Compaction and segregation of sister chromatids via active loop extrusion
Figures
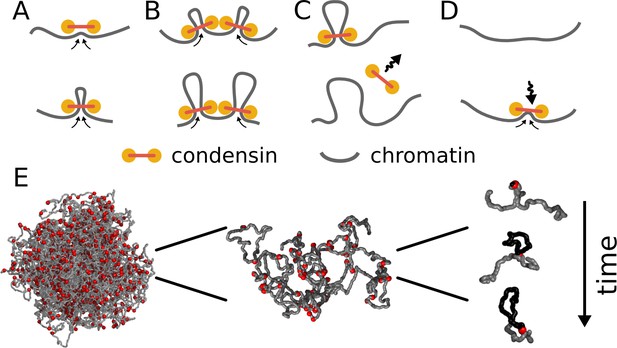
Model of loop extrusion by condensins.
Top row, the update rules used in simulations: (A) a condensin extrudes a loop by moving the two ends along the chromosome in the opposite directions, (B) collision of condensins bound to chromosomes blocks loop extrusion on the collided sides, (C) a condensin spontaneously dissociates and the loop disassembles; (D) a condensin associates at randomly chosen site and starts extruding a loop. Bottom row, (E) we use polymer simulations to study how combined action of many loop extruding condensins changes the conformation of a long chromosome.
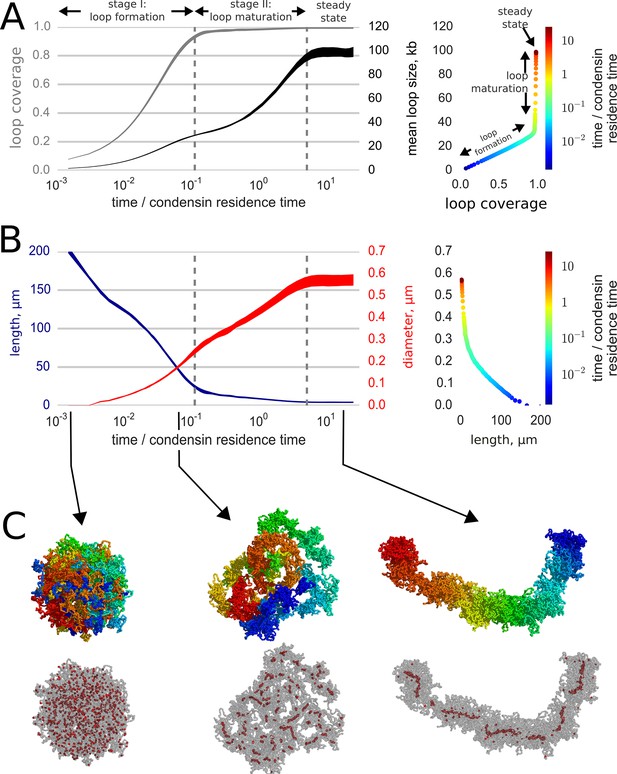
Loop-extruding condensins convert globular interphase chromosomes into elongated structures.
(A) Dynamics of loop array formation. Left, the fraction of the chromosome extruded into loops (loop coverage, gray) and the mean loop size (black). The time is measured in the units of condensin residence time. Data for each curve was obtained using 10 simulation replicas; the thickness of each line shows the difference between the 10th and 90th percentile at each time point. Right, a phase diagram showing loop coverage vs mean loop length. The curve is averaged over the simulation replicas and colored by log-time. Two stages of dynamics are separated by the two vertical dashed lines corresponding to 95% coverage and 95% of the steady state mean loop length. The distinction between the two stages of compaction is especially easy to see on the phase curve. (B) Dynamics of chromosome morphology. Left: chromosome length (blue) and the mean chromosome diameter (red) shown as a function of time; the thickness of each line shows the difference between the 10th and 90th percentile at each time point. Right, a phase diagram showing chromosome length vs diameter; the curve is averaged over the simulation replicas and colored by log-time. Note that most of chromosome linear compaction is achieved during the first phase, while the second is characterized by widening of the chromosome and further three-fold shortening. (C) Snapshots of polymer simulations. Top row: the conformations of chromosomes at the beginning of simulations (left), slightly before the transition between the loop formation and maturation stages (middle), and in the steady state (right). Color (rainbow: from red to blue) shows positions of individual monomers along the chain and demonstrates that loci in compacted chromosomes are arranged linearly according to their genomic location. Bottom row: same as above, with chromosomes shown in semitransparent gray and condensins as red spheres.
