
Flg22-induced ROS burst is compromised in xlg2 plants.
Flg22-induced ROS burst is compromised in xlg2 plants. Col-0, xlg2, and xlg2 transgenic lines complemented with the XLG2 transgene under control of the XLG2 native promoter were examined for flg22-induced ROS burst. RLU represent peak value of ROS production after flg22 treatment (mean ± SD; n ≥ 6; representative data from three independent experiments are shown).
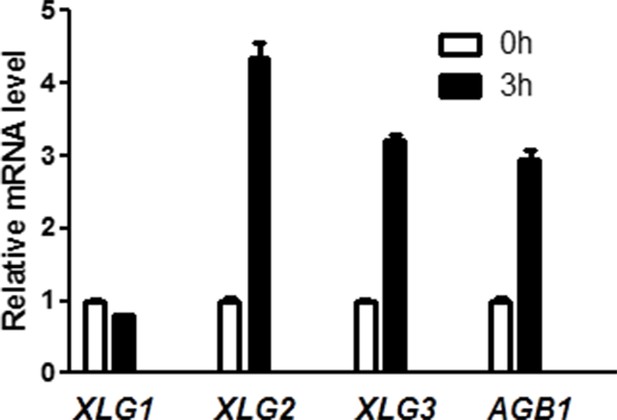
XLG2/3 and AGB1, but not XLG1, are transcriptionally induced by flg22.
qRT-PCR analyses of the indicated genes in WT plants 0 hr and 3 hr after infiltration with flg22. Representative data from three independent experiments are shown.
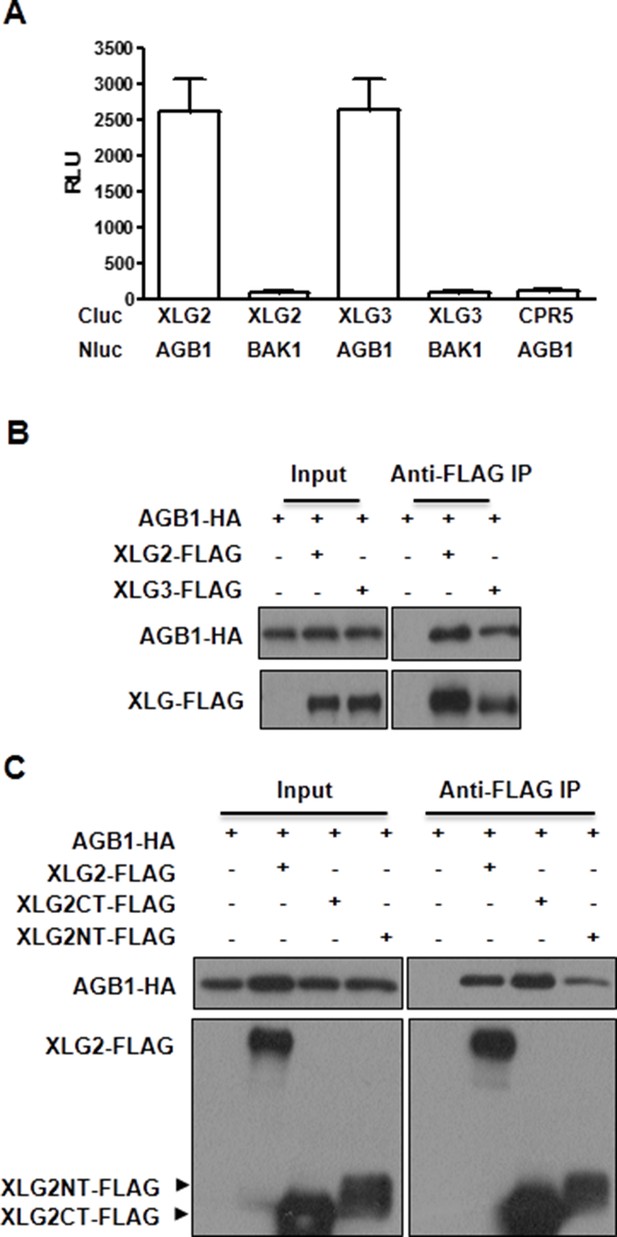
XLG2/3 interact with AGB1 through both N and C termini.
