The beetle amnion and serosa functionally interact as apposed epithelia
Figures
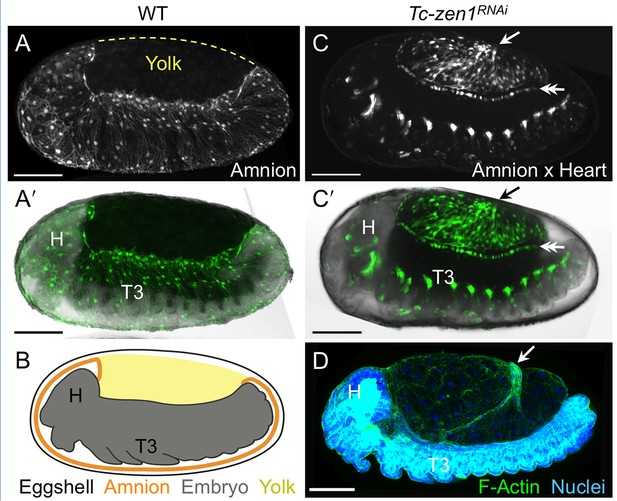
The enhancer trap line HC079 is an autonomous amniotic tissue marker.
Images are lateral, with anterior left and dorsal up, shown as maximum intensity projections or a mid-sagittal schematic. Visualization reagents are indicated. (A–B) In wild type (WT), EGFP expression is extraembryonic (EE) in a ventral domain that fully covers the embryo but not the yolk prior to rupture. See also Figure 1—figure supplement 1, Video 1. (C–D) Consistent with the WT EGFP domain being amniotic, the entire EE tissue expresses EGFP when serosal identity is eliminated after Tc-zen1RNAi. Here, the EE tissue does cover the yolk (dorsal to the cardioblast cell row: double-headed arrow), and, during the mid-withdrawal stage shown here, acquires a diagnostic ‘crease’ (arrow) (Panfilio et al., 2013). Scale bars are 100 µm. Abbreviations: H, head; T3, third thoracic segment.
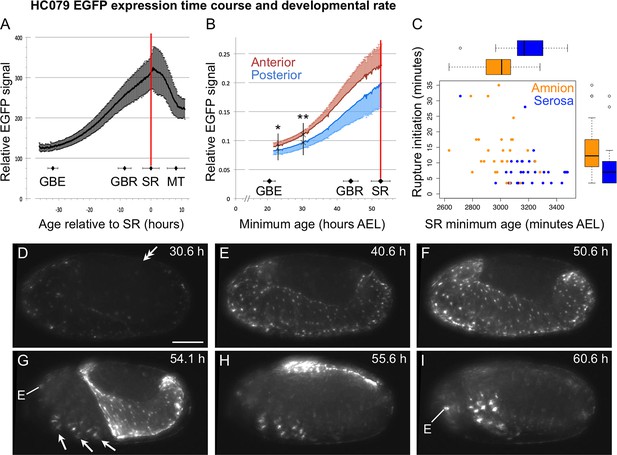
Developmental time course of EGFP expression in the enhancer trap line HC079.
(A–B) Here we provide a first characterization of the new enhancer trap line HC079. See Koelzer et al. (2014) for mapping technique, original staging definitions, acquisition parameters, and analysis method. HC079 was mapped to a gene-poor, intergenic region on chromosome 3. EGFP signal is first detected in the amnion during germband retraction, becoming increasingly bright throughout the tissue up to the time of serosal rupture, which heralds the onset of withdrawal morphogenesis. Relative EGFP signal quantification is plotted as the mean ± standard deviation during a 48-hr time-lapse recording, with corresponding values for landmark developmental stages (GBE, germband extension; GBR, germband retraction; SR, serosal rupture; MT, muscle twitches). Note that 'serosal rupture' is the staging term but in fact refers to first discernible rupture of both EE tissues at the beginning of withdrawal morphogenesis. Landmark stage values are also based on nGFP embryos that were recorded in parallel for calibration (combined n = 7–23 embryos per stage). Vertical red lines indicate the mean SR age. In A, relative EGFP signal is the mean grey value (n = 10), shown for minimum age (from a four-hr egg collection) relative to serosal rupture. In B, signal is the integrated density of the posterior and anterior egg halves (0–50% vs. 50–100% egg length) for embryos recorded in both lateral and ventral aspect (n = 5), with standard deviation shown on only one side of each plot for clarity. Here, the x-axis shows minimum absolute age in hours after egg lay (hours AEL). Vertical black lines and asterisks indicate the first time points at which the anterior and posterior EGFP values differ significantly (*: p<0.05, **: p<0.01 for paired, two-tailed t-tests). Although this difference in part reflects the relative area of visible amnion in lateral aspect (greater anteriorly), the trend still holds when only embryos in ventral aspect are considered (uniform relative amnion surface area). (C) In our examination of rupture initiation in the amnion (HC079) × heart (G04609) cross compared to the serosa (G12424) × heart (G04609) cross, we found that the amnion cross develops at a faster rate, shown here as the minimum age at the time of serosal rupture (x-axis; amnion cross: 3007 (median) ± 81 (MAD) min, n = 28; serosal cross: 3168 (median) ± 85 (MAD) min, n = 30; p = 4.942e-06, Mann-Whitney U test), shown in relation to the duration of rupture initiation (y-axis: same as main text Figure 4F). However, even when a normalization factor was introduced to adjust for developmental rate, rupture initiation is longer in the amnion background. (D–I) Selected still images of an example embryo in lateral aspect (maximum intensity projections of deconvolved data; anterior left, dorsal up), showing EGFP signal predominantly in the amnion as well as in the eye (E) and multiple ringed domains within the legs (arrows). After dorsal closure (I), weak EGFP signal is also seen segmentally and in the usual nervous system tissues in which the core 3xP3 promoter drives expression (Koelzer et al., 2014). Scale bar (shown in D) is 100 µm. The double-headed arrow in D marks the posterior tip of the abdomen, indicating that the embryo is midway through germband retraction at this time. See also Video 1.
