Nerve growth factor receptor negates the tumor suppressor p53 as a feedback regulator
Figures
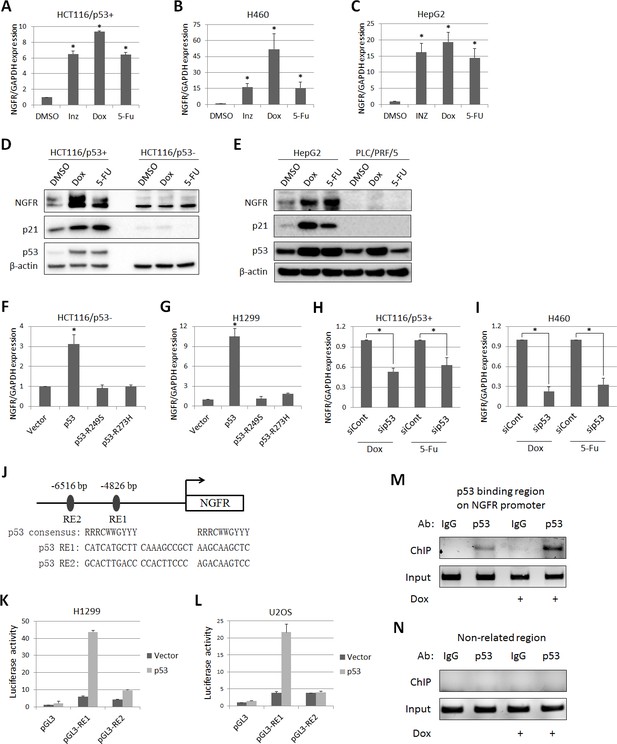
p53 transcriptionally induces NGFR expression in cancer cells.
(A,B,C) NGFR mRNA expression is elevated by p53-inducing agents. HCT116 p53+/+ (A) H460 (B) and HepG2 (C) cells were treated with Inauhzin, Doxorubicin or 5-Fluorouracil for 15 h, and NGFR expression was determined by q-PCR (Mean ± SEM, n = 3). Three biological replicates (independent experiments) and a two-tailed t-test were used for P value calculation, p*<0.01. Most q-PCR were performed by three biological replicates, each including three technical replicates. (D) NGFR protein expression is elevated by p53-inducing agents in colon cancer cell lines. HCT116 p53+/+ and HCT116 p53-/- cells were treated with Doxorubicin or 5-Fluorouracil for 15 hr followed by IB using antibodies as indicated. (E) NGFR protein expression is elevated by p53-inducing agents in liver cancer cell lines. HepG2 and PLC/PRF/5 cells were treated with Doxorubicin or 5-Fluorouracil for 15 hr followed by IB using antibodies as indicated. (F,G) NGFR mRNA expression is induced by ectopic wild-type, but not mutant, p53. HCT116 p53-/- (F) and H1299 (G) cells were transfected with wild-type or mutant p53 for 30 hr and NGFR expression was determined by q-PCR (Mean ± SEM, n = 3). Three biological replicates were used for p value, p*<0.01. (H,I) NGFR mRNA expression is inhibited by p53 knockdown. HCT116 p53+/+ (H) and H460 (I) cells were transfected with p53 or control siRNA for 72 hr, and Doxorubicin or 5-Fluorouracil was supplemented 15 hr before the cells were harvested for q-PCR (Mean ± SEM, n = 3). Three biological replicates were used for p value, p*<0.05. (J) A schematic of predicted p53 responsive elements in the NGFR promoter. (K,L) p53 induces luciferase activity through RE1. Luciferase assay was performed using H1299 and U2OS cells as described in Materials and methods (Mean ± SEM, n = 3 biological replicates). (M,N) p53 is associated with the NGFR promoter. HCT116 p53+/+ cells were treated with or without Doxorubicin for 15 hr followed by ChIP assay using anti-p53 or mouse IgG.