(A) Luciferase complementation assay for XLG2/3-AGB1 interactions in Nb plants. Nb leaves were infiltrated with Agrobacterium tumefaciens strains carrying the indicated constructs and luciferase activity was recorded 2 days later (mean ± SD; n≥6; representative data from three independent experiments are shown). (B) Co-IP assay for XLG2/3-AGB1 interaction in protoplasts. Three independent experiments were performed with similar results. (C) Both N and C termini of XLG2 interact with AGB1. XLG2NT-FLAG contains amino acids 1–458 whereas XLG2CT-FLAG contains amino acids 459–861. The indicated constructs were expressed in WT protoplasts, immunoprecipitated with agarose-conjugated FLAG antibody, and the immune complex was subject to immunoblot with specific antibodies.
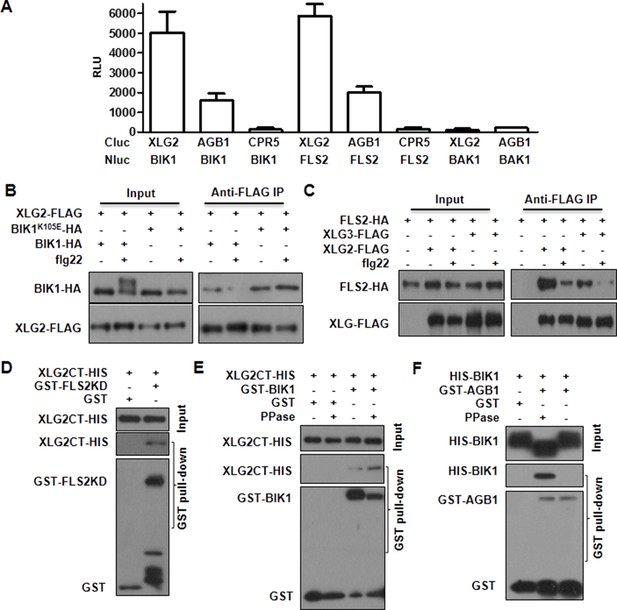
Flg22 regulates interactions between G proteins and the FLS2-BIK1 receptor complex.
(A) XLG2 and AGB1 interact with BIK1 and FLS2 in Nb plants. The indicated Nluc and Cluc constructs were transiently expressed in Nb plants for luciferase complementation assay. Relative luminescence unit (RLU) shows the strength of protein-protein interaction (mean ± SD; n ≥ 6. (B) XLG2 interacts with BIK1 in Arabidopsis protoplasts and the interaction is dynamically regulated by flg22. (C) XLG2/3 interact with FLS2 in Arabidopsis protoplasts and the interaction is dynamically regulated by flg22. The indicated constructs were co-expressed in WT protoplasts, and Co-IP assays were performed using agarose-conjugated anti-FLAG antibody. BIK1K105E carries a mutation in the ATP-binding site. (D) The C terminus of XLG2 directly interacts with FLS2 kinase domain. XLG2CT-HIS (amino acids 459–861) was incubated with GST or GST-FLS2KD (FLS2 kinase domain) for GST pull-down assay and detected by anti-HIS and anti-GST immunoblots. (E) XLG2 primarily interacts with non-phosphorylated BIK1. XLG2CT-HIS was incubated with GST or GST-BIK1 that was untreated or pre-treated with λ phosphatase (PPase), and GST pull-down assay was performed. (F) AGB1 interacts with the non-phosphorylated BIK1. Untreated or PPase-treated BIK1-HIS was incubated with GST or GST-AGB1, and GST pull-down assay was performed. Each experiment was repeated two (D–F) or three (A–C) times, and data of one representative experiment are shown.
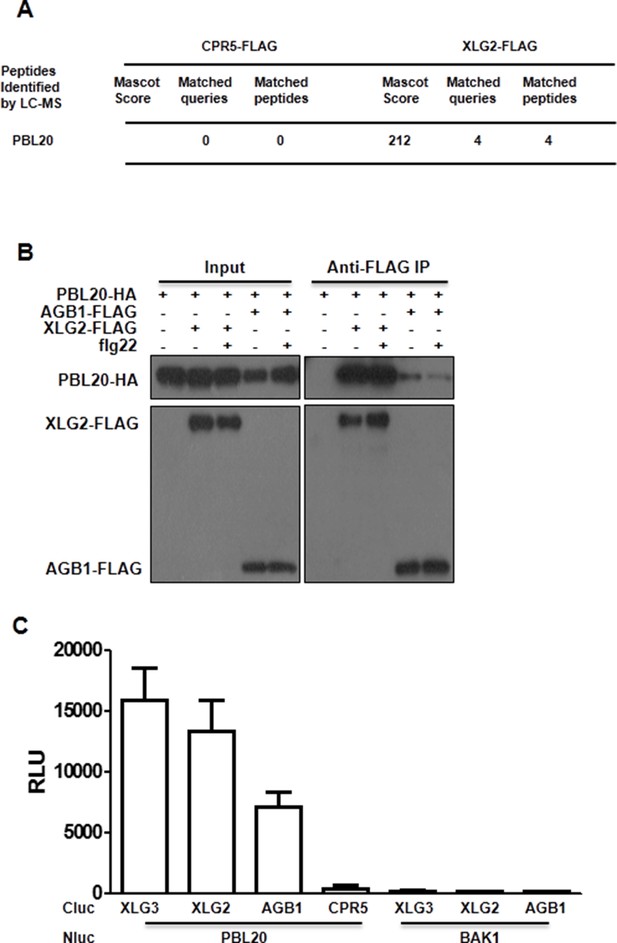
PBL20 interacts with G proteins.