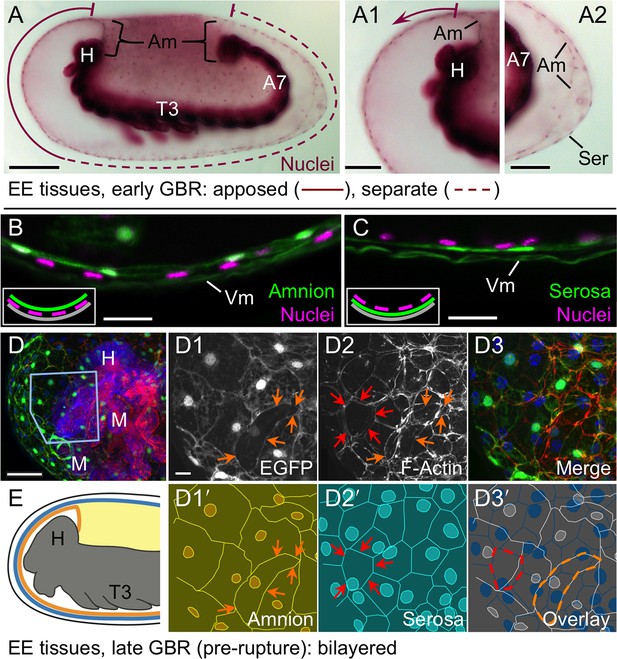
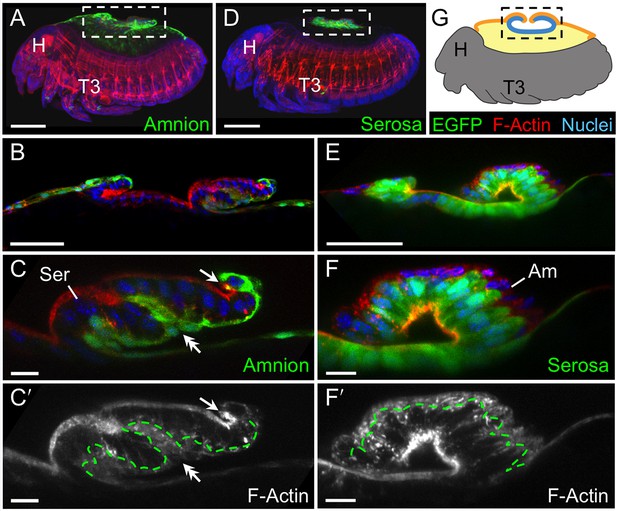
The amnion and serosa form a bilayer during the germband retraction stage.
