Polymerase θ is a robust terminal transferase that oscillates between three different mechanisms during end-joining
Figures
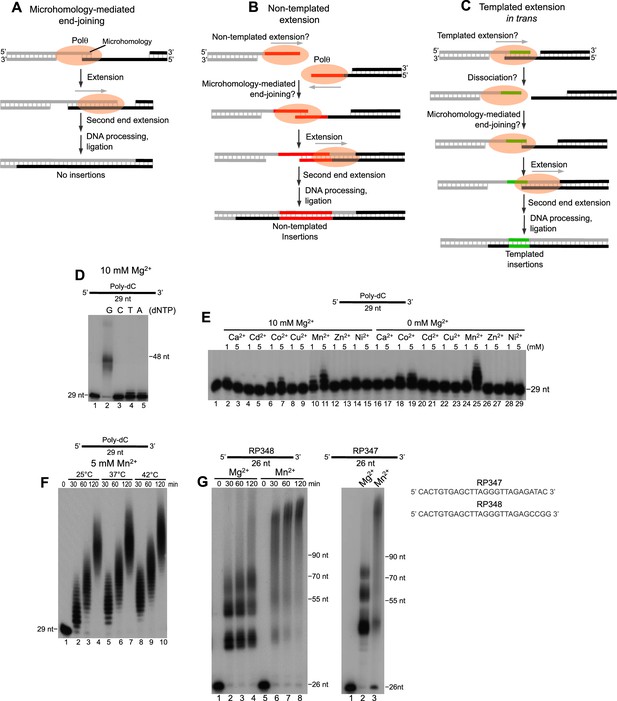
Polθ exhibits robust template-independent terminal transferase activity in the presence of manganese.
(A-C) Models of Polθ dependent DNA end-joining. (A) Polθ uses existing sequence microhomology to facilitate DNA end-joining. (B) Polθ is proposed to extend ssDNA by a template-independent mechanism, then use the newly generated sequence to facilitate DNA end-joining. (C) Polθ is proposed to extend ssDNA by using the opposing overhang as a template in trans, then after DNA synapse dissociation Polθ uses the newly generated sequence to facilitate DNA end-joining. (D) A denaturing gel showing Polθ extension of poly-dC ssDNA in the presence of indicated dNTPs and 10 mM Mg2+. (E, F) Denaturing gels showing Polθ extension of poly-dC ssDNA in the presence of dTTP and indicated divalent cation concentrations (E) and time intervals and temperatures (F). (G) Denaturing gels showing Polθ extension of indicated ssDNA in the presence of all four dNTPs and 10 mM Mg2+ or 5 mM Mn2+.
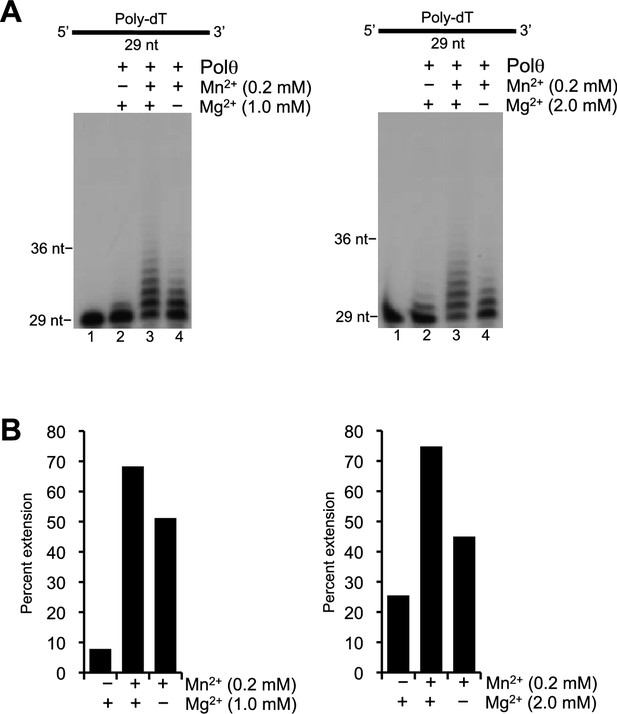
Polθ template-independent activity is stimulated by physiological concentrations of Mn2+ and Mg2+.
(A) Denaturing gels showing Polθ extension of poly-dT in the presence of dCTP with indicated concentrations of Mn2+ and Mg2+. (B) Plots of percent ssDNA extension observed in panel A. Percent extension was calculated by dividing the intensity of the sum of the extended products by the sum of the intensity of all DNA in each lane.
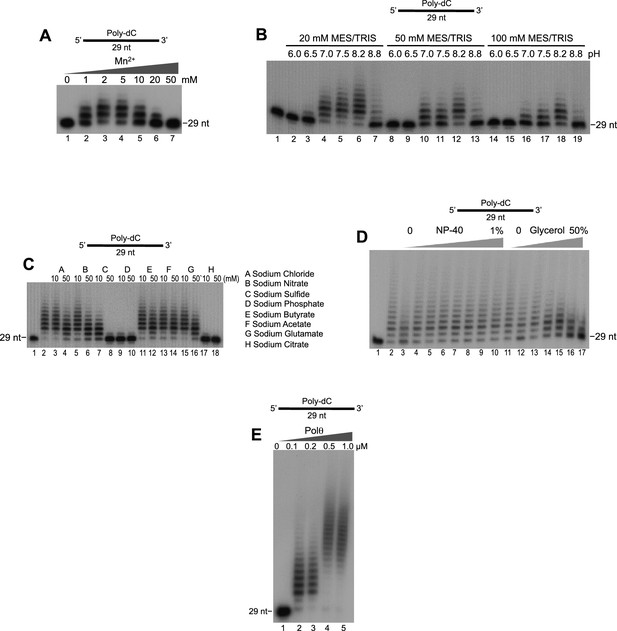
Optimization of Polθ-Mn2+ template-independent terminal transferase activity.
