A shunt pathway limits the CaaX processing of Hsp40 Ydj1p and regulates Ydj1p-dependent phenotypes
Figures
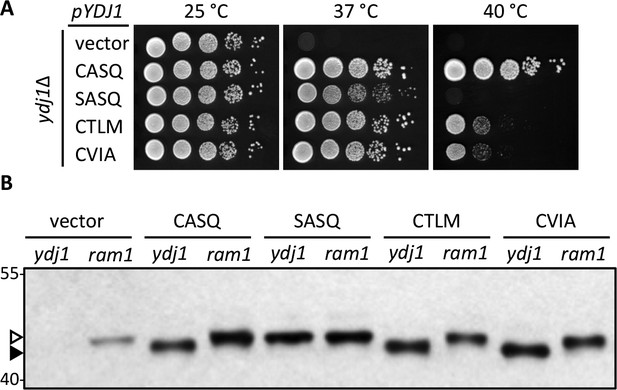
Alternate CaaX motifs on Ydj1p affect thermotolerance but not farnesylation.
(A) Yeast cultured in selective SC-Ura media were normalized for culture density and spotted as 10-fold serial dilutions onto YPD; the leftmost spot in each panel is undiluted. Plates were incubated at the indicated temperature as described in Materials and Methods. The strain used was yWS304 (ydj1∆); the CEN plasmids used were pRS316, pWS942, pWS1132, pWS1246, and pWS1286. Similar results were observed for Ydj1p CaaX variants expressed at the chromosomal level (see Figure 1—figure supplement 1). (B) Lysates from the indicated genetic backgrounds were prepared from cultures grown at 25°C in SC-Ura and analyzed by immunoblot with Ydj1p antiserum. The strains used were yWS304 (ydj1∆) and yWS1632 (ram1∆); the CEN plasmids used were the same as reported for panel A. The ram1 strain lacks farnesyl transferase activity and produces unmodified Ydj1p (open triangle) having slower mobility than farnesylated Ydj1p (closed triangle). Approximately 25% of the signal in the ram1 samples comes from the chromosomal copy of Ydj1p present in that strain (see Figure 1—figure supplement 2). Values to the left of the image indicate the migration of protein standards (kDa). The farnesylation of the CaaX variants is not temperature dependent (see Figure 1—figure supplement 3). The data in panel A are representative of multiple biological replicates (n= 2, full set of strains; n=7, all but vector strain). The data in panel B are representative of 2 biological replicates.
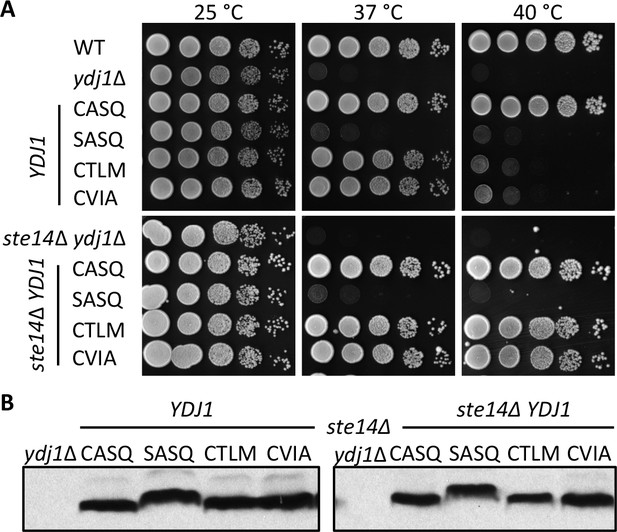
Thermotolerance profiles of chromosome-encoded Ydj1p CaaX mutants.