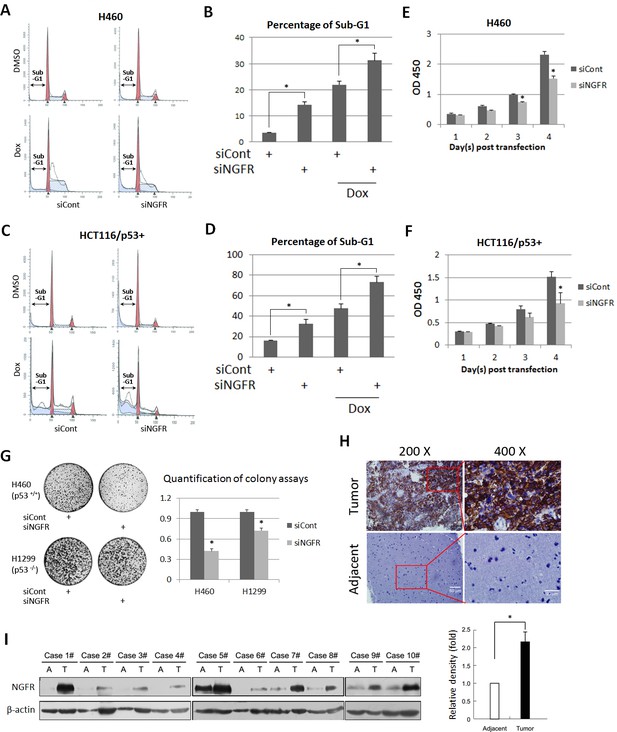
NGFR is required for cancer cell survival and clonogenicity, and highly expressed in glioma.
(A) NGFR knockdown induces apoptosis of H460 cells. Cells were transfected with NGFR or control siRNA for 72 to 96 hr, and Doxorubicin was supplemented 15 hr before the cells were harvested for flow cytometry analyses. (B) Quantification of Sub-G1 population in (A) (Mean ± SEM, n = 3). Three biological replicates were used for p value, p*<0.05. (C) NGFR knockdown induces apoptosis of HCT116 p53+/+cells. Cells were transfected with NGFR or control siRNA for 72 to 96 hr, and Doxorubicin was supplemented 15 hr before the cells were harvested for flow cytometry analyses. (D) Quantification of Sub-G1 population in (C) (Mean ± SEM, n = 2). Three biological replicates were used for p value, p*<0.05. (E, F) NGFR knockdown suppresses cell survival. H460 (E) and HCT116 p53+/+ (F) cells were transfected with NGFR or control siRNA and seeded in 96-well plate the next day (Day 1). Cell viability was evaluated every 24 hr (Mean ± SEM, n = 3). Three biological replicates were used for p value, p*<0.05. (G) NGFR knockdown inhibits clonogenicity. H460 and H1299 cells were transfected with NGFR or control siRNA and seeded in 10-cm plates the next day. Colonies were fixed by methonal and stained with crystal violet solution (left panel). Quantification of colonies is shown in the right panel (Mean ± SEM, n = 2). Two biological replicates were used for p value, p*<0.05. (H) IHC analyses of human glioma and the adjacent noncancerous tissues. (I) IB analyses of human glioma and the adjacent noncancerous tissues (left panel). Quantification of NGFR expression (right panel) (Mean ± SEM, n = 48, p*<0.01).
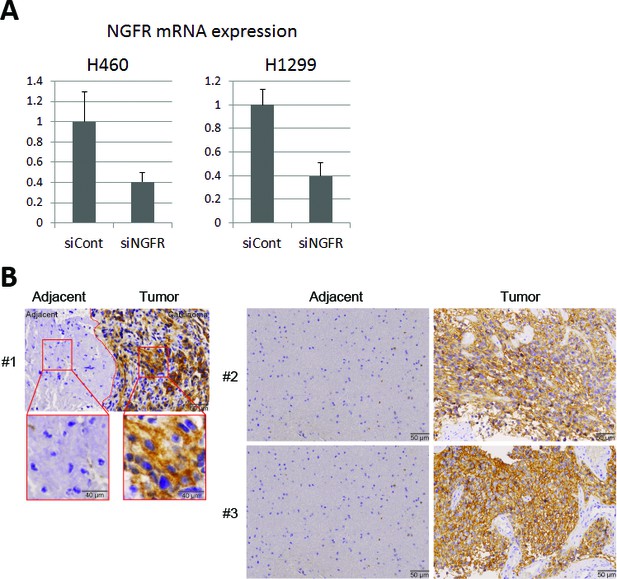
Representative expression of NGFR in lung cancer cell lines by siRNA knockdown and in glioma tissues.