(A) Identification of PBL20 as a XLG2-interacting protein. XLG2-FLAG was expressed in protoplasts, isolated by anti-FLAG immunoprecitation and subject to LC-MS/MS. No PBL20 peptides were identified in the control experiment using CPR5-FLAG as bait. (B) PBL20 interacts strongly with XLG2 and weakly with AGB1 in Arabidopsis protoplasts. Co-IP assay was performed using Arabidopsis protoplasts transfected with the indicated constructs. Three independent experiments were performed with similar results. (C) PBL20 interacts with XLG2 in Nb plants. Nb leaves were infiltrated with Agrobacteria containing the indicated constructs and luciferase activity was recorded 2 days later (mean ± SD; n≥6; representative data from two independent experiments are shown).
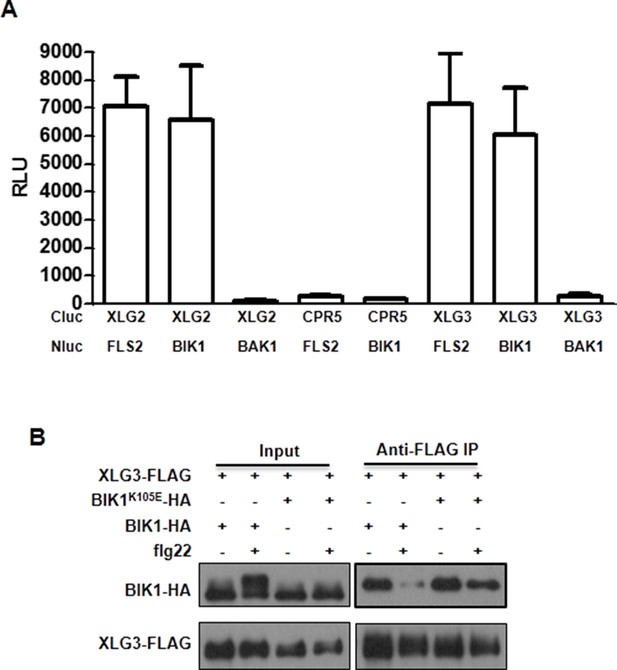
XLG3 interacts with BIK1.
(A) XLG3 interacts with BIK1 in Nb plants. Agrobacteria containing the indicated constructs were infiltrated into Nb leaves, and luciferase activity was recorded 2 days later (mean ± SD; n ≥ 6). (B) XLG3 interacts with BIK1 in protoplasts. Co-IP assay was performed using WT Arabidopsis protoplasts transfected with the indicated constructs. Representative data from three independent experiments are shown.