Images are lateral (A–C,E) or ventral-lateral (D), with anterior left and dorsal up. (A) During germband retraction, the amnion progressively comes together with the serosa, shown sagittally at an intermediate stage when the tissues are apposed anteriorly (A1) but still separate posteriorly (A2). Fuchsin preparation causes embryo shrinkage (Wigand et al., 1998), amplifying apparent amniotic cavity volume, but without altering tissue topography, which is consistent across dozens of stage-matched specimens. (B–D) Before rupture, the ventral EE tissue under the eggshell (autofluorescent vitelline membrane) is comprised of distinct serosal (outer) and amniotic (inner) layers. In sagittal sections (B,C), tissue-specific EGFP labels continuous tissue sheets, while nuclei (DAPI stain) of the apposed EE tissue remain EGFP-negative and in a separate layer (inset schematic). Maximum intensity projections (D) also show two epithelial layers, which can be distinguished by tissue-specific cellular morphologies. The box in the first panel indicates the magnified region in D1-D3. Amnion-specific EGFP illuminates the nuclei and cell boundaries in this tissue (D1,D1′). A phalloidin counterstain shows a complex network of F-Actin filaments, including weak signal for amniotic cell boundaries (orange arrowheads), and a distinct pattern of thicker filaments in a double-walled, polygonal arrangement that corresponds to serosal cell boundaries (D2,D2′, see Figure 2—figure supplement 1 for serosal EGFP labeled specimens and additional details). Finally, comparison of a nuclear counterstain (DAPI) with the EGFP nuclear signal distinguishes the nuclei of the two EE tissues (here, EGFP-negative nuclei are serosal, shown in D2′), providing the information for a schematic overlay of these distinct tissues (D3,D3′: outlined cells are the same as those indicated by arrows in the previous panels). (E) The EE bilayer is illustrated in mid-sagittal view of the anterior, according to the color scheme in Figure 1B and with the serosa shown in blue. Scale bars are 100 µm (A), 50 µm (A1-A2,D), and 10 µm (B,C,D1-D3). Abbreviations: A7, seventh abdominal segment; Am, amnion; GBR, germband retraction; M, mandible; Ser, serosa; Vm, vitelline membrane; and as defined in Figure 1.
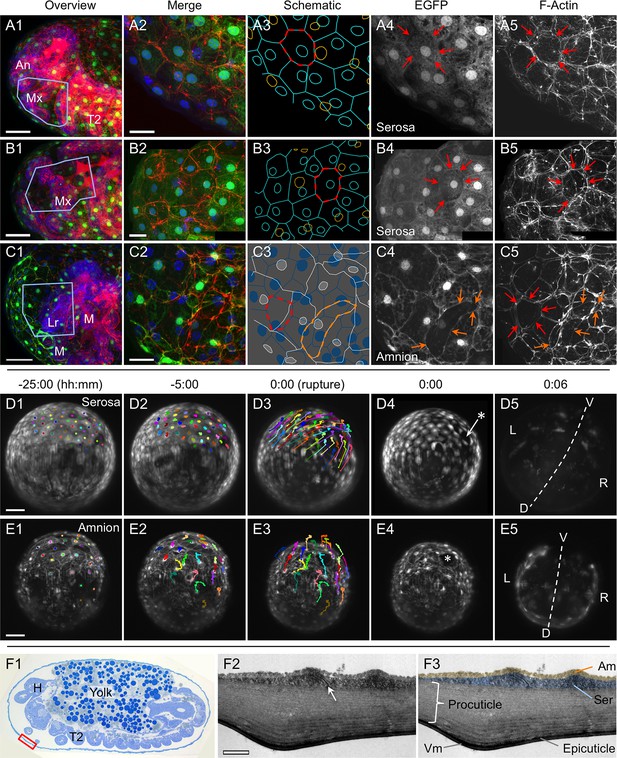
The amnion and serosa form a persistent bilayer comprised of two morphologically distinct epithelia, including in the anterior-ventral rupture competence zone.
Although EGFP intensity varies between cells, EGFP signal for a single EE tissue type labels all cells within a continuous epithelial sheet. Moreover, the amnion and serosa have strikingly different cellular morphologies, enabling cells of both tissues to be distinguished, with the serosa overlying the amnion. (A–C) Both EE tissues can be discerned as continuous epithelia within the rupture competence zone (shown after the completion of germband retraction, at 44–48 hr after egg lay). Shown are two examples with serosal EGFP (A–B) and one with amniotic EGFP (C, reproduced from main text Figure 2D, for comparison). 'Overview' images show lateral, ventral, and ventral-lateral views of maximum intensity projections (A1,B1,C1, respectively) with anterior left and the region of interest around the head appendages indicated by the blue boxed region (due to the curvature of the egg, only the regions actually visible in high-magnification projections of thinner z-stacks are indicated). 'Merge' images (A2,B2,C2) show maximum intensity projections of the indicated region, labeled for F-Actin (red, phalloidin), nuclei (blue, DAPI), and the specified EE tissue (green, EGFP). Note that embryonic-specific signal (F-Actin and DAPI signal for small cells not in contact with the tissue at the egg surface) was manually deleted from deeper optical sections with the freehand selection tool in ImageJ before rendering the projected images shown here. 