(A) NGFR knockdown validation in the colony formation experiment. (B) NGFR is highly expressed in human glioma samples. IHC analyses of human glioma and the adjacent noncancerous tissues reveals that NGFR is overexpressed in glimoas.
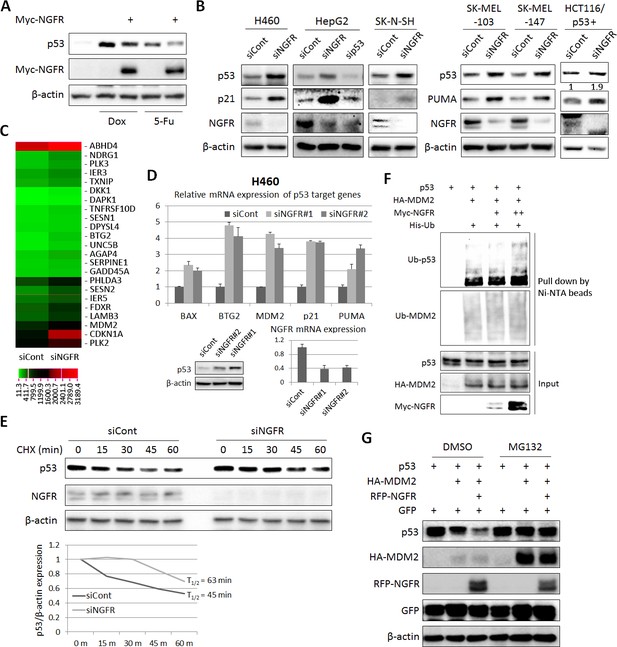
NGFR suppresses p53 activity by enhancing MDM2-mediated ubiquitination and proteasomal degradation.
(A) NGFR inhibits p53 activation by Doxorubicin or 5-Fluorouracil. H460 cells were transfected with NGFR for 30 hr and treated with Doxorubicin or 5-Fluorouracil for 15 hr before harvested for IB using antibodies as indicated. (B) NGFR knockdown induces p53 expression and activity. A panel of cancer cell lines were transfected with NGFR or control siRNA followed by IB using antibodies as indicated. The values in the rightmost panel indicate the p53/β-actin ratios. (C) NGFR knockdown induces p53 target gene expression. H460 cells were transfected with NGFR or control siRNA followed by RNA-sequencing analyses. Genes with over 1.5-fold increase in expression were shown (Three biological replicates were used for p value, p<0.05. (D) Two siRNAs targeting different sequences were used for knocking down NGFR in H460 cells. The expression of p53 target genes was assessed by q-PCR (Mean ± SEM, n = 2 biological replicates), while p53 expression was detected by IB. Validation of NGFR knockdown by the two siRNAs is shown in the right corner. (E) NGFR knockdown prolongs p53’s half-life. H460 cells transfected with NGFR or control siRNA for 72 hr were treated with 100 µg/ml of CHX and harvested at the time points as indicated. IB was performed using antibodies as indicated (upper panel) and quantification of p53/β-actin ratio is shown in the lower panel. (F) NGFR promotes MDM2-induced p53 ubiquitination. H1299 cells were transfected with combinations of plasmids encoding p53, HA-MDM2, Myc-NGFR or His-Ub and treated with MG132 6 hr before harvested for in vivo ubiquitination assay. Bound proteins and inputs were detected by IB using antibodies as indicated. (G) NGFR enhancesMDM2-mediated p53 proteasomal degradation. H1299 cells were transfected with combinations of plasmids encoding p53, HA-MDM2 or RFP-NGFR followed by IB using antibodies as indicated. MG132 was supplemented to the medium for 6 hr.