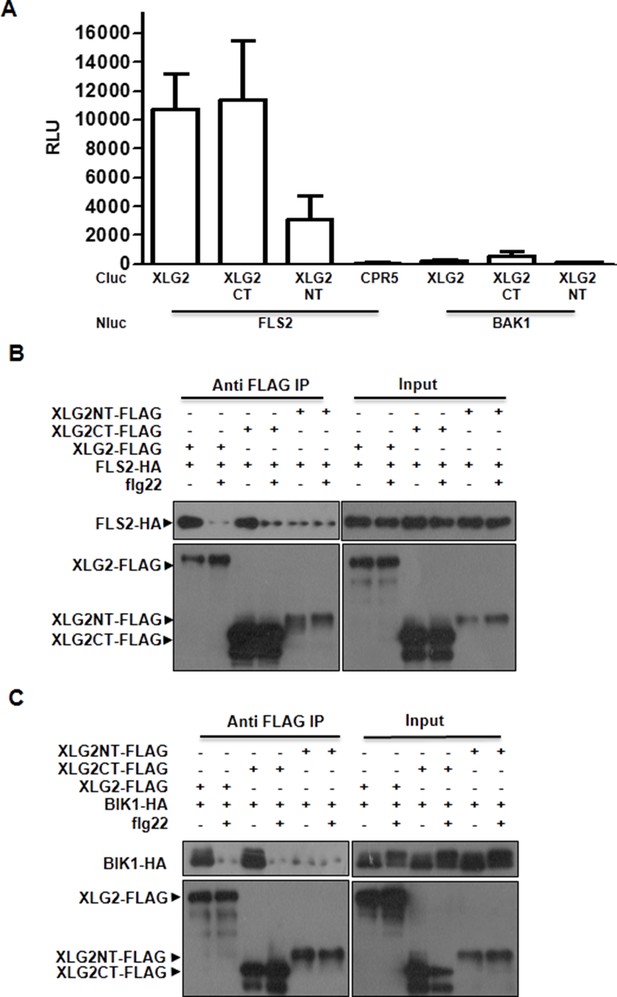
XLG2 interacts with FLS2 and BIK1 primarily through the C terminus.
(A) XLG2 C terminus interacts with FLS2 in Nb plants. Agrobacteria carrying the indicated constructs were infiltrated into Nb leaves, and luciferase activity was recorded 2 days later (mean ± SD; n≥6). (B) XLG2 C terminus interacts with BIK1. (C) XLG2 C terminus interacts with FLS2. WT Arabidopsis protoplasts were transfected with the indicated constructs, and total protein was subject to Co-IP assays. Two independent experiments were performed with similar results.
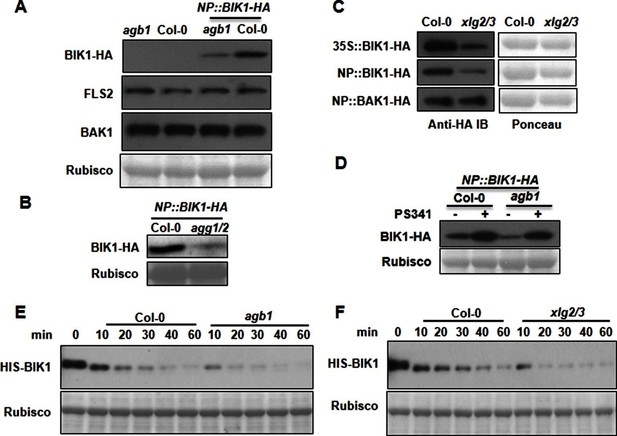
G proteins positively regulate immunity and BIK1 stability.
(A) AGB1 is required for accumulation of BIK1, but not FLS2 and BAK1. BIK1-HA was introduced into agb1 by crossing, homozygotes of the indicated genotypes in F3 generation were used for immunoblot analyses. (B) AGG1/2 are required for BIK1 stability. NP::BIK1-HA was introduced into agg1/2 by crossing, homozygous plants in F3 generation were subject to immunoblot analyses. (C) XLG2/3 are required for BIK1 accumulation. NP::BIK1-HA, 35S::BIK1-HA and NP::BAK1-HA plasmids were transiently expressed in WT and xlg2/3 protoplasts, and accumulation of BIK1 and BAK1 was determined by immunoblot analyses. (D) AGB1 regulates BIK1 accumulation through the proteasome pathway. One-week-old NP::BIK1-HA seedlings of WT (Col-0) or agb1 background were pretreated with DMSO (-) or 100 μM proteasome inhibitor PS341(+) for 8 hr before total protein was isolated for immunoblot analysis. (E) The agb1 extract shows accelerated degradation of BIK1 in vitro (F) The xlg2 xlg3 extract shows accelerated degradation of BIK1. Total extracts from WT (Col-0), agb1 and xlg2 xlg3 seedlings were incubated with HIS-BIK1 protein at 22°C for the indicated times, and equal amounts of sample were analyzed using anti-HIS immunoblot. Each experiment was repeated at least three times, and data from one representative experiment are shown.