'Schematic' images (A3,B3,C3) show cell and nuclear outlines. Cell outlines were determined by EGFP signal (for A3,B3,C3) and F-Actin signal (for C3, for serosal outlines only). Nuclear outlines were determined by EGFP signal or DAPI (for all EGFP-negative nuclei only). Serosal cells are shown in cyan or blue; amniotic nuclei and cells are shown in orange or white. Dashed outlines highlight individual serosal (red) and amniotic (orange) cells, which are also labeled by arrows of the same color in the single-channel micrographs for EGFP (A4,B4,C4) and F-Actin (A5,B5,C5). Strong F-Actin signal correlates with serosal cell shape (compare A4,A5 and B4,B5), such that F-Actin signal can be used to infer serosal cell shape even when that tissue is not labeled with EGFP (C3,C5). Furthermore, the F-Actin-inferred serosal cell outlines correspond to EGFP-negative nuclei in the amnion enhancer trap line (C3). Note that in order to preserve anterior-ventral EE tissue structure and topography, the specimens presented here (A–C) are still within the vitelline membrane. After fixation, embryos were cut in half (transversely) with a razor blade to allow the phalloidin and DAPI staining reagents to penetrate into the sample. (The EGFP is the endogenous signal.) (D–E). For over a day prior to rupture, no cell mixing or change in cell number occurs in either the serosa (D) or the amnion (E). Selected stills from time-lapse movies are shown at the times indicated relative to rupture (at 30°C, z-stacks acquired every 2 min), in anterior aspect and oriented with the ventral region uppermost, with light sheet illumination coming from above. Individual nuclei were tracked hourly (uniquely colored tracks), showing the static nature of serosal cells until the time of rupture, while amniotic nuclei move much more within the cells and the amniotic tissue itself shifts slightly anterior-dorsally. The initial opening at the time of rupture is indicated with an asterisk (D4,E4). Axes were determined based on weak embryonic signal (D5,E5; the withdrawing edge of the amnion is still visible in E5). Images shown here are maximum intensity projections with gamma corrections 0.5 (serosa) and 0.7 (amnion) as well as brightness correction to accommodate for changes in signal intensity of these lines during development (see Figure 1—figure supplement 1 and Koelzer et al., 2014). For these specimens, cellular details at the time of rupture are shown in main text Figure 4D–E. Acquisition details are specified in Supplementary file 1. (F) The bilayer arrangement of the amnion and serosa can also be seen in transmission electron micrographs of sagittal sections (examined after the completion of germband retraction, at 44–48 hr after egg lay; see also main text Figure 2B–C). The red box on the semithin section (1 µm thick, stained with toluidine blue) approximately indicates the anterior-ventral region shown in F2–F3. The arrow highlights an intercellular junction within the serosa (F2), and false coloring highlights the continuity of the two distinct epithelial layers (F3). Both EE tissues have very flat cells, with an apical-basal thickness of only ~200 nm in the amnion and ~450 nm in the serosa, in the aponuclear region shown here. Ultrathin sections (50–100 nm thick) were cut with an 'Ultra' diamond knife on a Leica Ultracut UCT microtome, and examined with a Zeiss EM 109 electron microscope. Scale bars are 50 µm (A1,B1,C1,D1–D5,E1–E5), 20 µm (A2–A5,B2–B5,C2–C5), and 1 µm (F2–F3). Abbreviations: An, antenna; Am, amnion; D, dorsal; H, head; L, left; Lr, labrum; M, mandible; Mx, maxilla; R, right; Ser, serosa; T2, second thoracic segment; V, ventral; Vm, vitelline membrane.
The extraembryonic bilayer moves as a single unit throughout late morphogenetic remodeling.
Images are lateral, with anterior left and dorsal up, shown as maximum intensity projections (A,D), sagittal optical sections (B,C,E,F), or as a mid-sagittal schematic (G). (A–G) The bilayered EE structure is maintained during early serosal compaction. The embryos shown here are at a stage when the serosa folds medially, such that mid-sagittal sections show anterior and posterior arms to the folding tissue (B,E,G). Higher magnification images of cellular structure focus on the posterior arms (C,F). Panels A–C and D–F each show a single embryo. Annotations: arrow, ruptured edge of both tissues; double-headed arrow, double bend in the tissues; dashed green lines delimit EGFP domains. Scale bars are 100 µm (A,D), 50 µm (B,E), and 10 µm (C,C′,F,F′). Abbreviations and schematic color scheme as in previous figures.
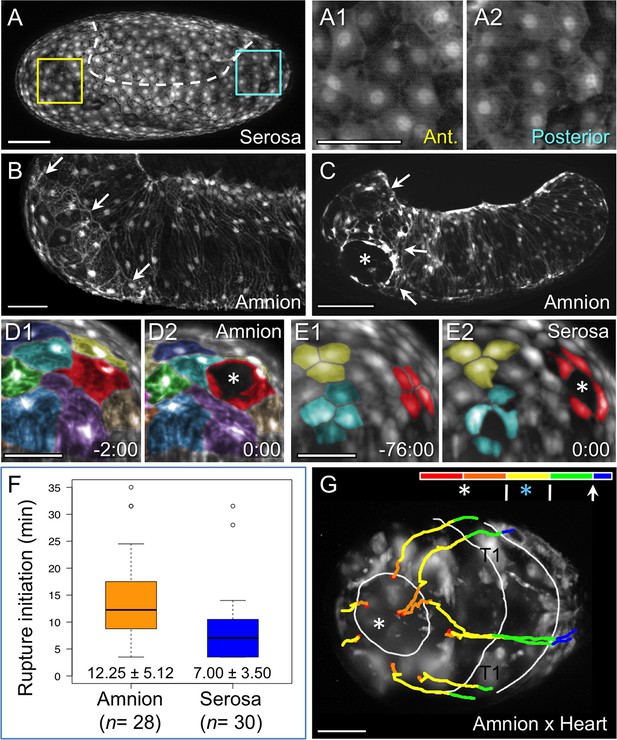
Rupture dynamics: amniotic initiation.
