Viral hijacking of a replicative helicase loader and its implications for helicase loading control and phage replication
Figures
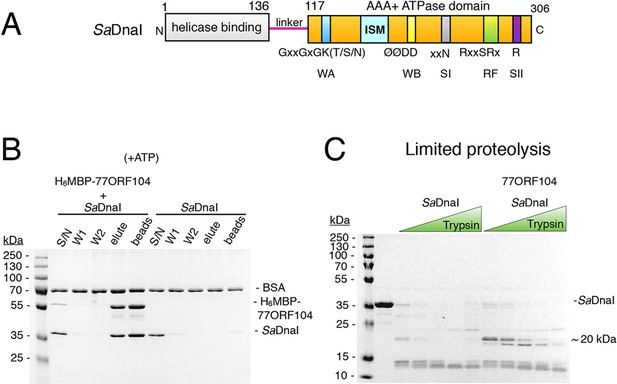
The phage 77 ORF104 protein binds the S. aureus DnaI ATPase domain.
(A) Domain organization of SaDnaI. The N-terminal helicase-binding domain is colored gray, the linker region is magenta, and the AAA+ domain is orange. Numbers refer to amino acid positions. AAA+ ATPase motifs are labeled: Walker-A (WA), Walker-B (WB), Sensor-I (SI), Box VII, Sensor-II (SII), and the initiator/loader specific motif (ISM). (B) SDS-PAGE analysis of amylose pull-downs between His6MBP-77ORF104 and full-length SaDnaI (with ATP). A DnaI alone control is shown. Supernatant – S/N, W1 and W2 – washes. (C) Limited trypsin proteolysis and SDS-PAGE analysis of full-length SaDnaI in the presence of 77ORF104. A ~20kDa band of SaDnaI is stabilized in the presence of 77ORF104.
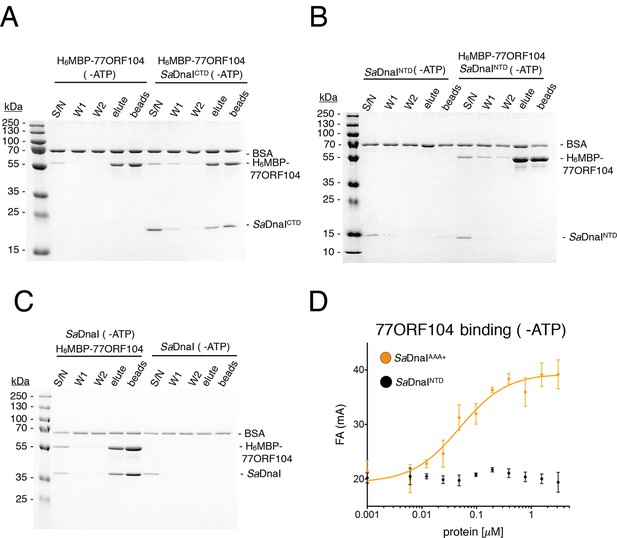
Binding of 77ORF104 to the SaDnaI AAA+ domain is nucleotide-independent.
SDS-PAGE analysis of amylose pull-downs between His6MBP-77ORF104 and (A) SaDnaICTD, (B) SaDnaINTD, or (C) full-length SaDnaI in the absence of ATP. DnaI and 77ORF104 alone controls are also shown. (D) Binding the phage 77 ORF104 protein to SaDnaIAAA+ and SaDnaINTD as measured by a change in fluorescence anisotropy (ΔFA – change in milli-anisotropy units). The X-axis denotes protein concentration. Data points and error bars derive from three-independent experiments. No measurable binding was observed for SaDnaINTD.