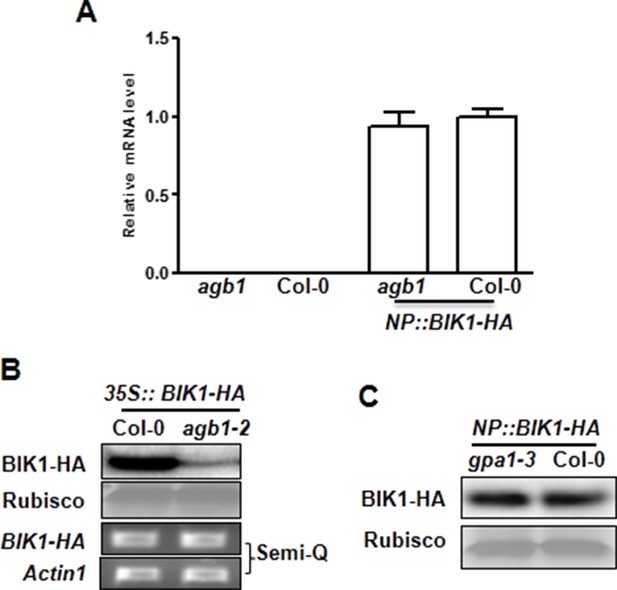
G proteins are required for BIK1 stability.
(A) agb1 plants are largely normal in BIK1 transcription. Total RNA was isolated from plants of the indicated genotypes, and quantitative real time PCR was carried out to determine BIK1-HA transcript levels. (B) Accumulation of BIK1 expressed from a transgene under control of the constitutive 35S promoter was similarly compromised in the agb1-2 mutant. 35S::BIK1-HA transgenic lines in WT and agb1-2 background with similar BIK1 transcript levels were identified by Semi-qPCR and used for detection of BIK1 accumulation. (C) Accumulation of BIK1 is not compromised in the gpa1-3 mutant. NP::BIK1-HA was introduced into gpa1-3 by crossing, and BIK1 accumulation was detected by anti-HA immunoblot. Three independent experiments were performed with similar results.
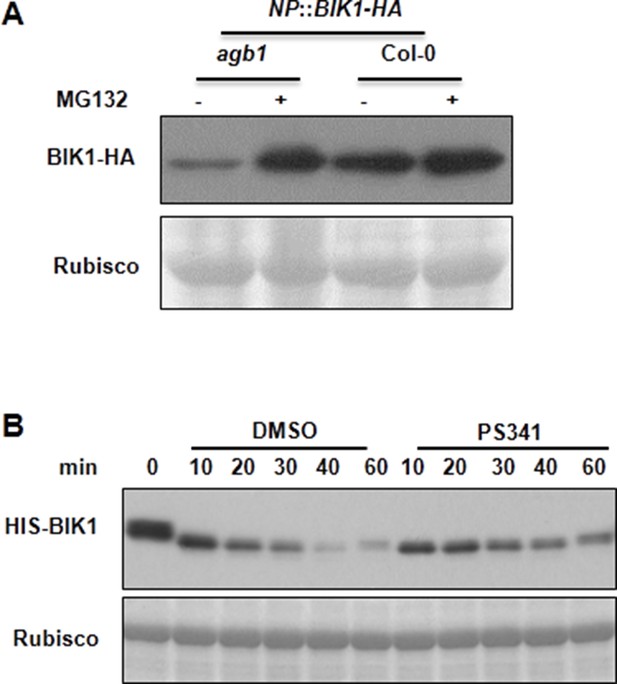
AGB1 regulates BIK1 stability through proteasome pathway.
(A) Treatment with MG132 allows accumulation of BIK1 in agb1 mutant seedlings. NP::BIK1-HA seedlings of WT (AGB1) and agb1 background were treated with DMSO (-) or 100 μM specific proteasome inhibitor MG132 (+) for 8 hr, BIK1 stability was detected by anti-HA immunoblot. (B) Treatment with PS341 inhibits BIK1 degradation in vitro. Total extracts from WT plants pretreated with DMSO or 100 μM PS341 were incubated with the HIS-BIK1 recombinant protein, and equal amounts of sample were withdrawn at the indicated times for anti-HIS immunoblot assays. Three independent experiments were performed with similar results.
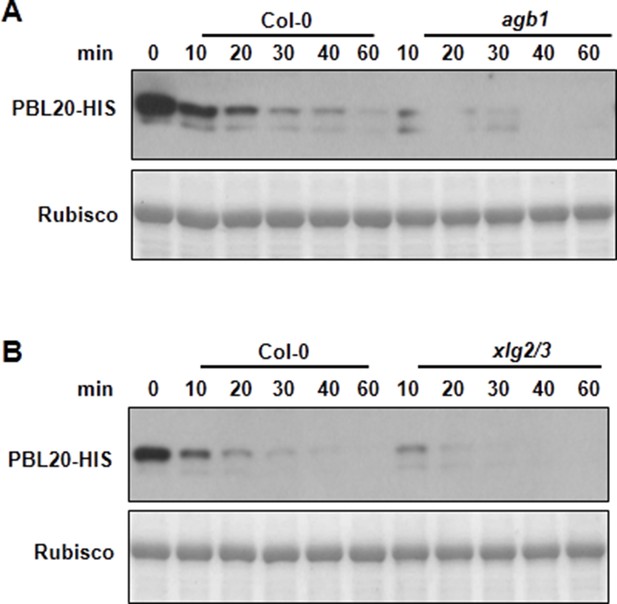
AGB1 and XLG2/3 attenuate PBL20 degradation.