Images are maximum intensity projections in lateral (A–C), anterior (D–E), or anterior-ventral (G) views. (A) The serosa maintains a homogeneous cellular morphology throughout the region apposed to the amnion (below the dashed white line, compare with Figure 1A′), shown in detail for anterior (Ant., A1) and posterior (A2) regions. (B–C) In contrast, amnion-EGFP signal before (B) and during (C) rupture highlights an anterior-ventral cap of morphologically distinct cells (arrows); the asterisk marks the tissue opening (see also Video 2). (D–E) Cell shape changes around the site of opening (asterisk, red cells) differ markedly between amniotic cell contortions (D) and more regular serosal cell shapes (E). Colors mark unique amniotic cells or groups of 3–4 neighboring serosal cells. The nature of opening and cellular connections is further described in the main text, and long-term cell tracking is shown in Figure 2—figure supplement 1. Time stamps are in minutes:seconds at 30°C, relative to rupture at 0:00. The entire interval from initial opening until the EE tissue fully cleared the head was 8 min (D) and 4 min (E). (F) Box plot showing that the rupture initiation interval, as defined in the Materials and methods section, is longer in the amnion (values are median ± median absolute deviation, at 27.5–28ºC). (G) Tissue opening in a heterozygote embryo permits tracking of the ruptured EE tissue relative to embryonic anatomical landmarks ('T1' labels the proximal region of the first leg pair). The colored time scale shows duration of track segments over 36.7 min at 19.5 ± 1°C. Along the time scale and superimposed on the embryo are the site of rupture (white asterisk) and the withdrawing EE tissue edge (white lines, line with arrowhead marks time point shown). Tracks are shown for selected nuclei. For comparison, rupture is also indicated along the time scale for a morphologically stage matched embryo with serosal EGFP (blue asterisk; see also Figure 4—figure supplement 1, Video 3), which shows a shorter rupture initiation interval (interval from the asterisk to the right end of the colored time scale), consistent with the data in panel F. Scale bars are 100 µm (A,C) and 50 µm (A1–A2,B,D1–D2,E1–E2,G). Panel B shows the same embryo as in Figure 1A.
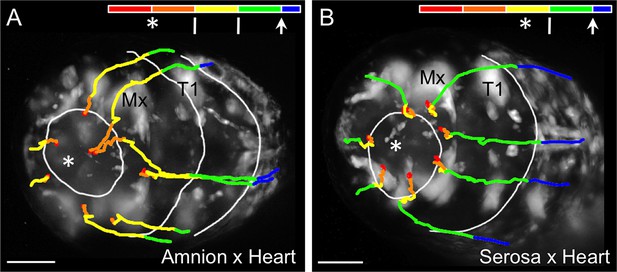
Comparison of early tissue opening in the amnion and serosa.
(A–B) The embryos are heterozygotes expressing EGFP in the specified extraembryonic tissue and in the embryonic landmark domains of the 'heart' enhancer trap line. They are shown in anterior-ventral views as maximum intensity projections of single time points taken from z-stacks acquired every 20 s with a light sheet microscope. Light sheet illumination is coming from above in both recordings (acquisition details are specified in Supplementary file 1). Image A is reproduced from main text Figure 4G for side-by-side comparison with image B. The colored time scale shows the duration of track segments over 36.7 min at 19.5 ± 1°C. The embryos are stage matched for the degree of EE tissue withdrawal by the end of the 36.7-min interval, when the EE tissue has cleared the proximal portion of the first leg pair (T1). Along the time scale and superimposed on the embryo are the site of rupture (white asterisk) and the withdrawing EE tissue edge (white lines, line with arrowhead marks time point shown). In the main text, statistical analysis of the duration of 'rupture initiation' was based on embryos viewed in lateral aspect and was defined as the time from first discernible opening to when the EE tissues cleared the embryo’s head dorsally (see Materials and methods, main text Figure 4F). Since dorsal head structures are not visible in the anterior-ventral movies shown here, the approximate equivalent duration is through the time when the maxillae are cleared (Mx): the interval from 9:00 to 20:20 in the amnion (11:20 duration) and from 20:00 to 27:20 in the serosa (7:20 duration). Tracks are shown for selected nuclei. Scale bars are 50 µm. See also Video 3.