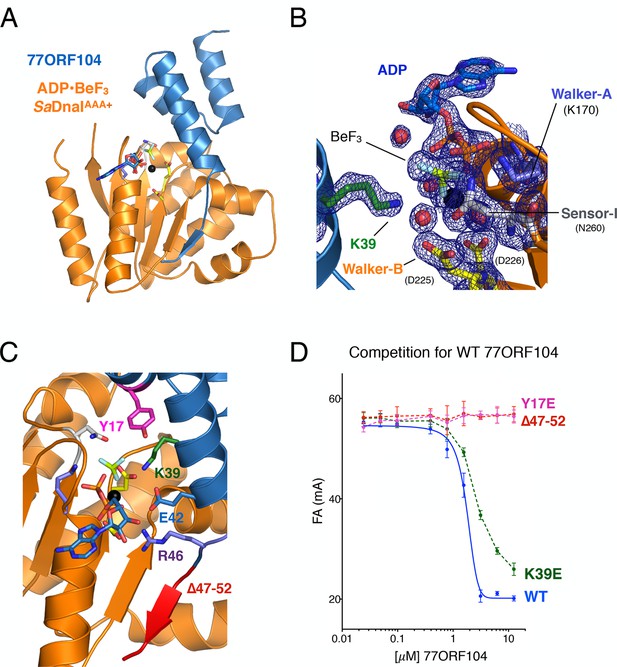
Structure of the ADP•BeF3-bound SaDnaIAAA+•77ORF104 complex and biochemical validation of observed interactions.
(A) Overall structure of ADP•BeF3-bound SaDnaIAAA+ complexed with 77ORF104. SaDnaIAAA+ is colored orange and 77ORF104 sky-blue. ADP (dark blue), BeF3 (limon-teal) and a magnesium ion (black), are shown within the ATP binding site of SaDnaIAAA+. (B) Close-up view of the SaDnaI AAA+ ATP binding site. Conserved motifs are colored: Walker-A (Lys170) (blue), Walker-B (Asp225 and Asp226) (yellow), and Sensor-I (Asn260) (grey). Lys39 from 77ORF104 is colored green, a magnesium ion in black and liganding waters in red. Refined 2Fo-Fc electron density is shown for a portion of the region, contoured at 1.6 σ. (C) Analysis of the ADP•BeF3-bound SaDnaIAAA+•77ORF104 interface. Several residues from 77ORF104 that participate in the interaction are shown as sticks and labeled. Three elements selected for mutagenesis studies are colored pink (Tyr17), green (Lys39), and red (residues 47–52). (D) Competition assay showing the ability of different 77ORF104 mutants to compete away wild-type, N-terminally labeled 77ORF104 from interacting with SaDnaIAAA+. Competition is evident by a decrease in fluorescence anisotropy (ΔFA – change in milli-anisotropy units) as labeled protein is displaced by the unlabeled 77ORF104 competitor.
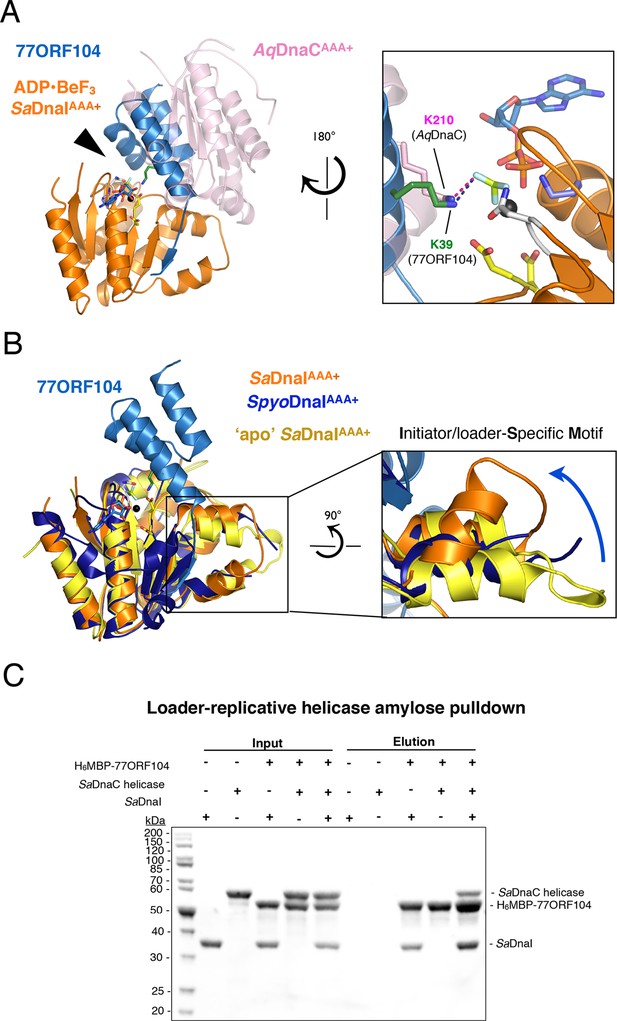
77ORF104 alters the local conformation of the SaDnaI ISM but does not block interactions with the SaDnaC replicative helicase.