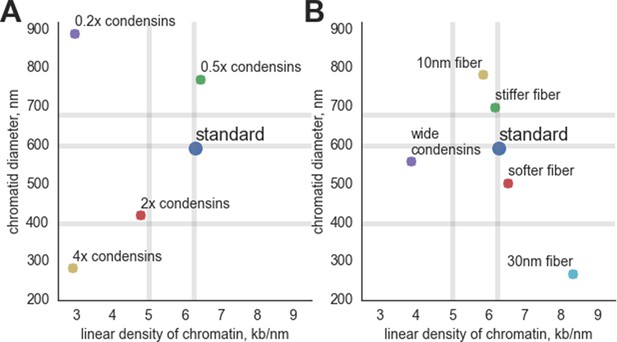
The geometrical parameters of compacted chromosomes in simulations with altered parameters.
(A) Simulations with altered condensin abundance: '0.2x condensin', '0.5x condensin', '2x condensins' and '4x condensins' - simulations with 200, 500, 2000 and 4000 condensins, correspondingly. (B) Simulations with altered parameters of the chromatin fiber and condensins: 'Stiffer fiber' – simulations with 2x bending energy of the fiber, 'softer fiber' – simulations with 0.5x bending energy; 'wide condensins' – simulations with increased spacing between condensins at loop bases; '10 nm fiber' - simulations with reduced linear DNA of the fiber; '30 nm fiber' – simulations with increased linear DNA density of the fiber and increased fiber thickness. The vertical gray lines show the experimental observations on the linear density of DNA along the human prophase chromosome I (6.25 kb/nm) and chromosome XXII (5.0 kb/nm) (Yunis, 1981). The horizontal gray lines show the experimental data on the diameter of prophase chromosomes: 400 nm (Kireeva et al., 2004), 600 nm (Sarkar et al., 2002) and 679 nm (el-Alfy and Leblond, 1989).
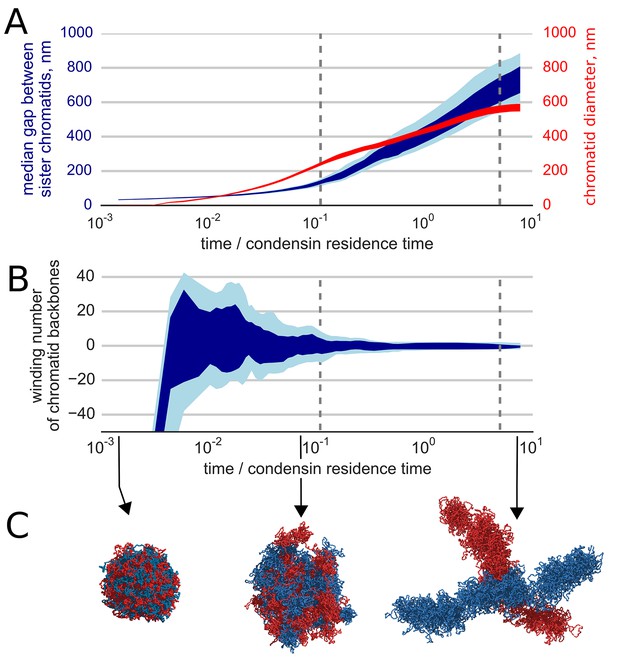
Segregation and disentanglement of sister chromatids.