(A-E) Denaturing gels showing Polθ-Mn2+ extension of poly-dC ssDNA in the presence of dTTP with indicated [Mn2+] (A), buffers (B), salts (C), detergent and glycerol (D), and Polθ concentration (E). Panels B–D included 5 mM Mn2+.
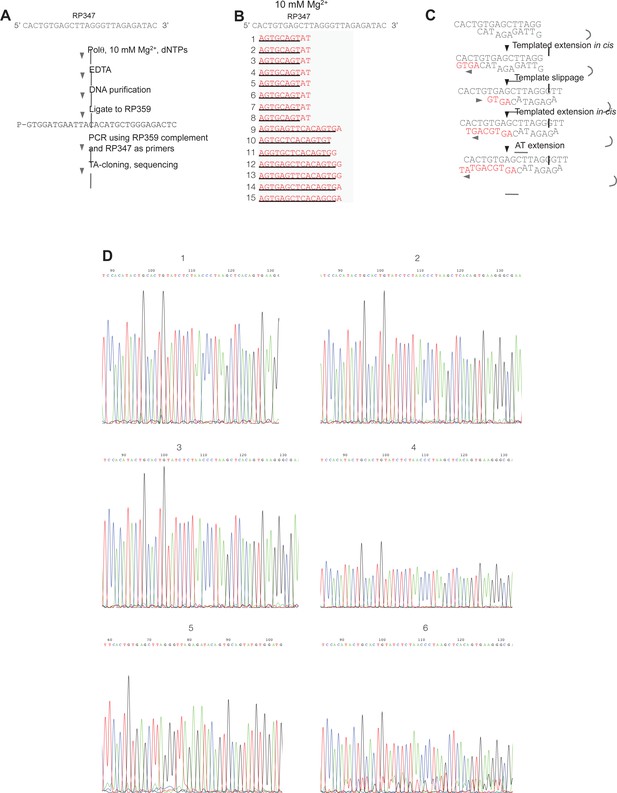
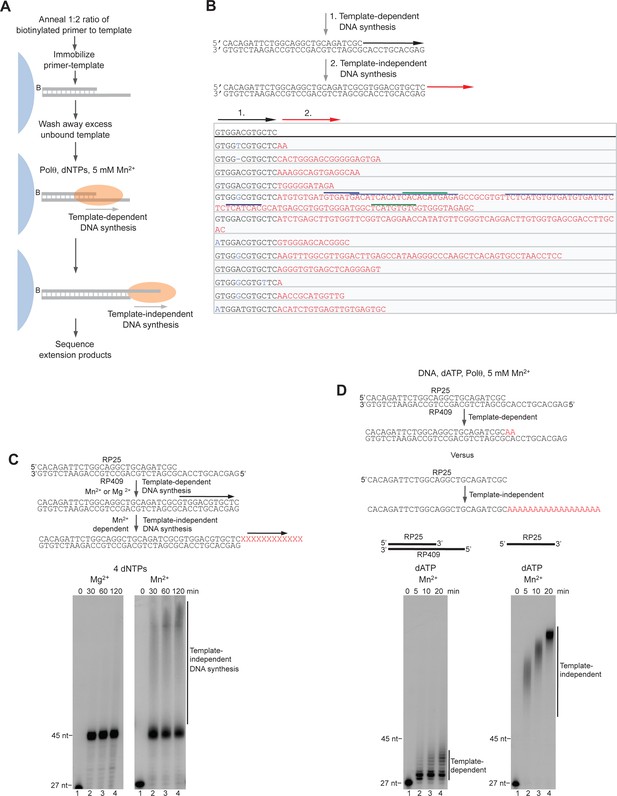
Sequence analysis of Polθ-Mg2+ template-dependent terminal transferase activity.
(A) Schematic of method used to sequence Polθ-Mg2+ extension products. (B) Sequences of extension products generated by Polθ in the presence of 10 mM Mg2+, all four dNTPs, and ssDNA RP347. Initial sequence of RP347 ssDNA is indicated at top. Sequences of extension products are shown in a 5’-3’ direction. Black underline, sequence copied from template. (C) Model of how Polθ-Mg2+ repeatedly generates products 1–8 from RP347 ssDNA via snap-back replication. (D) Representative sequence traces of products 1–8 demonstrate non-identical sequencing reactions and files. Certain sequences are represented as complements due to their particular orientation during TA-cloning.
Polθ oscillates between three different modes of terminal transferase activity.