(A) Superposition of a nucleotide-stabilized Aquifex aeolicus DnaCAAA+ (AqDnaCAAA+) dimer (PDB ID 3ECC, [Mott et al., 2008]) onto the SaDnaIAAA+•77ORF104 structure. The docking results in a steric clash between 77ORF104 and the neighboring protomer of the AqDnaCAAA+ dimer. The inset (rotated 180˚) shows a close-up view of the ATP binding center, highlighting how the dimer-partner of the Aquifex aeolicus DnaCAAA+ domain projects a positive amino acid (Lys210) into the AAA+ active site in a manner similar that seen for Lys39 from 77ORF104. (B) Induction of a local conformational change to the SaDnaI ISM by 77ORF104 can be seen in a comparative structural analysis with other DnaI homologs. The inset (rotated 90˚) shows a close-up view of ISM helices in the 77ORF104-inhibited complex aligned to apo SaDnaIAAA+ and Streptococcus pyogenes DnaI (SpyoDnaIAAA+) (PDB ID 2QGZ, Seetharaman et al., to be published). The blue arrow indicates the direction of ISM bending by the phage inhibitor protein. (C) 77ORF104 associates with SaDnaI when the helicase loader is bound to the host SaDnaC helicase. SDS-PAGE analysis of amylose pull-down experiments using purified His6MBP-tagged 77ORF104, SaDnaI, and the SaDnaC replicative helicase. The positions of each protein are indicated on the right; inputs are shown on the left half of the gel. His6MBP-tagged 77ORF104 co-precipitates with SaDnaI alone and SaDnaI bound to the SaDnaC replicative helicase, but not with the SaDnaC helicase alone. Pull-down experiments were performed in the absence of nucleotide. Performing the experiment in the presence of nucleotide generated the same result (not shown), indicating nucleotide is not required for SaDnaI to associate with SaDnaC.
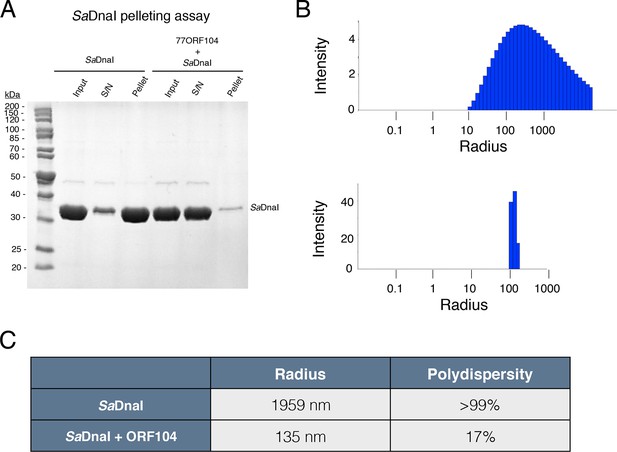
77ORF104 directly interferes with the self-association of SaDnaI.
(A) SDS-PAGE analysis of centrifugal SaDnaI assembly assay. Lanes from reaction containing SaDnaI with or without 77ORF104 are shown and labeled. S/N – supernatant. (B) Dynamic light scattering plots of SaDnaI alone (upper) and SaDnaI in the presence of equimolar 77ORF104 (lower). (C) Table representing some the physical parameters calculated from DLS measurements of DnaI alone and in the presence of the ORF104 protein.