(A) Yeast cultured in YPD were normalized for culture density and spotted as 10-fold serial dilutions onto YPD as described in Figure 1. The strains used in the top panel were yWS42 (WT), yWS304 (ydj1∆), yWS2109 (CASQ), yWS2110 (SASQ), yWS2111 (CTLM), and yWS2112 (CVIA). The strains used in the bottom panel were yWS1635 (ste14∆ ydj1∆), yWS2117 (CASQ), yWS2118 (SASQ), yWS2119 (CTLM), and yWS2120 (CVIA). (B) Lysates of strains reported in panel A were prepared and analyzed as described for Figure 1, except that cells were cultured in YPD. The data in panels A and B are representative of two biological replicates (n= 2).
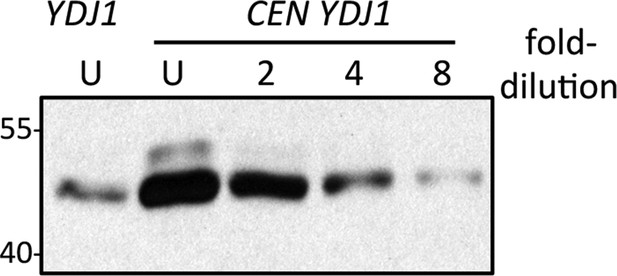
Comparison of genomic and plasmid-based expression of Ydj1p in the ram1 strain.
Yeast expressing Ydj1p from the genome (YDJ1) or a plasmid expression vector (CEN YDJ1) was cultured at 25°C in SC-Ura. Equal amounts of cells were used to prepare lysates. Equal percentages of each undiluted (U) sample along with a two-fold serial dilution series were analyzed by SDS-PAGE and Ydj1p immunoblot. The data shown is representative of three biological replicates. Band intensities for all replicates were quantified using both Photoshop and ImageJ software, and the values used to estimate the relative over-expression of plasmid-based Ydj1p relative to genomic. The strain used was yWS1632 (ram1∆) containing either pRS316 (CEN URA3) or pWS942 (CEN URA3 YDJ1).
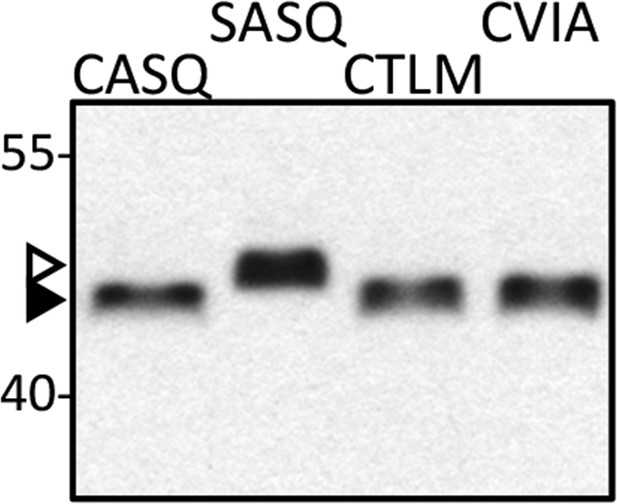
Isoprenylation status of Ydj1p CaaX variants at high temperature.
Lysates prepared from cultures grown at 40°C in YPD (CASQ, CTLM, CVIA) or SC-Ura at 25°C (SASQ) were analyzed by immunoblot with Ydj1p antiserum. The strain used was yWS1635 (ste14∆ ydj1∆); the plasmids used were the same as reported in Figure 1. For unknown reasons, total protein yields are consistently lower in the 40°C samples. For the immunoblot, 2.5-times the volume of lysate from the 40°C samples was loaded in order to equalize the signal observed for Ydj1p SASQ. Unmodified Ydj1p (SASQ; open triangle) has slower mobility than farnesylated Ydj1p (closed triangle). Values to the left of the image indicate the migration of protein standards (kDa). The data are representative of 4 technical replicates.