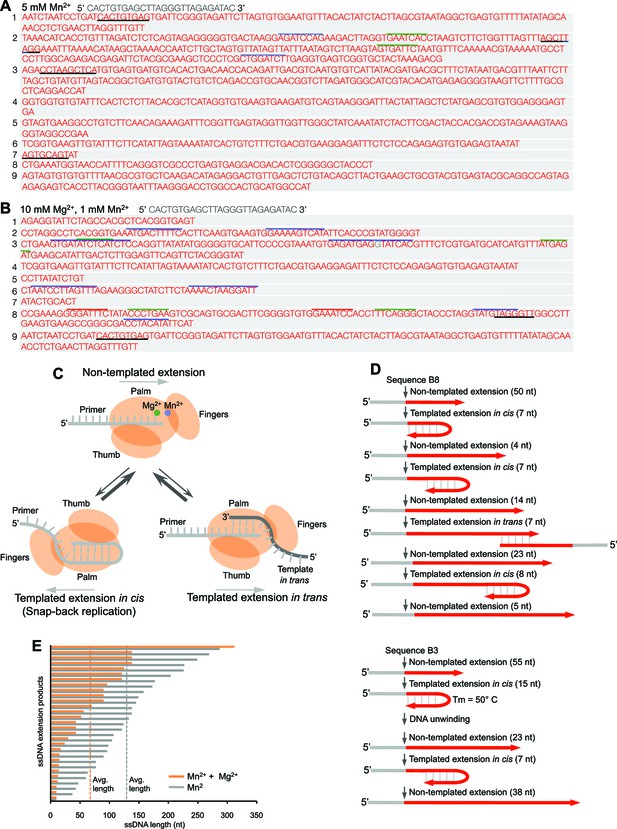
(A,B) Sequences of Polθ ssDNA extension products in the presence of indicated divalent cations (A, 5 mM Mn2+; B, 10 mM Mg2+, 1 mM Mn2+). Initial ssDNA sequences are indicated at top. Black underlines, sequences copied from either original template or complementary sequences generated from original template; matching colored lines, complementary sequences due to snap-back replication. (C) Models of Polθ terminal transferase activities. (Top) Polθ preferentially exhibits template-independent activity in the presence of Mg2+ and Mn2+. Polθ also performs templated ssDNA extension in cis (bottom left) and in trans (bottom right), and oscillates between these three mechanisms. (D) Models of Polθ terminal transferase activity based on sequences 3 and 8 from panel B. (E) Plot showing lengths of ssDNA products generated by Polθ in the presence of indicated divalent cations.
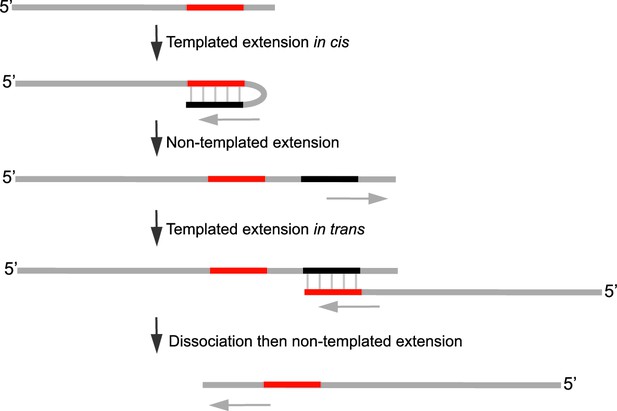
Model of Polθ-Mn2+ terminal transferase activity involving template copying in cis and in trans.
Model of how Polθ generates sequence tracts identical to the initial template in the presence of Mn2+. Red, original sequence copied; black, complement of red sequence. The black complementary sequence may also be generated via templated extension in trans.
Control experiments for Polθ-Mn2+ template-independent activity.
(A) Scheme of experimental conditions. (B) Model of sequential activity of Polθ-Mn2+ on a primer-template (top). Sequences generated by Polθ-Mn2+ during primer-extension in solid-phase in the presence of 5 mM Mn2+. Black sequence, template-dependent; red sequence, template-independent; blue sequence, misincorporation; dash, frameshift mutation. Colored lines, complementary sequences generated by snap-back replication. (C) Models of Polθ activity on a primer-template in the presence of Mg2+ and Mn2+ (top). Denaturing gels showing Polθ primer-extension products in the presence of 10 mM Mg2+ (left) and 5 mM Mn2+ (right). (D) Models of Polθ-Mn2+ activity on a primer-template and ssDNA in the presence of dATP (top). Denaturing gels showing template-dependent (left) and template-independent (right) Polθ-Mn2+ activities on a primer-template (left) and primer (right), respectively, in the presence of 5 mM Mn2+ and dATP.
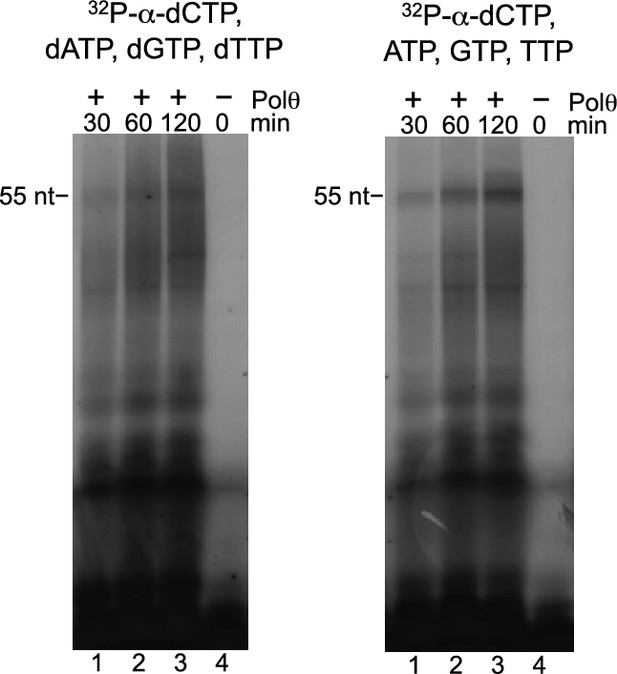
Polθ-Mn2+ exhibits de novo DNA and RNA synthesis activities.