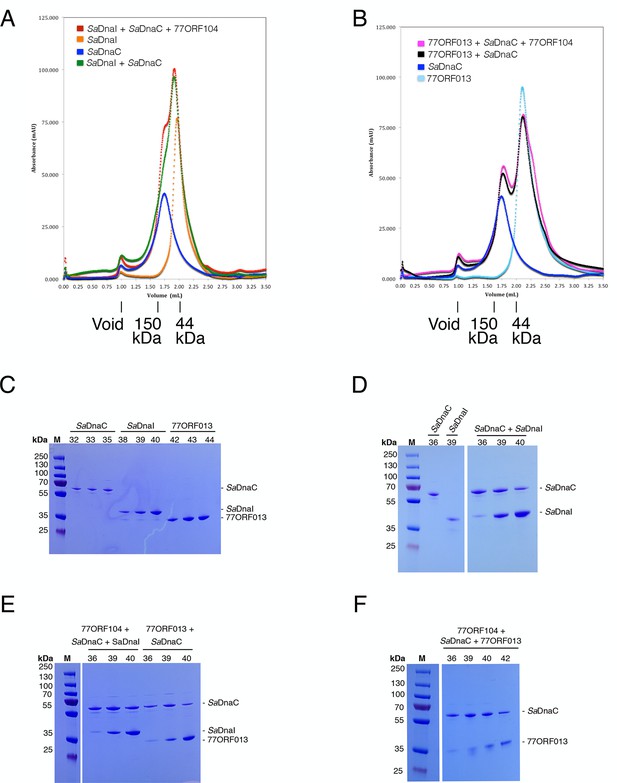
Analytical gel filtration results of SaDnaC, SaDnaI, 77ORF013 and 77ORF104, both alone and in various combinations.
The relative positions of marker proteins are indicated. (A) Analytical gel filtration chromatographs of SaDnaC and SaDnaI alone, SaDnaC and SaDnaI together, and SaDnaC and SaDnaI with 77ORF104. (B) Analytical gel filtration chromatographs of SaDnaC and 77ORF013 alone, SaDnaC and 77ORF013 together, and SaDnaC and 77ORF013 with 77ORF104. (C) SDS-PAGE analysis of selected peak fractions for SaDnaC, SaDnaI and 77ORF013 alone. (D) SDS-PAGE analysis of selected peak fractions for SaDnaC, SaDnaI and SaDnaC + SaDnaI. (E) SDS-PAGE analysis of selected peak fractions for 77ORF104 + SaDnaC + SaDnaI and 77ORF013 + SaDnaC. (F) SDS-PAGE analysis of selected peak fractions for 77ORF104 + SaDnaC + 77ORF013.
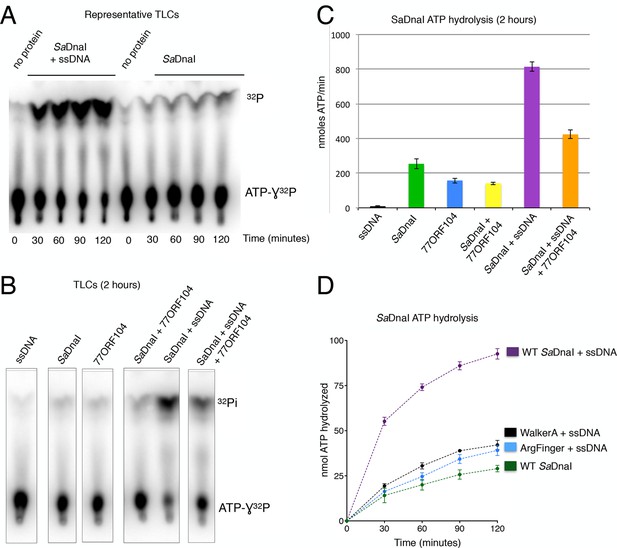
77ORF104 inhibits ssDNA stimulation of ATP hydrolysis by SaDnaI.
(A) Representative TLC image shown for SaDnaI ATP hydrolysis in the presence (left) or absence (right) of M13ssDNA. (B) M13ssDNA stimulates ATP hydrolysis by SaDnaI while 77ORF104 inhibits this effect. (C) Representative TLC image of SaDnaI ATP hydrolysis experiments in the presence and absence of wild-type 77ORF104 and/or M13ssDNA. (D) Effects of ATPase mutations on observed hydrolysis activity. Walker-A (K170A) and arginine finger (R288A) mutations in SaDnaI show reduced stimulation of activity, comparable to that seen in the absence of ssDNA.