(A) Accelerated PBL20 degradation in agb1 extracts. (B) Accelerated PBL20 degradation in xlg2 xlg3 extracts. Total extracts from WT, agb1 and xlg2/3 were incubated with the PBL20-HIS recombinant protein, and equal amounts of sample were withdrawn at the indicated times for anti-HIS immunoblot analyses. Two independent experiments were performed with similar results.
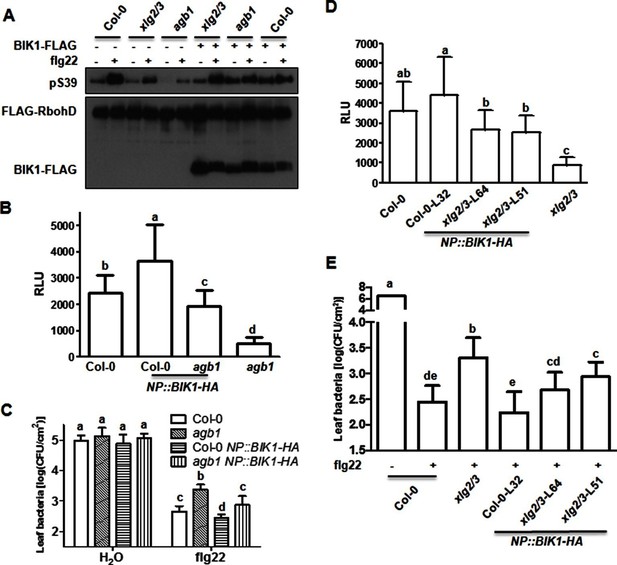
BIK1 level accounts for G protein-mediated regulation of FLS2 immunity.
(A) Transient expression of BIK1 in agb1 and xlg2/3 mutant protoplasts restores RbohD phosphorylation. FLAG-RbohD and BIK1-HA constructs are transiently expressed in protoplasts from WT (Col-0), agb1 and xlg2/3. FLAG-RbohD protein was affinity purified and detected by anti-FLAG and anti-pSer39 immuoblots. (B) BIK1 transgene restores flg22-induced ROS burst in agb1. (C) BIK1 transgene partially restores flg22-induced resistance to Pst in agb1. NP::BIK1-HA was introduced into agb1 by crossing, transgenic lines of agb1 or Col-0 background in the F3 generation were used for the assays. (D) BIK1 transgene partially restores flg22-induced ROS burst in xlg2 xlg3 mutant. (E) BIK1 transgene partially restores flg22-induced resistance to Pst. The NP::BIK1-HA transgene was introduced into WT (Col-0-L32) and xlg2 xlg3 (xlg2/3-L64 and xlg2/3-L51) plants by Agrobacterium-mediated transformation. Independent T2 transgenic lines were used for the assays. Peak relative luminescence unit (RLU) values were shown for ROS assays (B and D) and leaf bacterial populations 2 days after bacterial inoculation were shown for flg22-protection assays (C and E). Bars in B-E represent mean ± SD (n ≥ 6; p<0.05, Student’s t-test; different letters indicate significant difference). Each experiment was repeated two (A) or three (B–E) times, and data of one representative experiment are shown.
-
Figure 4—source data 1
Raw data and exact p value of Figure 4B—E.