Denaturing gels showing de novo nucleic-acid synthesis by Polθ in the presence of 5 mM Mn2+ and indicated nucleotides.
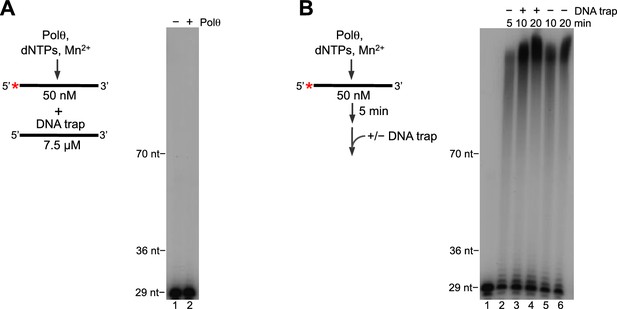
Polθ-Mn2+ exhibits processive terminal transferase activity.
(A) Scheme of experiment (left). Denaturing gel showing inhibition of Polθ-Mn2+ terminal transferase activity by a ssDNA trap (right). (B) Scheme of experiment (left). Denaturing gel showing a time course of Polθ-Mn2+ terminal transferase activity in the presence and absence of ssDNA trap (right). Assays (A,B) included 5 mM Mn2+.

Polθ-Mn2+ oscillates between different terminal transferase activites in the presence of a DNA trap.
(A) Scheme of experiment performed in solid-phase. (B) Bar graph depicting ssDNA product lengths generated by Polθ in the presence (orange) and absence (grey) of excess ssDNA with 10 mM Mg2+ and 1 mM Mn2+. (C,D) Sequences generated by Polθ incubated with the indicated ssDNA substrate in the presence (D) and absence (C) of excess ssDNA trap with 10 mM Mg2+, 1 mM Mn2+, and all four dNTPs. Black underlines, sequences identical or complementary to initial ssDNA substrate; red underlines, sequences complementary to ssDNA trap; colored lines above text, complementary sequences within individual ssDNA products.
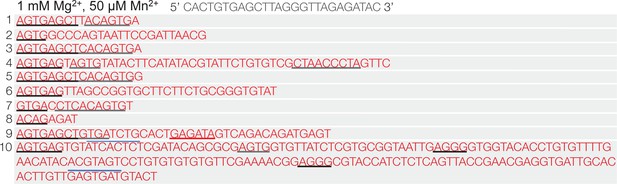
Polθ oscillates between templated and non-templated terminal transferase activities in the presence of physiological concentrations of Mg2+ and Mn2+.
Sequences generated by Polθ during ssDNA extension in the presence of 1 mM Mg2+ and 50 µM Mn2+. Black and grey inderlines, sequence complementary to initial ssDNA substrate; red underline, sequence identical to initial ssDNA substrate; blue lines, complementary sequence generated by snap-back replication; red text without lines, random insertions. Initital ssDNA sequence indicated at top.
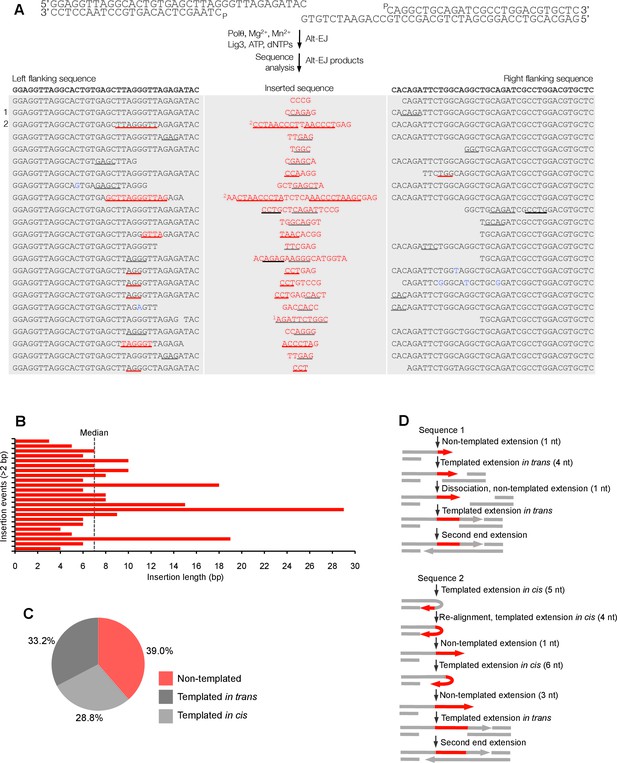
Polθ oscillates between three different modes of terminal transferase activity during alternative end-joining in vitro.
(A) Scheme for reconstitution of Polθ mediated alt-EJ in vitro (top). Sequences of alt-EJ products generated by Polθ in vitro using 10 mM Mg2+ and 1 mM Mn2+ (bottom). Red text, insertions; black text, original DNA sequence; black and grey underlines, sequences copied from original template; red underlines, complementary sequences due to snap-back replication; red sequence without underlines, random insertions; superscript 1, suggests sequences were copied from a template portion that was subsequently deleted during alt-EJ; superscript 2, suggests sequences were copied from the template in more than one way. Original DNA sequences indicated at top. Blue type, mutations. (B) Plot of insertion tract lengths generated in panel A. (C) Chart depicting percent of individual nucleotide insertion events due to non-templated extension, templated extension in cis and templated extension in trans. t test indicates no significant difference between percent of non-templated and templated in cis insertions. (D) Models of Polθ activity based on end-joining products 1 and 2 from panel A.