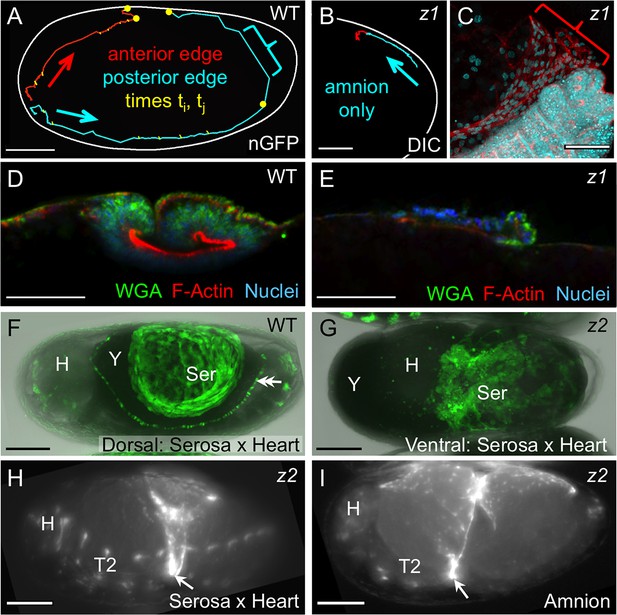
Withdrawal dynamics: serosa-driven progression.
Images are lateral (A–E,H), dorsal-lateral (F,I) or ventral (G), with anterior left and dorsal up, shown as sagittal optical sections (A–B,D–E) or maximum intensity projections (C,F–I). (A−E) Tracking and histological staining of withdrawing EE tissue edges. The WT EE tissue edge squeezes, then rapidly clears, the abdomen. In A, the blue bracket marks a 1-minute interval within a 6.7-hr total interval for the entire red/anterior and cyan/posterior tracks, at 21°C. The distance of the bracketed track segment from the vitelline membrane (white outline) reflects the degree of squeezing of the abdomen. After Tc-zen1RNAi (z1), an amnion-only posterior edge contracts slowly over an uncompressed abdomen. In B, the entire track represents a 3.9-hr interval at 21°C. Note that the track is close to the vitelline membrane outline. Slow amnion-only progression is in part due to ruffling of the tissue. The red track segment in B corresponds to the period of tissue ruffling shown morphologically in the region of the red bracket in C (same staining reagents as in D). Furthermore, there is no apical F-actin enrichment in the folding tissue (E, compare with D and Figure 3E–F). (F–G) While WT serosal contraction leads to a dorsally condensed tissue (F), in strong Tc-zen2RNAi (z2) phenotypes, the serosa condenses ventrally over the unopened amnion and the confined embryo (G). The double arrowhead labels the cardioblasts; 'Y' marks the opaque yolk. (H–I) In weaker Tc-zen2RNAi (z2) phenotypes the EE tissues can rupture and tear ectopically, leaving a 'belt' of EE tissue that squeezes the embryo (arrows). Scale bars are 100 µm (A,F–I) and 50 µm (B–E). Abbreviations as in previous figures.
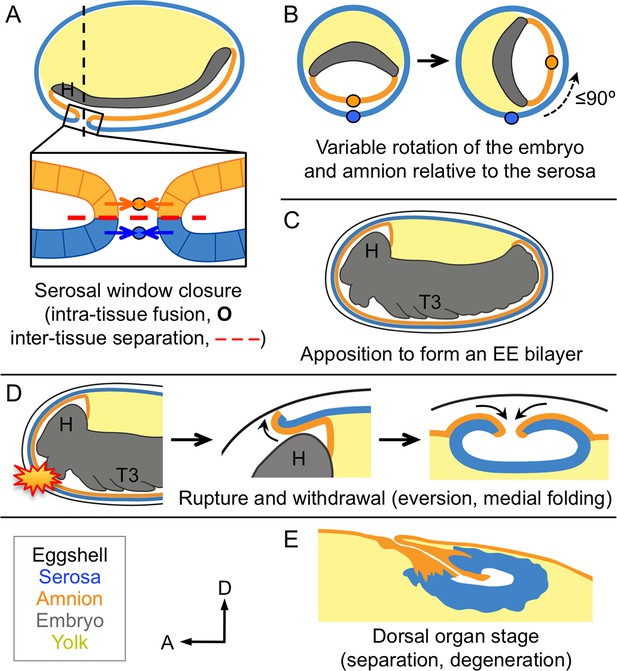
Changing amnion-serosa interactions during Tribolium extraembryonic morphogenesis.