- https://doi.org/10.7554/eLife.13568.017
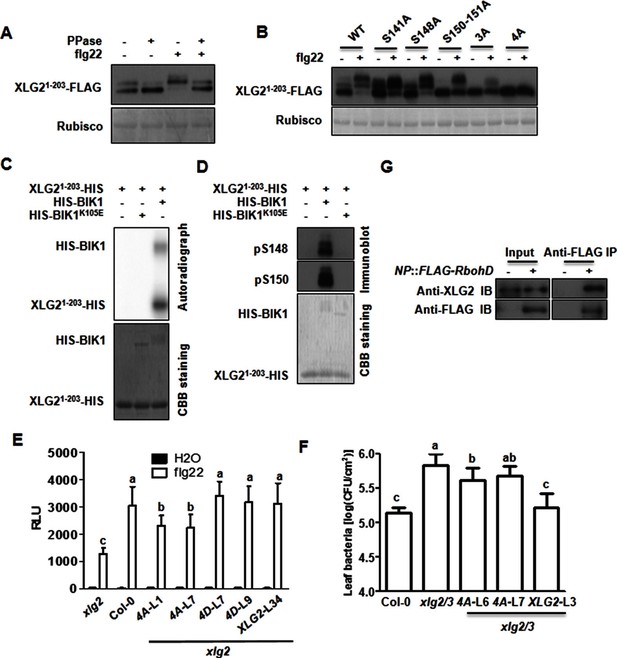
Phosphorylation of XLG2 by BIK1 regulates flg22-induced ROS.
(A) Flg22-induces phosphorylation of XLG2 in the N terminus. Protoplasts expressing XLG21-203-FLAG were treated with flg22. The total protein was treated with (+) or without (-) λ protein phosphatase (PPase) prior to anti-FLAG immunoblot analysis. (B) Flg22-induced phosphorylation of XLG2 in protoplasts primarily occurs in Ser141, Ser148, Ser150 and Ser151. Different mutated form of XLG21-203-FLAG constructs were transiently expressed in WT protoplast, treated with flg22 and the migration of XLG21-203-FLAG were examined by anti-FLAG immunoblot. (C) BIK1 phosphorylates XLG2 N terminus in vitro. XLG21-203-HIS was incubated with HIS-BIK1 and HIS-BIK1K105E in the presence of 32P-γ-ATP and analyzed by autoradiography. CBB, coomassie brilliant blue. (D) BIK1 phosphorylates XLG2 at Ser148 and Ser150 in vitro. XLG21-203-HIS was incubated with HIS-BIK1 and HIS-BIK1K105E in kinase reaction buffer. Protein phosphorylation was detected by anti-pSer148 and pSer150 immunoblots. (E) XLG2 phosphorylation is required for flg22-induced ROS. xlg2 mutant plants were transformed with WT (NP::XLG2-L34), non-phosphorylatable (4A-L1 and 4A-L7), or phospho-mimicking (4D-L7 and 4D-L9) forms of XLG2 under control of the native XLG2 promoter. Independent T2 lines were examined for flg22-induced ROS burst and peak relative luminescence unit (RLU) values are shown. (mean ± SD; n ≥ 6; p<0.05, Student’s t-test; different letters indicate significant difference). (F) XLG2 phosphorylation is required for Pst resistance. xlg2/3 double mutant plants were transformed with WT (XLG2-L3) or non-phosphorylatable (4A-L6 and 4A-L7) form of XLG2 under control of the native XLG2 promoter. Independent T2 lines were inoculated with Pst, and bacterial populations in leaves were measured 3 days post inoculation. (mean ± SD; n ≥ 6; p<0.05, Student’s t-test; different letters indicate significant difference). (G) XLG2 interacts with RbohD in Arabidopsis plants. rbohD plants were transformed with the FLAG-RbohD transgene under control of the RbohD native promoter. The resulting plants were used for Co-IP assay. Each experiment was repeated two (C, G) or three (A, B, D–F) times, and data of one representative experiment are shown.
-
Figure 5—source data 1
- https://doi.org/10.7554/eLife.13568.019
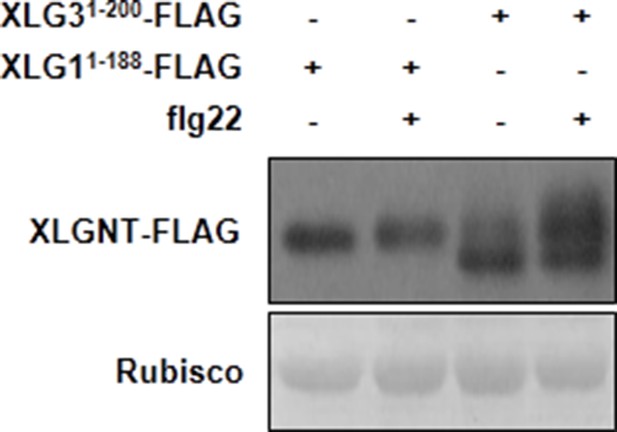
The N terminus of XLG3, but not XLG1, is phophorylated upon flg22-treatment.
WT protoplasts were transfected with XLG11-188-FLAG or XLG31-200-FLAG, treated with flg22, and protein was analyzed by anti-FLAG immunoblot.