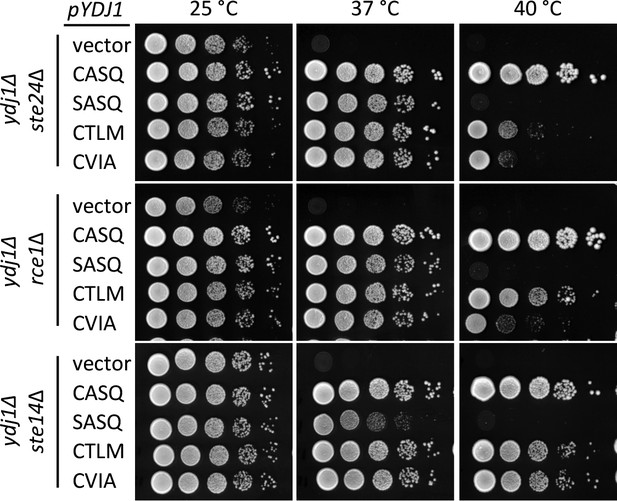
Post-isoprenylation processing negatively impacts Ydj1p variants having alternative CaaX motifs.
Data were collected as described in Figure 1. The strains used were yWS1693 (ydj1∆ ste24∆), yWS1689 (ydj1∆ rce1∆), and yWS1635 (ydj1∆ ste14∆); the CEN plasmids used are the same as described in Figure 1 except that the vector used for yWS1693 and yWS1689 was pRS416. The data for each genetic background are representative of 2 biological replicates.
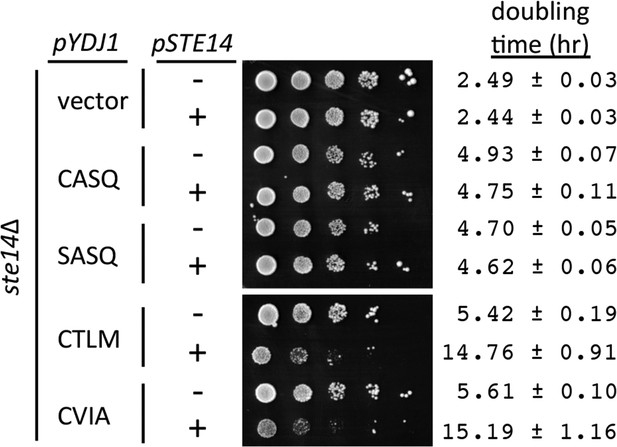
Over-expression of Ydj1p CaaX variants results in STE14 dependent slow growth.
Yeast cultured in selective SC-Ura,Leu media were normalized for culture density and spotted as 10-fold serial dilutions onto selective SC-Ura,Leu; the leftmost spot in each panel is undiluted. Plates were incubated at 30°C. Growth rates of the strains were determined in SC-Ura,Leu liquid media at 30°C. The strain used was SM1188 (ste14∆), which was transformed with two plasmids. The plasmids used for over-expression of Ydj1p were pWS948, pWS972, pWS1247, and pWS1291; pSM703 was the representative empty vector. The plasmid used for expression of Ste14p was pSM1316; pRS315 was used for the vector (-) condition. The serial dilution data are representative of 2 biological replicates. The doubling times are averages of 4 biological replicates; error ranges represent the 95% confidence interval (see Figure 3—source data 1).
-
Figure 3—source data 1
Spreadsheet with raw values for A600 vs. time of yeast over-expressing Ydj1p CaaX variants in the presence or absence of STE14.
Data points included in the analysis are from time segments of exponential growth.