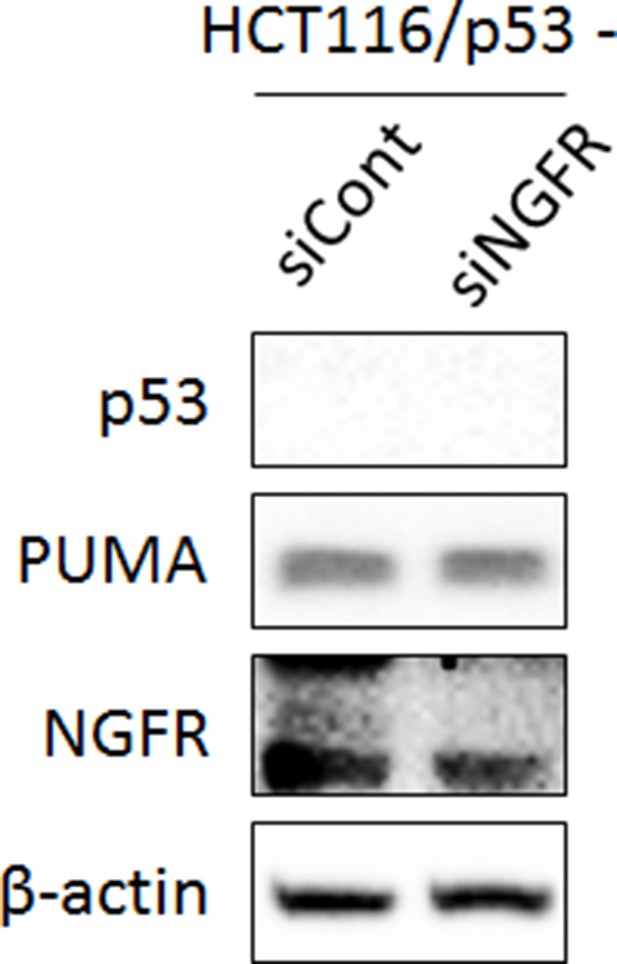
Knockdown of NGFR does not affect PUMA expression in the p53-null HCT116 cells.
HCT116 p53-/- cells were transfected with control or NGFR siRNA for 72 hr followed by IB using antibodies as indicated.
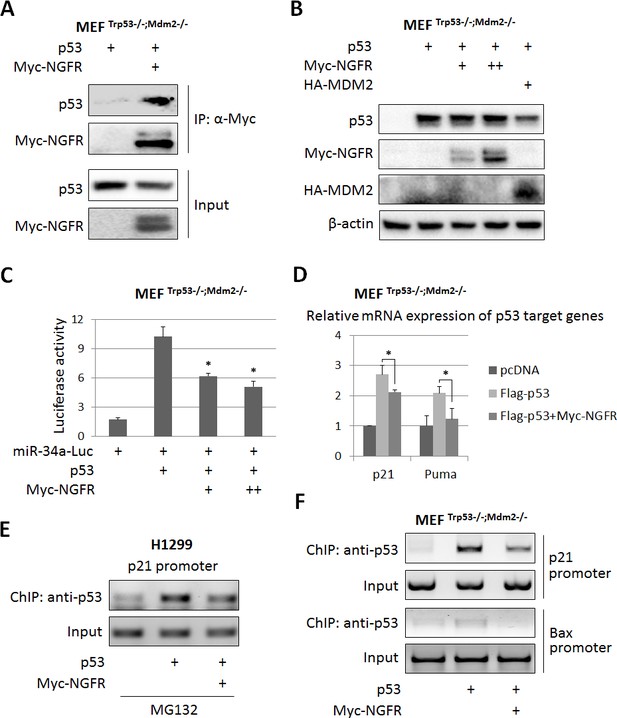
NGFR inactivates p53 independently of MDM2.
(A) NGFR interacts with p53 in the absence of MDM2. MEF p53-/-;Mdm2-/- cells were transfected with plasmids encoding Myc-NGFR or p53 followed by co-IP-IB assays using antibodies as indicated. (B) NGFR does not affect p53 protein expression in the absence of MDM2. MEF p53-/-;Mdm2-/- cells were transfected with combinations of plasmids encoding Myc-NGFR, HA-MDM2 or p53 followed by IB using antibodies as indicated. (C) NGFR represses p53-induced luciferase activity independently of MDM2. MEF p53-/-;Mdm2-/- cells were transfected with plasmids as indicated in the figure and luciferase assay was performed as described in Materials and methods (Mean ± SEM, n = 3). Three biological replicates were used for p value, p*<0.05. (D) NGFR inhibits p53-induced target gene expression independently of MDM2. MEF p53-/-;Mdm2-/- cells were transfected with plasmids as indicated in the figure followed by q-PCR analysis (Mean ± SD, n = 3). Three technical replicates were used for p value, p*<0.05. (E) NGFR impedes p53 association with the p21 promoter in H1299 cells. Cells were transfected with plasmids as indicated and treated with MG132 for 6 hr before harvested for ChIP-PCR analyses. (F) NGFR impedes p53 association with the p21 and Bax promoters independently of MDM2. MEF p53-/-;Mdm2-/- cells were transfected with plasmids as indicated followed by ChIP-PCR analyses.