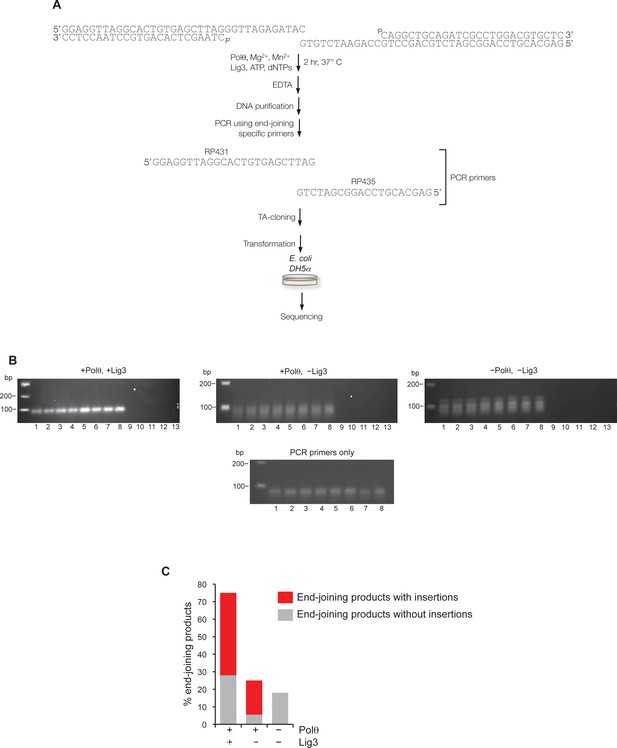
Supporting information for Polθ mediated alt-EJ in vitro.
(A) Schematic of alt-EJ reaction and subsequent procedures used for amplification and sequencing of end-joining products. Control alt-EJ reactions were performed with 10 mM Mg2+ and 1 mM Mn2+. (B) Non-denaturing gels showing the products of PCR reactions containing either purified DNA from alt-EJ reactions performed in the presence of Polθ and Lig3 (top left), Polθ alone (top middle), and in the absence of Polθ and Lig3 (top right), or no DNA with primers only (bottom middle). Products in the top middle and top right gels are due to primer-dimer events as shown in the primers only control (bottom middle gel). Lanes 1-8 represent PCR reactions performed at the following respective temperatures: 61°C, 60.8°C, 60.4°C, 59.9°C, 59.2°C, 58.6°C, 58.2°C, 58°C. Lanes 9–13 represent PCR reactions performed in the absence of PCR primers RP435 and RP431 and at the following respective temperatures: 61°C, 60.4°C, 59.9°C, 59.2°C, 58.2°C. The absence of PCR products in lanes 9–13 show that Taq polymerase cannot amplify original pssDNA templates via end-joining or other mechanisms. (C) Plot showing percent of end-joining products observed in cloning vectors following end-joining reactions containing the indicated proteins. Red, end-joining products with insertions; grey, end-joining products without insertions. n = 64 (+Polθ, +Lig3), n = 72 (+Polθ, –Lig3), n =12 (–Polθ, –Lig3). End-joining products in the absence of Polθ and Lig3 are likely due to infrequent byproducts of PCR.
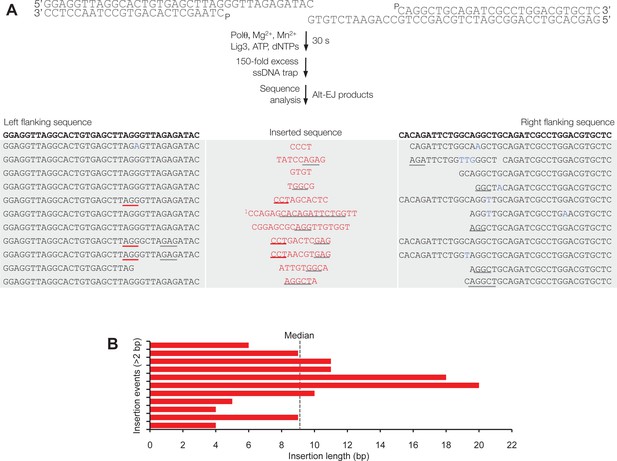
Polθ acts processively during alt-EJ in vitro.
(A) Scheme for reconstitution of Polθ mediated alt-EJ in vitro with ssDNA trap (top). Sequences of alt-EJ products generated by Polθ in vitro using 10 mM Mg2+ and 1 mM Mn2+ (bottom). Red text, insertions; black text, original DNA sequence; grey underlines, sequences copied from original template; red underlines, complementary sequences due to snap-back replication; red sequence without underlines, random insertions; superscript 1, suggests sequences were copied from a template portion that was subsequently deleted during alt-EJ. Original DNA sequences indicated at top. Blue type, mutations. (B) Plot of insertion tract lengths generated in panel A.
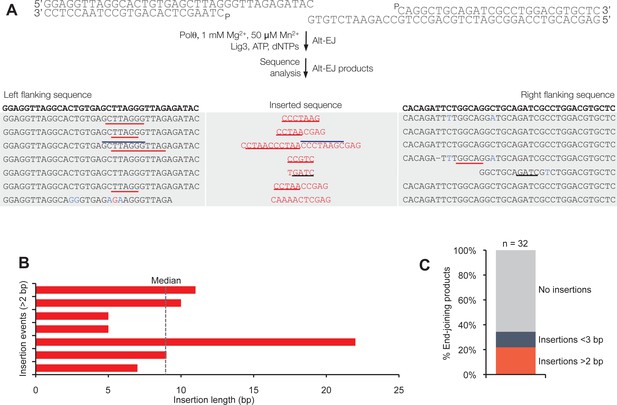
Polθ generates insertions during alt-EJ in the presence of low concentrations of Mg2+ and Mn2+.