- https://doi.org/10.7554/eLife.15899.008
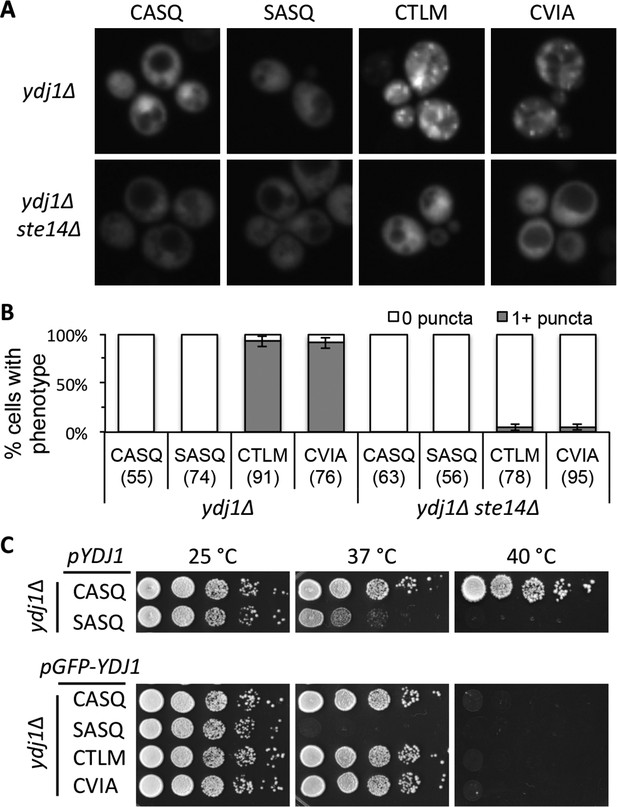
Ydj1p variants with alternative CaaX motifs are mislocalized.
(A) The indicated GFP-Ydj1p CaaX variants were expressed in yWS304 (ydj1∆) or yWS1635 (ydj1∆ ste14∆). Images of multiple fields were collected under fluorescence optics, with images representative of the majority phenotype being shown. The CEN plasmids used were pWS1389-1392, encoding CaaX variants CASQ, SASQ, CTLM and CVIA, respectively. (B) Quantification of phenotypes observed in panel A. Values in parentheses associated with each motif indicate the total number of cells classified over the course of two independent experiments. Bars indicate the average number of cells observed to have the indicated phenotype. (C) Thermotolerance of strains expressing GFP-Ydj1p CaaX variants compared to untagged versions. The differences are not due to farnesylation defects (see Figure 4—figure supplement 1). Data were collected as described in Figure 1. The strain used was yWS304 (ydj1∆); the CEN plasmids used were pWS942, pWS1132, and pWS1389-1392. Data in panel A are representative of 4 biological replicates; data in panel B were calculated from 2 biological replicates with error bars representing the range observed between replicates (see Figure 4—source data 1); data in panel C are representative of 3 biological replicates.
-
Figure 4—source data 1
Spreadsheet with raw values used for percent calculations of puncta in GFP-Ydj1p CaaX variant strains.
- https://doi.org/10.7554/eLife.15899.010
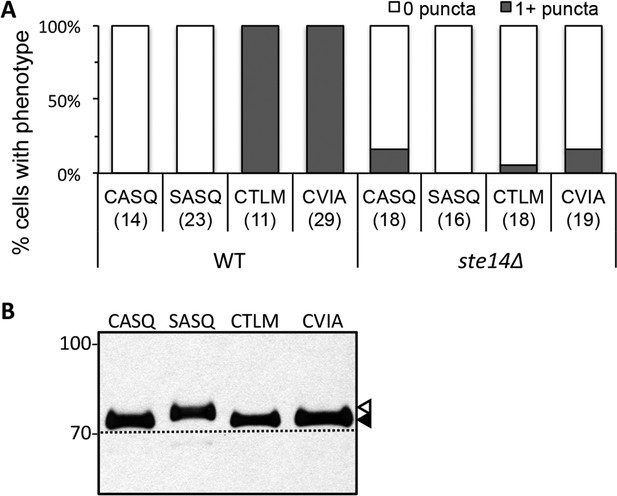
Localization and farnesylation of GFP-Ydj1p variants with alternative CaaX motifs.