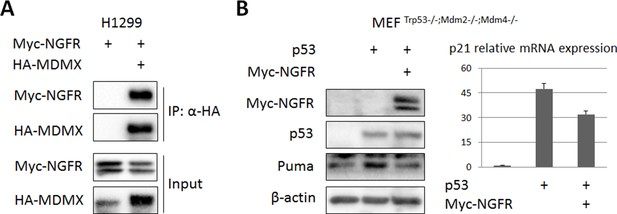
MDMX, though binds to NGFR, is not required for NGFR-mediated p53 inactivation.
(A) MDMX binds to NGFR. H1299 cells were transfected with HA-MDMX and Myc-NGFR followed by co-IP-IB assays using antibodies as indicated. (B) NGFR inactivates p53 independently of MDM2 and MDMX. MEF p53-/-; Mdm2-/-; Mdmx-/- cells were transfected with plasmids encoding each of Myc-NGFR and p53 follow by IB analysis using antibodies as indicated (left panel) and Q-PCR analysis of p21 expression (right panel) (Mean ± SD, n = 3 technical replicates).
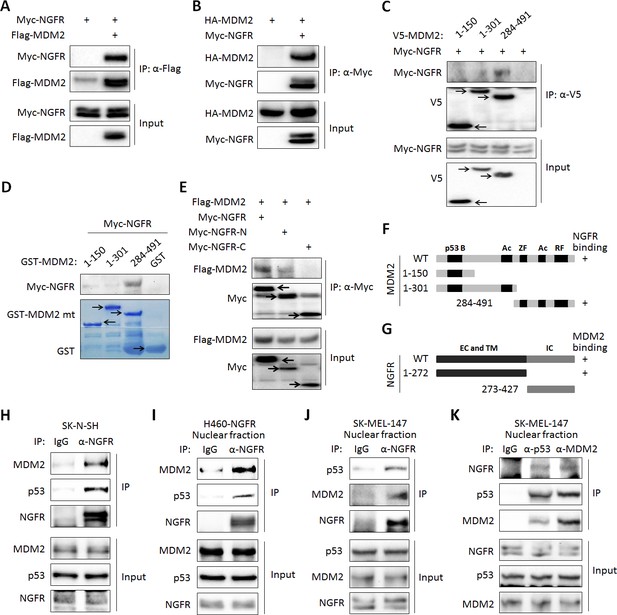
NGFR interacts with MDM2 in the nucleus.
(A,B) NGFR interacts with MDM2. H1299 cells were transfected with plasmids encoding Myc-NGFR, Flag-MDM2 or HA-MDM2 followed by co-IP-IB assays using antibodies as indicated. (C) Mapping the NGFR binding domain on MDM2 by co-IP-IB assays. H1299 cells were transfected with the plasmid encoding V5-tagged MDM2 fragment, aa 1–150, aa 1–301 or aa 284–491, along with the Myc-NGFR-encoded plasmid. Co-IP-IB assays were performed using antibodies as indicated. (D) Mapping the NGFR binding domain on MDM2 by GST-pull down assay. The prokaryotic expressed GST-tagged MDM2 fragment, aa 1–150, aa 1–301 or aa 284–491, or GST protein alone was incubated with cell lysates overexpressing Myc-NGFR. Bound proteins were detected by IB using anti-NGFR or coomassie staining. (E) Mapping the MDM2 binding domain on NGFR. H1299 cells were transfected with the plasmid encoding Myc-tagged NGFR fragment, aa 1–272 or aa 273–427, along with the Flag-MDM2-encoded plasmid. Co-IP-IB assays were performed using antibodies as indicated. (F) A schematic of NGFR binding region on MDM2. (G) A schematic of MDM2 binding region on NGFR. (H) Endogenous interaction of NGFR and MDM2 in SK-N-SH cells. Cells treated with Doxorubicin for 12 hr and MG132 for 6 hr were harvested for co-IP-IB assays using antibodies as indicated. (I,J,K) NGFR interacts with both MDM2 and p53 in the nucleus. Nuclear fractions from NGFR-stably expressed H460 (I) and SK-MEL-147 cells (J,K) were subjected to co-IP-IB assays using antibodies as indicated.