(A) Scheme for reconstitution of Polθ mediated alt-EJ in vitro with 1 mM Mg2+ and 50 µM Mn2+ (top). Sequences of Polθ–mediated alt-EJ products with insertions >2 bp (bottom). Red text, insertions; black text, original DNA sequence; black underlines, sequences copied from original template; red and blue lines, complementary sequences due to snap-back replication; red sequence without lines, random insertions. (B) Plot of insertion tract lengths illustrated in panel A. (C) Plot showing percentage of Polθ–mediated alt-EJ products with and without insertions. n = 32.
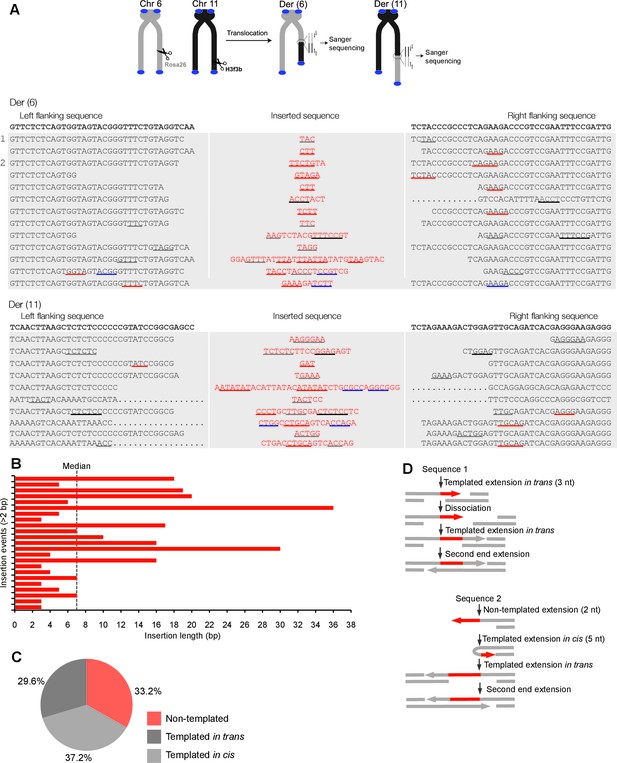
Polθ oscillates between three different modes of terminal transferase activity during alternative end-joining in vivo.
(A) Scheme for Polθ mediated alt-EJ of site-specific DSBs in mouse embryonic stem cells (top). Sequences of alt-EJ products generated by Polθ in cells (bottom). Red text, insertions; black text, original DNA sequence; black and grey underlines, sequences copied from original template; red and blue underlines, complementary sequences due to snap-back replication; red sequence without underlines, random insertions; Original DNA sequences indicated at top;... , large deletions. (B) Plot of insertion tract lengths generated in panel A. (C) Chart depicting percent of individual nucleotide insertion events due to non-templated extension, templated extension in cis and templated extension in trans. t test indicates no significant difference between percent of non-templated and templated in cis insertions. (D) Models of Polθ activity based on end-joining products 1 and 2 from panel A.
Large insertions copied from remote donor locations.
Scheme for Polθ mediated alt-EJ of site-specific DSBs in mouse embryonic stem cells (top). Insertion sequences of alt-EJ products generated by Polθ in cells (bottom three panels). Probable remote donor sites listed at right based on sequence similarity. The large templated insertions copied from remote donor locations are likely due to strand invasion into duplex DNA followed by D-loop extension and dissociation.
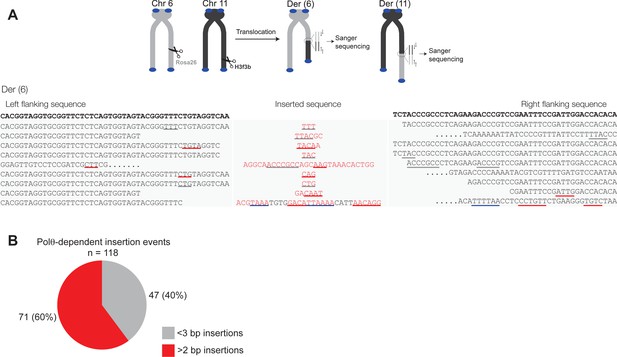
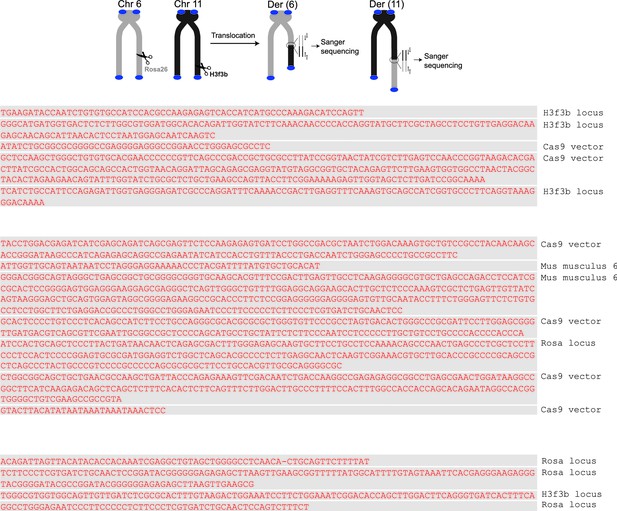
Additional sequence analysis of alternative end-joining products generated in vivo.