(A) The indicated GFP-Ydj1p CaaX variants were expressed in IH1783 (wildtype) or SM1188 (ste14∆) and analyzed as described for Figure 4 to obtain a quantified measure of phenotypes. The data are from a single experiment. (B) Lysates containing GFP-Ydj1p were prepared from yWS304 (ydj1∆). Strains were grown at 25°C in SC-Ura and analyzed by immunoblot with Ydj1p antiserum. Unmodified Ydj1p (SASQ; open triangle) has slower mobility than farnesylated Ydj1p (closed triangle); a dashed line is provided as a reference. Values to the left of the image indicate the migration of protein standards (kDa). The plasmids used for GFP-Ydj1p expression in all panels are the same as those reported in Figure 4. The data in panel A are from 1 biological replicate (see Figure 4—source data 1); panel B is representative of 4 technical replicates.
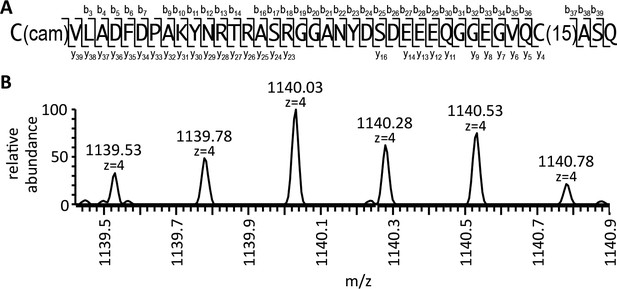
Biophysical analysis of C-terminal peptide derived from Ydj1p.
(A) His6-Yd1jp was expressed in yWS304 (ydj1∆), isolated by immobilized metal affinity chromatography, and analyzed by mass spectrometry after digestion with endoproteinase GluC. The plasmid used was pWS1307. The sequence indicates the detected b- and y- fragments of the C-terminal peptide observed with MS/MS (see Figure 5—figure supplement 1). C(cam) is carbamidomethyl cysteine; C(15) is isoprenylated cysteine. (B) Full MS of the detected C-terminal peptide, with m/z value and mass accuracy of the species indicated. The same result was observed in two independent His6-Ydj1p samples.
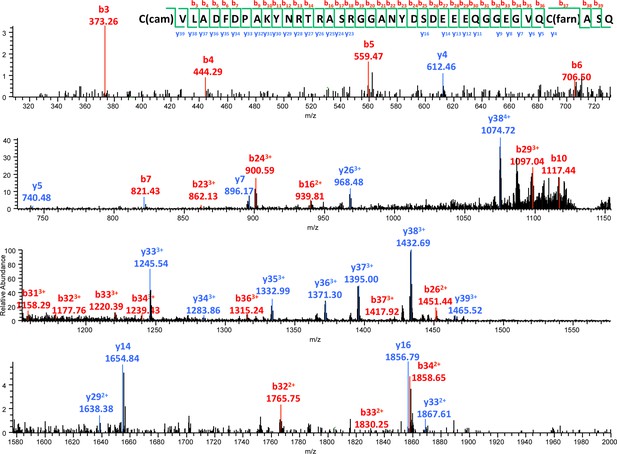
MS/MS spectrum of the farnesylated C-terminal peptide.
b- and y- fragments of the peptide are labeled with their m/z values and charge states. The m/z value and mass accuracy of the precursor are also indicated.
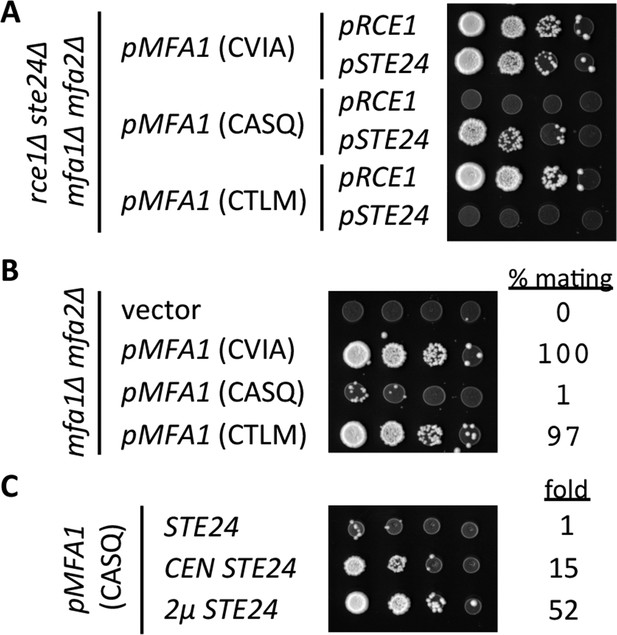
Impact of different CaaX motifs on a-factor bioactivity.