Schematics are sagittal, with anterior left and dorsal up (A,C–E), or transverse with dorsal up (B), illustrating: initial separation during formation of the amnion and serosa as distinct epithelial covers (A), relative rotation in many embryos (B: shown at the position of the dashed line in A, see also Figure 6—figure supplement 1), EE apposition at the retracted germband stage (C), the successive stages of EE tissue withdrawal (D), and final tissue structure during degeneration at the dorsal organ stage (E, see also Figure 6—figure supplement 2). Amnion-serosa apposition persists during withdrawal morphogenesis (D), which involves EE tissue rupture (starburst), curling over of the resulting tissue edge as it everts (shown for the anterior-dorsal edge), and early serosal compaction as the EE edges fold medially. Abbreviations as in previous figures.
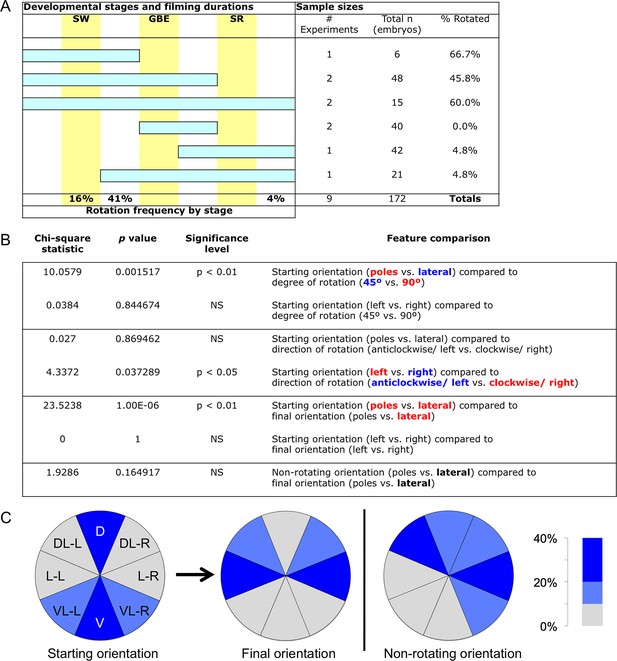
Frequency of embryonic longitudinal rotation during development.
(A) Sample sizes and film durations (blue bars) relative to landmark developmental stages (yellow bars: SW, serosal window closure; GBE, germband extension complete; SR, serosal rupture; staging as in Koelzer et al. (2014)). Embryos were analyzed from multiple time-lapse data sets of at least 15.75-hr duration at 30°C, recorded with an inverted DeltaVision RT (Applied Precision) microscope. Pooled data included embryos in the nGFP and various enhancer trap backgrounds (G04609-heart, G12424-serosa, KT650-serosa, HC079-amnion), including heterozygote crosses of HC079 with each of G04609, G12424, and nGFP. (Background autofluorescence was sufficient for scoring early development in the absence of specific GFP signal.) 'Frequency by stage' refers to the beginning of rotation; all embryos beginning during (16%) or after (41%) the SW stage completed rotation before the GBE stage. Rarely, a moderate degree of rotation occurred during dorsal closure (4%). (B). Chi-square statistics for specified comparisons. For significant results, text color-coding indicates the nature of the interaction. Longitudinal rotation ranged from approximately 20 to 100 degrees, with two-thirds of embryos rotating approximately 90°. Embryos laying on their dorsal or ventral surface ('poles') were more likely (69%) to rotate 90°, while embryos in lateral aspect (85%) tended to only rotate 45°. While overall there was no bias in direction of rotation, embryos initially laying on their right sides tended to rotate anticlockwise (70%) while embryos on their left sides rotated clockwise (78%). Direction of rotation was from the perspective of the embryo’s posterior (i.e., clockwise is right, anticlockwise is left). These directional biases are applicable across the full range from ventral-lateral through dorsal-lateral, such that rotation tended to result in more strictly lateral final orientations (64%). Meanwhile, most non-rotating embryos began in a lateral orientation (89%). (C) Pie charts representing the eight 45°-sectors of the egg circumference (D, dorsal; DL-L and -R, dorsal-lateral-left and -right; L-L and -R, lateral-left and -right; VL-L and -R, ventral-lateral-left and -right; V, ventral), color-coded for frequency of occurrence according to the heat map.
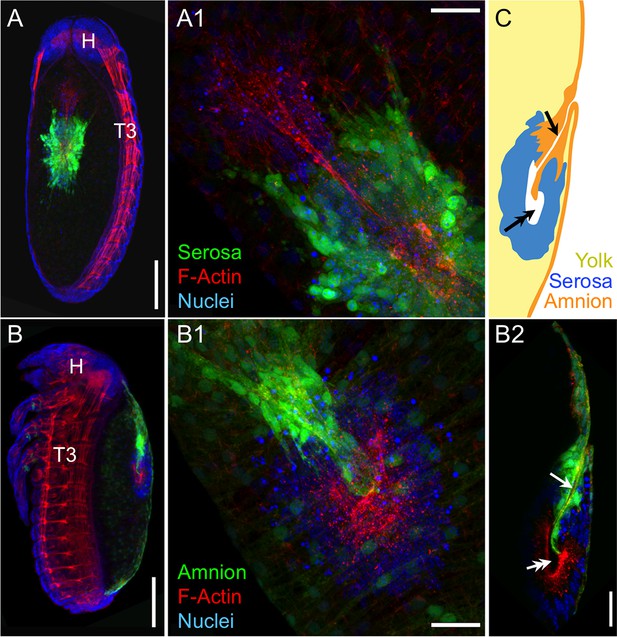
Separation during degeneration: the amnion and serosa resolve into two distinct dorsal organ structures.