(A) Scheme for Polθ mediated alt-EJ of site-specific DSBs in mouse embryonic stem cells (top). Sequences of alt-EJ products generated by Polθ in cells (bottom). Red text, insertions; black text, original DNA sequence; grey underlines, sequences copied from original template; red and blue underlines, complementary sequences due to snap-back replication; red sequence without underlines, random insertions; Original flanking DNA sequences indicated at top in bold;... , large deletions. (B) Pie chart of insertion tract lengths generated in vivo. n = 118.
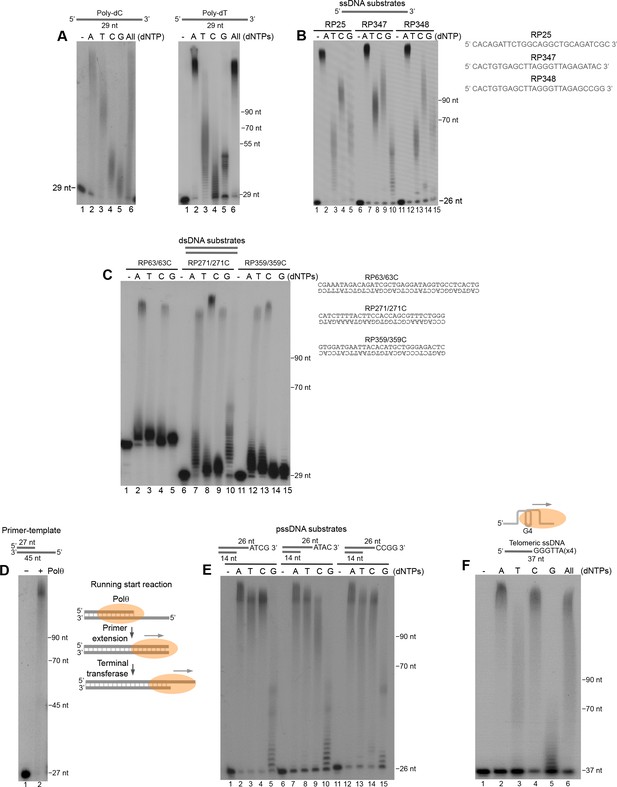
Polθ exhibits preferential terminal transferase activity on pssDNA.
(A) Denaturing gels showing Polθ extension of poly-dC (left) and poly-dT (right) ssDNA with 5 mM Mn2+ and the indicated dNTPs. (B) Denaturing gel showing Polθ extension of the indicated ssDNA with 5 mM Mn2+ and indicated dNTPs. (C) Denaturing gel showing Polθ extension of the indicated dsDNA with 5 mM Mn2+ and indicated dNTPs. (D) Denaturing gel showing Polθ extension of a primer-template with 5 mM Mn2+ and all four dNTPs. Model of Polθ-Mn2+ activity on a primer-template (right). (E) Denaturing gel showing Polθ extension of the indicated pssDNA with 5 mM Mn2+ and indicated dNTPs. (F) Denaturing gels showing Polθ extension of ssDNA modeled after telomere sequence with 5 mM Mn2+ and the indicated dNTPs.
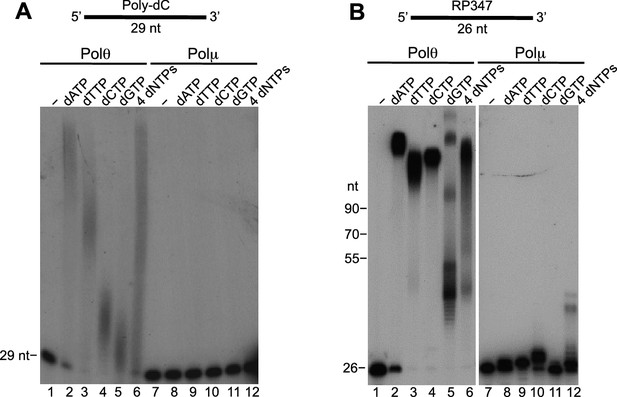
Comparison of Polθ and Polμ terminal transferase activities with Mn2+.
(A, B) Denaturing gels showing Polθ and Polμ extension of poly-dC (A) and RP347 ssDNA (B) in the presence of Mn2+ and the indicated nucleotides.
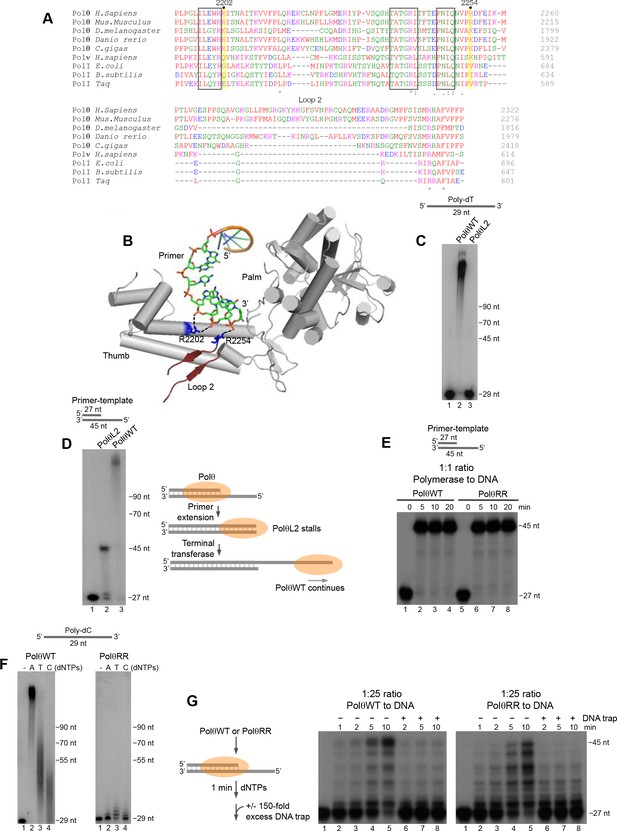
Conserved residues contribute to Polθ processivity and template-independent terminal transferase activity.