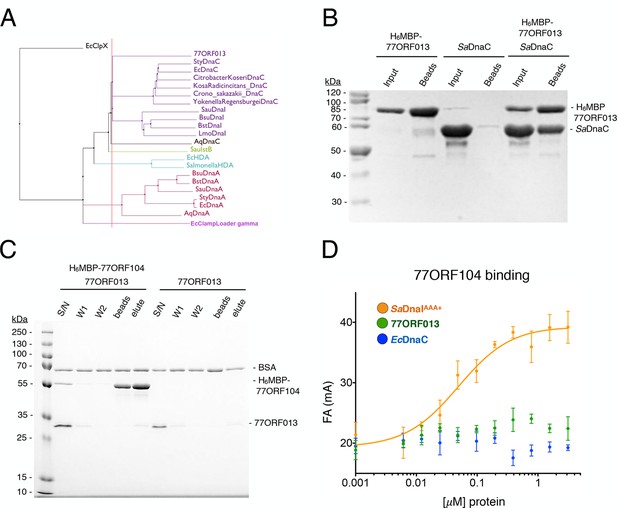
Phage 77 encodes a bacterial helicase loader homolog (ORF013) that binds to the host SaDnaC replicative helicase.
(A) Phylogenetic tree of the phage encoded ORF013 gene with various initiator/loader clade AAA+ ATPases. 77ORF013 clusters with DnaC/DnaI family helicase loaders; a distantly related AAA+ ATPase (E. coli ClpX) was included for the purpose of rooting the tree. (B) SDS-PAGE analysis of amylose pull-down experiments using His6MBP-tagged phage loader 77ORF013 and the S. aureus DnaC replicative helicase. The 77ORF013 helicase loader homolog binds to the host SaDnaC helicase. (C) SDS-PAGE analysis of amylose pull-downs using His6MBP-tagged 77ORF104 and 77ORF013 helicase loader homolog. 77ORF013 does not associate with the 77ORF104 inhibitor protein. (D) Binding to phage 77ORF104 inhibitor protein to SaDnaIAAA+, 77ORF013 and E. coli DnaC as measured by a change in fluorescence anisotropy (ΔFA – change in milli-anisotropy units). The X-axis represents protein concentration. Data points and error bars derive from three-independent experiments. No measurable binding was observed for the phage 77 ORF013 or E. coli DnaC. SaDnaIAAA+ is shown as a positive control.
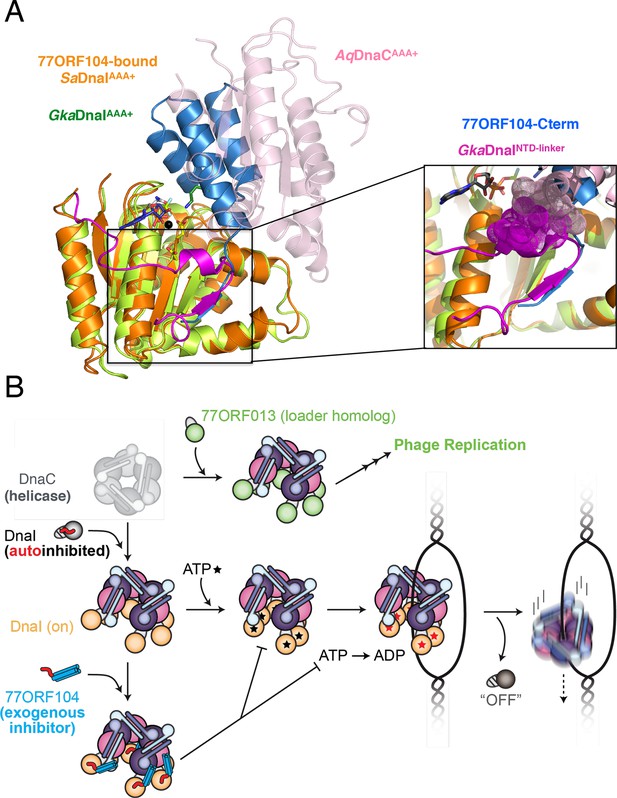
Insights into bacterial helicase loader mechanism and regulation derived from this study.