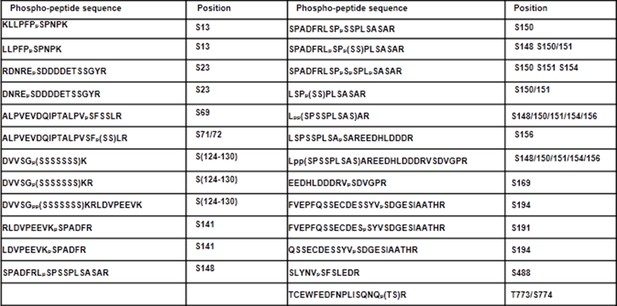
Phospho-sites in XLG2 isolated from flg22-treated protoplasts.
List of phospho-peptides identified. Protoplasts prepared from WT plants were transfected with XLG2-FLAG and treated with flg22 for 10 min, affinity-purified and subjected to LC-MS/MS for phospho-sites identification.
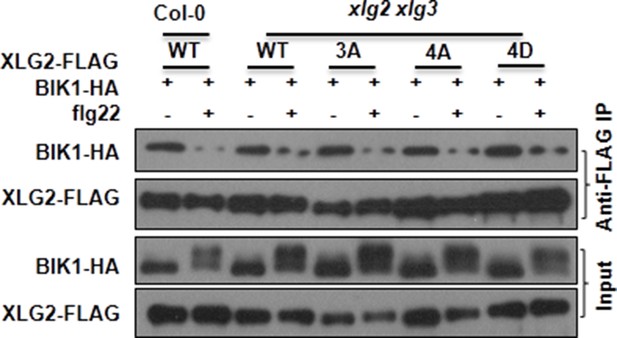
Mutations that block or mimic XLG2 phosphorylation do not impact BIK1 stability and XLG2-BIK1 interaction.
WT or xlg2 xlg3 protoplasts were transfected with BIK1-HA along with WT, non-phosphorylatable (3A, 4A), phospho-mimicking (4D) forms of XLG2-FLAG constructs, treated with (+) or without (-) flg22, and total protein was subject to Co-IP assays and immunoblot analysis. Three independent experiments were performed with similar results.
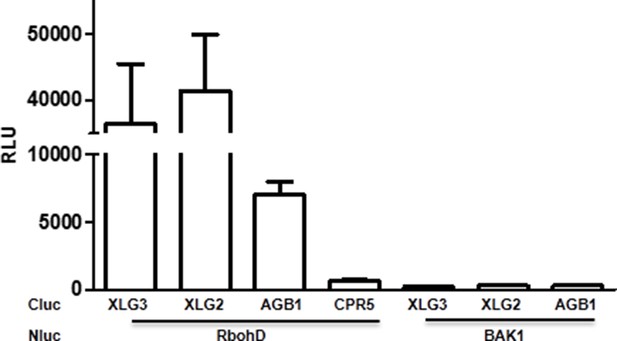
XLG2/3 interact with RbohD in Nb plants.
Agrobacteria containing the indicated constructs were infiltrated in to Nb leaves, and luciferase activity was recorded 2 days later (mean ± SD; n≥6; representative data from 2 independent experiments are shown).
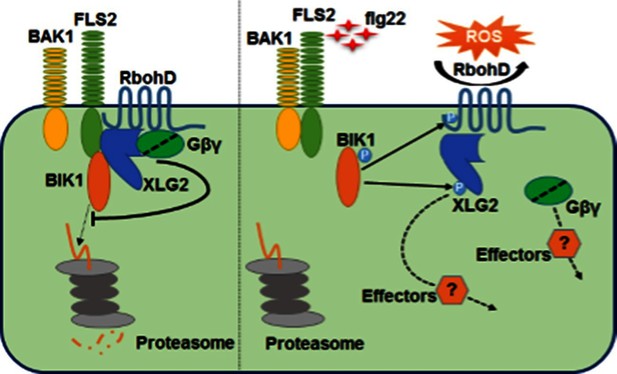
Model for G protein-coupled FLS2 signaling.
In the pre-activation state, the heterotrimeric G proteins composed of XLG2/3, AGB1, and AGG1/2 interact with the FLS2-BIK1 complex. Stimulation by flg22 induces BAK1-FLS2 interaction and activation of the receptor complex. This leads to the activation of the G proteins and phosphorylation of XLG2 in the N terminus. The activated G proteins dissociate from the receptor complex and regulate RbohD and other downstream effectors to positively modulate immune responses.