(A) The median separation between condensin-rich axes of two sister chromosomes (blue) and the mean chromosome diameter (red) as a function of time, measured in condensin residence times, as above. The blue band shows the range between 25th and 75th percentile across 10 simulation replicas at each time point; the light blue shows 10%--90% range. (B) Entanglement of sister chromatids measured by the linking number of the condensin cores of sister chromatids; the blue bands show variability between simulation replicas, same as above. (C) Conformations of two sister chromatids in the polymer simulations (shown in red and blue) illustrate the observed segregation and disentanglement.
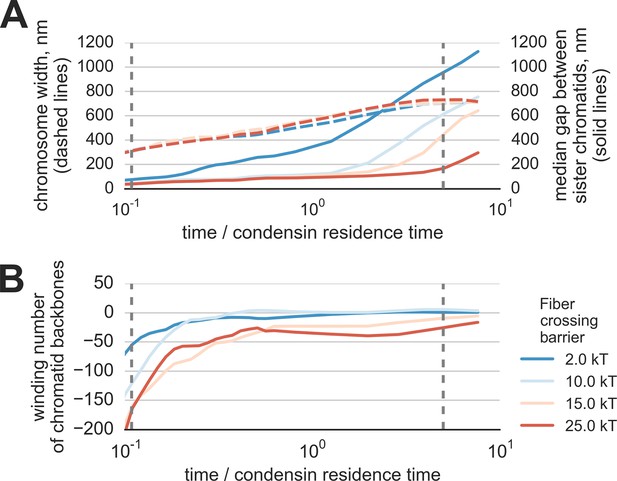
Segregation and disentanglement of sister chromatids in simulations with reduced Topo II concentration.
(A) The median separation between condensin-rich axes of two sister chromosomes (solid lines) and the mean chromosome diameter (red) as a function of time, measured in condensin residence times, as above. Depletion of topo II was simulated by a simultaneous increase of the energy barrier of fiber overlap (shown by the line color) and a 3x increase in of the radius of repulsion. (B) Entanglement of sister chromatids in simulations of topo II depletion, measured by the linking number of the condensin cores of sister chromatids.
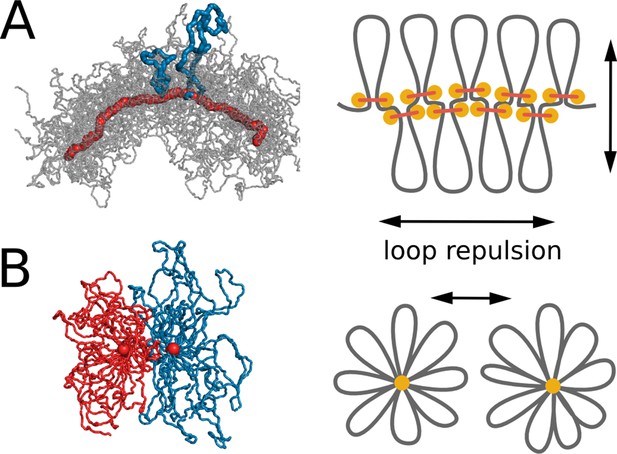
Steric repulsion between chromatin loops shapes the compacted chromosomes and leads to segregation of sister chromatids.
(A) Left, conformation of a single chromosome with chromatin fiber shown in semitransparent gray and condensins shown in red; two loops are highlighted with blue. Right: the diagram showing that loop repulsion straightens chromosomal cores and extends individual loops radially. (B) Left, cross section of two parallel loop brushes placed at a short distance from each other. Right, the diagram shows that steric interactions between loops of sister chromatids lead to chromatid repulsion and segregation.
Videos
An illustration of the model of loop extrusion by condensins.