(A) MATa yeast co-expressing the indicated a-factor species and CaaX protease were normalized for cell density then serially diluted in the presence of 10-fold or higher excess MATa cells. The mixtures were spotted onto media selective for diploid growth. Plasmid-derived strains of yWS164 (rce1∆ ste24∆ mfa1∆ mfa2∆) were created using appropriate combinations of CEN plasmids pSM1275, pSM1093, pWS610, and pWS612-13. (B) Mating mixtures were prepared and analyzed as described for panel A. Equal portions of each mating mixture were also analyzed for mating efficiency as described in the Materials and Methods section, where the condition involving wildtype MFA1 was set to 100%. Plasmid-derived strains of SM2331 (mfa1∆ mfa2∆) were created using pRS415 and the a-factor encoding plasmids described for panel A. (C) Mating mixtures were prepared and analyzed as described for panel B. The value for fold refers to the relative mating efficiency observed for each condition, where the condition involving chromosome-encoded STE24 was set as the reference. Plasmid-derived strains were created by transformation of SM3689 (rce1∆ mfa1∆ mfa2∆) and yWS164 with pWS612 and either pRS316, pSM1093, or pSM1194. The data in panel A are representative of 3 biological replicates; the serial dilutions in panels B and C are representative of 2 technical replicates with reported values derived from the averages of 2 technical replicates. Transfer of the CASQ motif to Ras2p also results in an ‘uncleaved’ phenotype (see Figure 6—figure supplement 1).
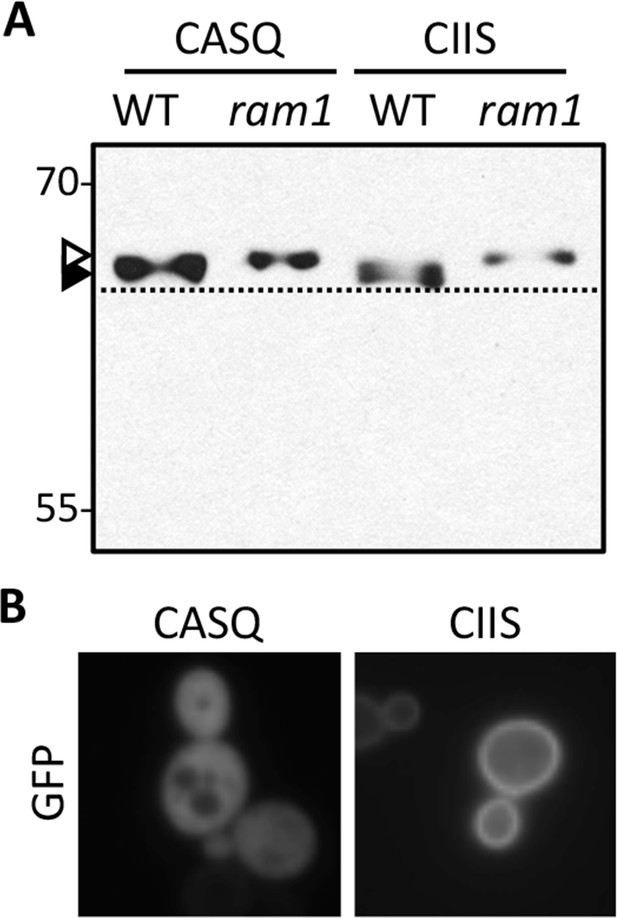
Impact of different CaaX motifs on GFP-Ras2p farnesylation and localization.