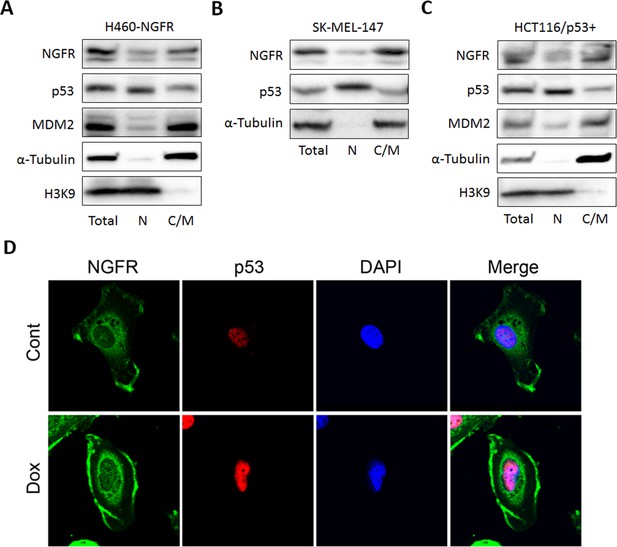
Full-length of NGFR localizes in both nucleus and cytoplasm.
(A,B,C) NGFR-stably expressed H460 (A), SK-MEL-147 (B) and HCT116 p53+/+ cells (C) were fractionated into nuclear and cytoplasmic/membrane fractions which were subsequently subjected to IB using antibodies as indicated. (D) SK-MEL-147 cells were treated with or without Dox for 20 hr and fixed in −20°C for overnight. The fixed cells were subjected to immunostaining using antibodies as indicated. The images were acquired by a confocal microscope.
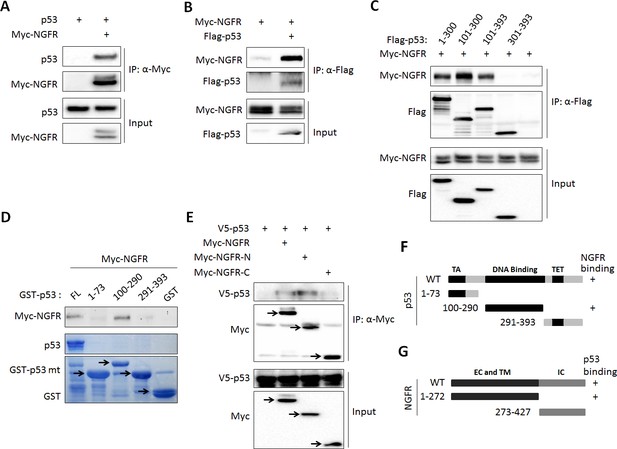
NGFR interacts with p53 in the nucleus.
(A, B) NGFR interacts with p53. H1299 cells were transfected with plasmids encoding Myc-NGFR, Flag-p53 or p53 followed by co-IP-IB assays using antibodies as indicated. (C) Mapping the NGFR binding domain on p53 by co-IP-IB assays. H1299 cells were transfected with the plasmid encoding flag-tagged p53 fragment, aa 1–300, aa 101–300, aa 101–393 or aa 301–393, along with the Myc-NGFR-encoded plasmid. Co-IP-IB assays were performed using antibodies as indicated. (D) Mapping the NGFR binding domain on p53 by GST-pull down assay. The prokaryotic expressed GST-tagged full-length p53 or p53 fragment, aa 1–73, aa 100–290 or aa 291–393, or GST protein alone was incubated with cell lysates overexpressing Myc-NGFR. Bound proteins were detected by IB using anti-NGFR or coomassie staining. (E) Mapping the p53-binding domain on NGFR. H1299 cells were transfected with the plasmid encoding Myc-tagged NGFR fragment, aa 1–272 or aa 273–427, along with the V5-p53-encoded plasmid. Co-IP-IB assays were performed using antibodies as indicated. (F) A schematic of NGFR binding region on p53. (G) A schematic of p53 binding region on NGFR.