(A) The N- and C-terminal domain linker of DnaC/I helicase loaders occupies a region bound by the phage 77 inhibitor protein. The monomeric ATPase domain of G. kaustophilus DnaI (GkaDnaI AAA+) (PDB ID 2W58, [Tsai et al., 2009b]) is shown superposed onto a nucleotide-assembled dimer of both the A. aeolicus DnaC AAA+ domain (a construct that lacks the linker, PDB ID 3ECC, [Mott et al., 2008])) and the SaDnaIAAA+ domain (as seen in complex with 77ORF104). The C-terminal tail of 77ORF104 forms a β-strand that occupies the same location as the GkaDnaIAAA+ linker (magenta), although the strand runs in the opposite direction. (Inset) Close-up view of the linker/tail binding site. Several amino acids in the linker are shown as space filling (magenta) to illustrate how this element would sterically clash with a second, incoming helicase loader ATPase domain to prevent self assembly (pink spheres, from AqDnaC). (B) Schematic summarizing the auto-regulation of DnaC/I helicase loaders and the ability of phage 77-family viruses to inhibit host helicase loading and co-opt host replicative helicases in S. aureus and related Gram-positive bacteria. The bacterial helicase loader linker region forms an intra-molecular interaction with its associated AAA+ domain to auto-repress inopportune loader/loader interactions; upon binding to the replicative helicase, the linker is proposed to undock from this region to allow self-assembly of the loader ATPase folds and subsequently assist with loading of the replicative helicase onto single-stranded origin DNA. The phage 77 ORF104 inhibitor protein binds to cognate loaders such as SaDnaI, repressing loader self-association, ATPase activity, and presumably its ability to properly participate in the helicase loading reaction. Phage 77 is also found to encode a helicase loader homolog (ORF013) that directly binds to the host replicative helicase, likely as a means to re-direct the host’s replication machinery toward replication of the viral genome.
Tables
Data collection, phasing and refinement statistics for apo SaDnaIAAA+ and ADP•BeF3-SaDnaIAAA+•77ORF104 structures.
| Construct: | 'apo' SaDnaIAAA+ | 77ORF104-SaDnaIAAA+ |
|---|---|---|
| Data Collection | ||
| Beamline | BNL NSLS X25 | BNL NSLS X25 |
| Wavelength | 1.500 | 0.979 |
| Space group | P212121 | P6522 |
| Cell edges (Å) | 113.10, 126.26, 183.34 | 73.17, 73.17, 189.72 |
| Cell angles (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 |
| Resolution range (Å) | 48.1–2.6 (2.74– 2.6) | 44.8–1.9 (1.97–1.9)a |
| Unique reflections | 81,324 (11,704) | 24,589 (2,390) |
| Completeness (%) | 99.9 (99.5) | 100.00 (100.00) |
| Rmerge | 0.16 (4.00) | 0.165 (1.55) |
| Rmeas | 0.17 (4.33) | 0.17 (1.65) |
| Rpim | 0.066 (1.66) | .054 (.57) |
| Redundancy | 13.3 (13.2) | 18.5 (15.7) |
| I/σ(I) | 3.4 (0.2) | 14.82 (1.44) |
| Wilson B-Factor | 29.6 | 26.6 |
| CC 1/2 | 0.996 (0.684) | 0.999 (0.693) |
| Phasing | ||
| # of sites | NA | 4 |
| FOMb | NA | 0.90 (0.54) |
| Refinement | ||
| Resolution limits (Å) | 48.13 (2.6) | 47.4 (1.9) |
| Rwork(Rfree)b | 22.7 (26.5) | 18.0 (21.8) |
| No. protein residues | 1975 | 223 |
| No. solvent/ligand molecules | 67/9 | 209/32 |
| RMSD Bond, Å | 0.003 | 0.012 |
| RMSD Angle, ° | 0.540 | 1.335 |
| Protein geometry | ||
| Ramachandran preferred/outliers (%) | 95.87/0.61 | 99.5/0 |
| Rotamer outliers (%) | 0.48 | 0 |
-
aNumbers in parentheses refer to the highest resolution shell.
-
bFive percent of the total number of reflections were used to calculate Rfree.