(A) Sequence alignment of Polθ and related A-family Pols. Conserved positively charged residues (2202, 2254) and loop 2 in Polθ are highlighted in yellow and grey, respectively. Black boxes indicate conserved motifs. * = identical residues,: = residues sharing very similar properties,. = residues sharing some properties. Red, small and hydrophobic; Blue, acidic; Magenta, basic; Green, hydroxyl, sulfhydryl, amine. (B) Structure of Polθ with ssDNA primer (PDB code 4X0P) (Zahn et al., 2015). Residues R2202 and R2254 are indicated in blue. Dotted blue lines indicate ionic interactions. Loop 2 is indicated in dark red. Thumb and palm subdomains are indicated. (C) Denaturing gel showing PolθWT and PolθL2 extension of ssDNA with 5 mM Mn2+ and all four dNTPs. (D) Denaturing gel showing PolθWT and PolθL2 extension of a primer-template with 5 mM Mn2+ and all four dNTPs. Model of PolθWT-Mn2+ and PolθL2-Mn2+ activities on a primer-template (right). (E) Denaturing gel showing a time course of PolθWT and PolθRR extension of a primer-template in the presence of 10 mM Mg2+ and all four dNTPs. (F) Denaturing gel showing PolθWT (left) and PolθRR (right) extension of poly-dC ssDNA with 5 mM Mn2+ and the indicated dNTPs. (G) Schematic of assay (left). Denaturing gel showing PolθWT and PolθRR extension of an excess of radiolabeled primer-template with all four dNTPs and 10 mM Mg2+ either in the presence or absence of 150-fold excess unlabeled DNA trap.
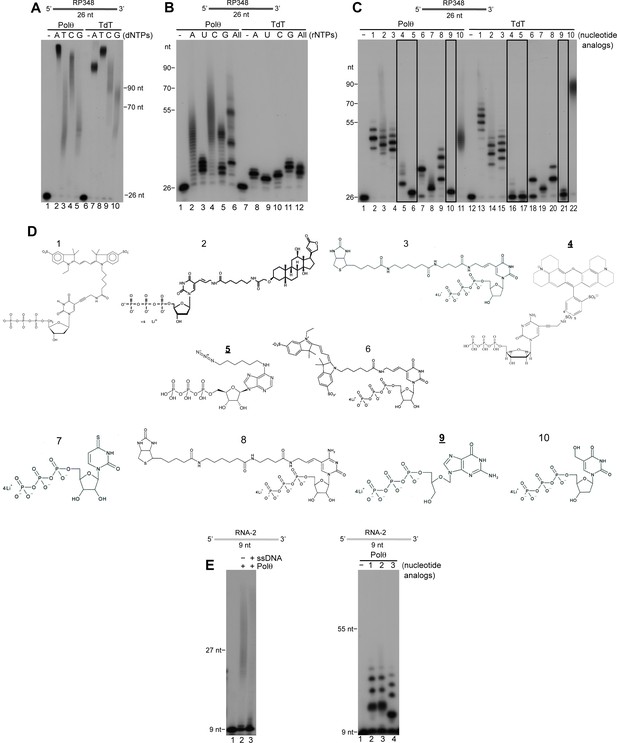
Comparison of Polθ and TdT terminal transferase activities.
(A) Denaturing gel showing Polθ-Mn2+ (lanes 1–5) and TdT (lanes 6–10) extension of ssDNA with the indicated dNTPs. (B) Denaturing gel showing Polθ-Mn2+ (lanes 1–6) and TdT (lanes 7–12) extension of ssDNA with the indicated ribonucleotides (rNTPs). (C) Denaturing gel showing Polθ-Mn2+ (lanes 1–11) and TdT (lanes 12–22) extension of ssDNA with the indicated nucleotide analogs illustrated in panel (d). Boxed lanes indicate nucleotides analogs that are exclusively transferred by Polθ-Mn2+. (D) Nucleotide analogs: 1, cy3-dUTP; 2, Digoxigenin-11-dUTP; 3, Biotin-16AA-dUTP; 4, Texas Red-5-dCTP; 5, N6 -(6-Azido)hexyl-ATP; 6, Cyanine 3-AA-UTP; 7, 4-Thio-UTP; 8, Biotin-16AA-CTP; 9, Ganciclovir Triphosphate; 10, 5-Hydroxymethyl-2’-deoxyuridine-5’-Triphosphate. Underlined nucleotide analogs (4,5,9) are exclusively transferred by Polθ-Mn2+. (E) Denaturing gel showing Polθ-Mn2+ extension of RNA with all four dNTPs in the presence (lane 3) and absence (lane 2) of unlabeled ssDNA (left panel). Denaturing gel showing Polθ-Mn2+ extension of RNA with the indicated nucleotide analogs (right panel). Polθ-Mn2+ extension assays (A-C,E) included 5 mM Mn2+.