When a condensin (red sphere) binds to chromatin (grey fiber), it binds to two adjacent loci and then slides the two contact points in the opposite directions, effectively extruding a loop (highlighted in orange). The diagram on the bottom shows the position of the two loci brought together by the condensin over time (red arc). Condensins occasionally unbind and then rebind to a randomly chosen site, starting a new loop. When two condensins (loops shown in orange and purple) meet on the chromosome, they stop extrusion on the collided sides. Condensins can also bind within already extruded loops. If the growth of the outer loop is blocked by other loops or barriers (not shown), the inner condensin can re-extrude it, forming a single loop reinforced by multiple condensins (the number above the arc). Such a loop is stabilized against unbinding of individual condensins and disassembles only upon unbinding of the last condensin. Available at https://www.youtube.com/watch?v=xikxZ0yF1yU&list=PLnV-JMzdy0ZMOvyK6Z1bzU7C2Jt8Brbit&index=1
A representative polymer simulation of compaction of 30 Mb chromosome by loop extruding condensins in presence of strand-passing topo II.
Top left, fiber color corresponds to the position of each monomer along the genome. Top right, fiber shown in semitransparent gray, condensins are shown as red spheres. Bottom, the diagram of the crosslinks (red arcs) formed by condensins. The numbers above the arcs show the number of condensins stacked at the bases of reinforced loops. We sped up the second half of the video 20 times to illustrate the slow phase of loop maturation and the long-term stability of the compacted state. Available at https://www.youtube.com/watch?v=_Vc7__xfnfc&list=PLnV-JMzdy0ZMOvyK6Z1bzU7C2Jt8Brbit&index=2
A polymer simulation of segregation and disentanglement of two sister chromatids by loop extruding condensins in presence of strand-passing topo II.
The two sister chromatids (30 Mb each) are shown in red and blue. Initially, the two chromatids were compacted and twisted around each other. Second half of the video is sped up 20x. Available at https://www.youtube.com/watch?v=stZR5s9n32s&index=3&list=PLnV-JMzdy0ZMOvyK6Z1bzU7C2Jt8Brbit
Sixteen simulation replicas of chromatid segregation and disentanglement.
Available at https://www.youtube.com/watch?v=UXXLnGGit-8&index=4&list=PLnV-JMzdy0ZMOvyK6Z1bzU7C2Jt8Brbit
Polymer simulation of segregation and disentanglement of two sister chromatids with altered model parameters.
Top row: left, simulations with a more flexible fiber (0.5x bending energy); right – a stiffer fiber (2x bending energy); bottom row, left - simulations with wider condensins at loop bases; right – simulations with disabled excluded volume interactions. Available at https://www.youtube.com/watch?v=YhgSHppJXVI&index=5&list=PLnV-JMzdy0ZMOvyK6Z1bzU7C2Jt8Brbit
Polymer simulation of segregation and disentanglement of two sister chromatids with modified condensin abundance.
Top row, left - 2000 condensins per chromatid, middle - 500 condensins per chromatid; right - 200 condensins; bottom row, left - 4000 condensins, left - 100 condensins; right, 50 condensins. Available at https://www.youtube.com/watch?v=4img9nmQrtU&index=6&list=PLnV-JMzdy0ZMOvyK6Z1bzU7C2Jt8Brbit
A polymer simulation of segregation and disentanglement of two sister chromatids upon topo II depletion.
Depletion of topo II was simulated by a simultaneous increase of the energy barrier of fiber overlap and the increased radius of repulsion, which decreased, but not fully prevented chain crossings. Available at https://www.youtube.com/watch?v=YO3Qx8liK3s&list=PLnV-JMzdy0ZMOvyK6Z1bzU7C2Jt8Brbit&index=7
A polymer simulation of segregation and disentanglement of sister chromatids in the presence of cohesins connecting the chromatids.
We simulated cohesins as crosslinks between the chromosomes (shown with the blue lines on the bottom arc diagram) that can be pushed along each of the chromatids by loop-extruding condensins. Available at https://www.youtube.com/watch?v=vmwNbz41pqM&list=PLnV-JMzdy0ZMOvyK6Z1bzU7C2Jt8Brbit&index=8