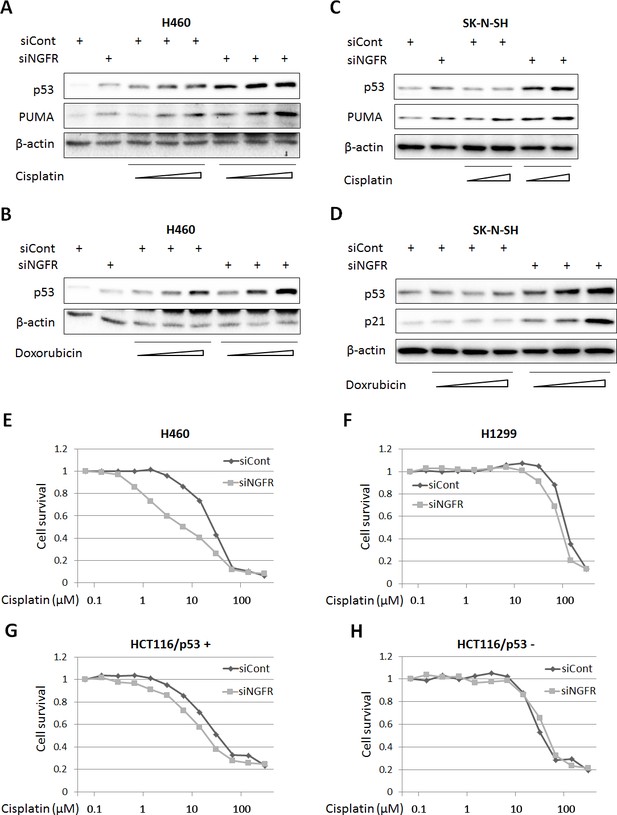
NGFR confers cancer cells resistance to chemotherapeutic agents.
(A,B) NGFR knockdown enhances Cisplatin or Doxorubicin-triggered p53 activation in H460 cells. Cells were transfected with NGFR or control siRNA for 72 hr and Cisplatin (A) or Doxorubicin (B) was supplemented in the medium 12 hr before the cells were harvested for IB using antibodies as indicated. (C,D) NGFR knockdown enhances Cisplatin (C) or Doxorubicin (D) -triggered p53 activation in SK-N-SH cells.The same experiments as shown in (A,B) were performed except that the SK-N-SH cell line was used instead. (E,F) NGFR knockdown sensitizes H460 (E) but not H1299 cells (F) to Cisplatin treatment. Cells were transfected with NGFR or control siRNA and seeded in 96-well plates the next day. Cisplatin was supplemented 48 hr before cell viability was determined using CCK-8 as described in Materials and methods. (G,H) NGFR knockdown sensitizes HCT116 p53+/+ (G) but not HCT116 p53-/- cells (H) to Cisplatin treatment. The same experiments described in (E,F) were performed except for using different cell lines.
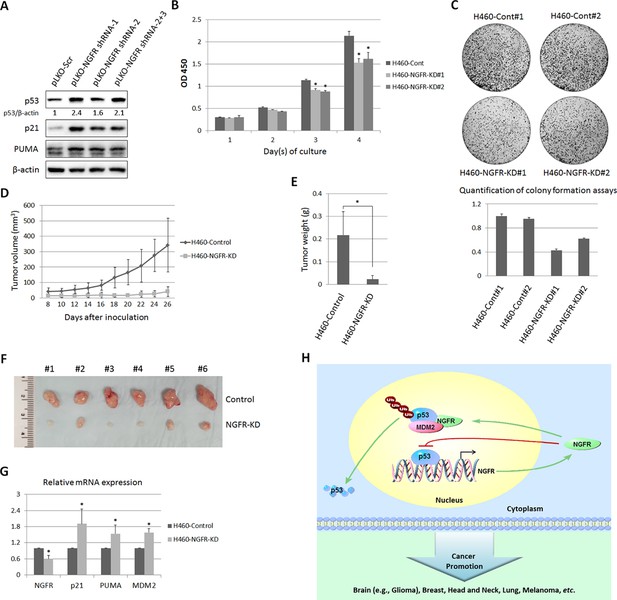
NGFR is required for tumor growth in vivo.