Images are dorsal (A,A1,B1) or lateral (B,B2,C), with anterior up (A,B,B2,C), or upper-left (A1,B1), shown as maximum intensity projections (A,A1,B,B1) or sagittal optical sections (B2,C). Tissue-specific EGFP labels two distinct dorsal organ structures, each characterized by apical F-actin enrichment around a hollow center or cleft (arrow: amniotic; double-headed arrow: serosal). Scale bars are 100 µm (A,B) and 25 µm (A1,B1,B2). After the stage shown here, the two dorsal organs will separate: the serosa remains medial, while the amniotic dorsal organ migrates anteriorly to a position behind the head before undergoing final degeneration (Panfilio et al., 2013).
Videos
Time course of EGFP expression in the Tribolium enhancer trap line HC079.
The embryo is shown in lateral aspect with anterior left and dorsal up. Strengthening EGFP signal is specific to the amnion during germband retraction and up to the point of EE tissue rupture and withdrawal morphogenesis. In the later stages, additional embryonic expression domains occur in the eyes and legs. Throughout the movie, wandering yolk globules also exhibit low levels of EGFP, a feature observed for other enhancer trap lines from this screen (Koelzer et al., 2014). The time stamp specifies minimum age from a four-hour egg collection, in hours after egg lay (h AEL). Maximum intensity projections from a deconvolved z-stack (5 µm step size for a 60 µm stack) recorded every ten minutes over a 48-hr time-lapse at 30ºC, acquired with an inverted DeltaVision RT microscope (Applied Precision). Scale bar is 100 µm. Selected still images are shown and further described in Figure 1—figure supplement 1.
Tribolium extraembryonic tissue rupture and withdrawal shown with amnion-specific EGFP (enhancer trap line HC079).
The embryo is shown in lateral aspect with anterior left and dorsal up, with amnion-specific EGFP expression, as well as restricted late embryonic expression domains in the legs and body segments (see also Figure 1—figure supplement 1). The movie spans late amnion morphogenesis, from 3.3 hr before rupture through late dorsal closure. Of particular note are the brighter, rounder amniotic cells in an anterior-ventral cap, in which rupture occurs. During withdrawal, the amnion everts (turns inside out), such that the surface that had faced inward toward the embryo is flipped outward to face the vitelline membrane: this is particularly apparent from 35 to 49 min after rupture as the ruptured tissue edge folds over. Time is shown relative to tissue rupture at 0 min. Images are maximum intensity projections with a gamma correction of 0.7 from a z-stack (7 µm step size for a 77 µm stack) recorded every seven minutes over a 9.9-hr time-lapse at 24°C, acquired with an Axio Imager.Z2 with ApoTome.2 structured illumination (Zeiss). Scale bar is 100 µm. Figure 4C shows a still at 21 min after rupture.
Extraembryonic rupture filmed at high temporal resolution.
Embryos are shown in anterior-ventral aspect with anterior left, and labeled with EGFP from a heterozygote cross of enhancer trap lines labeling the amnion (line HC079) or serosa (line G12424) with selected embryonic domains in the head, segments, and legs (line G04609, 'heart'). During the movie, the EE tissues rupture and withdraw to the extent that the head and first leg pair are exposed. Selected EE nuclei were tracked, where track color segments correspond to fixed units of time (red, orange, yellow, and green: 8.3 min; blue: final 3.3 min). Elapsed time is shown. Images are maximum intensity projections from a z-stack (2.58 µm step size for a 175 or 200 µm stack) recorded every 20 s at 19.5 ± 1°C and shown for a 36.7-minute period (see Supplementary file 1 for further acquisition details). Single-sided light sheet illumination is from the top, resulting in deterioration of the signal in the lower part of the image due to scattering. The final frame shows time point 36:40 with the approximate position of mouthparts labeled for orientation: An, antennae; Lr, labrum; Mx, maxillae; Lb, labium; T1, first thoracic segment. Scale bar is 50 µm. See also Figure 4—figure supplement 1 for a visual summary.
Additional files
-
Supplementary file 1
Acquisition parameters for featured mDSLM light sheet experiments.
- https://doi.org/10.7554/eLife.13834.017