(A) Lentiviral-based knockdown of NGFR induces p53 pathway. H460 cells were transduced with lentivirus expressing NGFR or control shRNA for 72 hr followed by IB using antibodies as indicated. (B) H460 cells stably expressing NGFR shRNA show suppressed cell survival. H460 cells stably expressing NGFR or control shRNA were seeded in 96-well plates and cell viability was evaluated every 24 hr (Mean ± SEM, n = 3). Three biological replicates were used for p value, p*<0.01. (C) H460 cells stably expressing NGFR shRNA exhibit restrained clonogenic capacity. H460 cells stably expressing NGFR or control shRNA were seeded on 10-cm plates and colonies were fixed by methonal and stained with crystal violet solution (upper panel). Quantification of colonies is shown in the lower panel. (D) H460 cells stably expressing NGFR shRNA reveal less xenograft tumor volume in average (Mean ± SEM, n = 6). (E) H460 cells stably expressing NGFR shRNA reveal less xenograft tumor weight in average (Mean ± SEM, n = 6). Six pairs of tumors were used for p value, p*<0.01. (F) Representative xenograft tumors reveal that NGFR knockdown dramatically suppressed tumor growth in vivo. (G) The p53 pathway is activated by NGFR knockdown in the xenograft tumors. The expression of p21, PUMA and MDM2 was determined by q-PCR (Mean ± SEM, n = 3). Three pairs of tumors were used for p value, p*<0.01. (H) A model for NGFR regulation of the MDM2-p53 loop in cancer. NGFR inactivates p53 through two mechanisms: 1) by directly associating with MDM2 and enhancing MDM2-mediated p53 ubiquitination and proteasomal degradation, and 2) by repressing p53 transcriptional activity through direct interaction with its DNA binding domain, consequently leading to tumorigenesis.
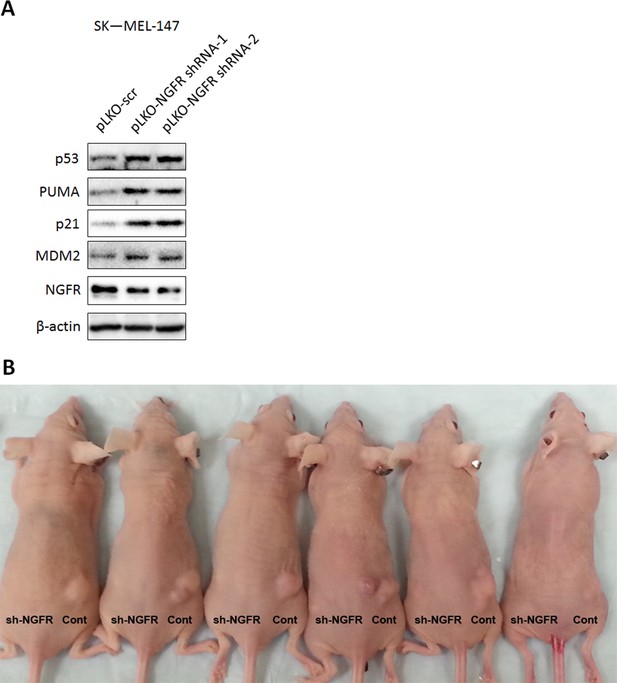
Lentivirus-mediated knockdown of NGFR activates p53 in a melanoma cell line and suppresses in vivo tumor growth.
(A) p53 pathway is activated in NGFR-shRNA stably expressed SK-MEL-147 cells. SK-MEL-147 cells infected with lentivirus expressing scrambled or NGFR shRNA were subjected to IB using antibodies as indicated. (B) Representative xenograft tumors in mice. NGFR knockdown suppresses tumor growth in vivo, original image related to Figure 8F.

NGFR is apt to be amplified in breast cancers sustaining wild-type p53.
TCGA database was searched and the data were modified from the cBioPortal for Cancer Genomics (http://www.cbioportal.org/; Gao et al., 2013; Cerami et al., 2012).