(A) GFP-Ras2p with indicated CaaX motifs were analyzed for farnesylation status by immunoblot. The natural CaaX motif for Ras2p is CIIS. Lysates were prepared from cultures grown in at 25°C in SC-URA with raffinose and glycerol (2% each) as carbon sources. 2% galactose was added during the final 5 hr of growth to induce GFP-Ras2p expression. Due to the poor galactose utilization by the ram1∆ strain, it was necessary to load 3-times more lysate from the ram1∆ samples to equalize the GFP-Ras2p signal. The strains used were yWS42 (WT) and yWS1632 (ram1∆); the plasmids used were pWS270 (CIIS) and pWS546 (CASQ). The ram1∆ strain lacks farnesyl transferase activity and produces unmodified Ydj1p (open triangle) having slower mobility than farnesylated Ydj1p (closed triangle); a dashed line is provided as a reference. (B) The indicated GFP-Ras2p CaaX variants were expressed in IH1783 as described previously [Herskowitz and Jensen, 1991]. Images of multiple fields were collected under fluorescence optics, with images representative of the majority phenotype being shown. GFP-Ras2p (CIIS) displays typical plasma membrane localization. The data in panel A are representative of 2 biological replicates and 4 technical replicates; the images in panel B are representative of 2 biological replicates.
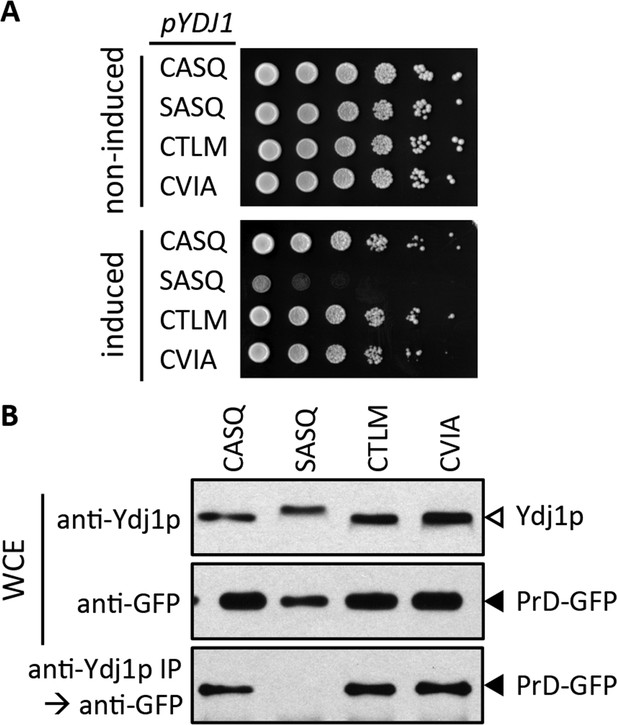
Impact of different CaaX motifs on Ydj1p client interactions.
(A) Yeast containing an inducible PrD expression plasmid and the indicated Ydj1p CaaX variants were spotted as a serial dilution series on media containing glucose (non-induced) or galactose (induced) and incubated at 30°C. The strains used are plasmid derivatives of yWS2078 ([RNQ+] ydj1∆) created using pWS1430 (PrD) and pWS1326-1329 (Ydj1p). (B) Yeast co-expressing PrD-GFP and the indicated Ydj1p CaaX variants were evaluated for interactions by coIP. Whole cell extracts (WCE) were prepared, and 10 µg of each lysate was analyzed by SDS-PAGE and immunoblot using the indicated antibody, while 100 µg of each lysate was immunoprecipitated using Ydj1p antibody and subsequently evaluated for recovery of PrD-GFP with GFP antibody. The strains used are similar to those described for panel A except that pWS1431 (PrD-GFP) was used as the source of PrD.
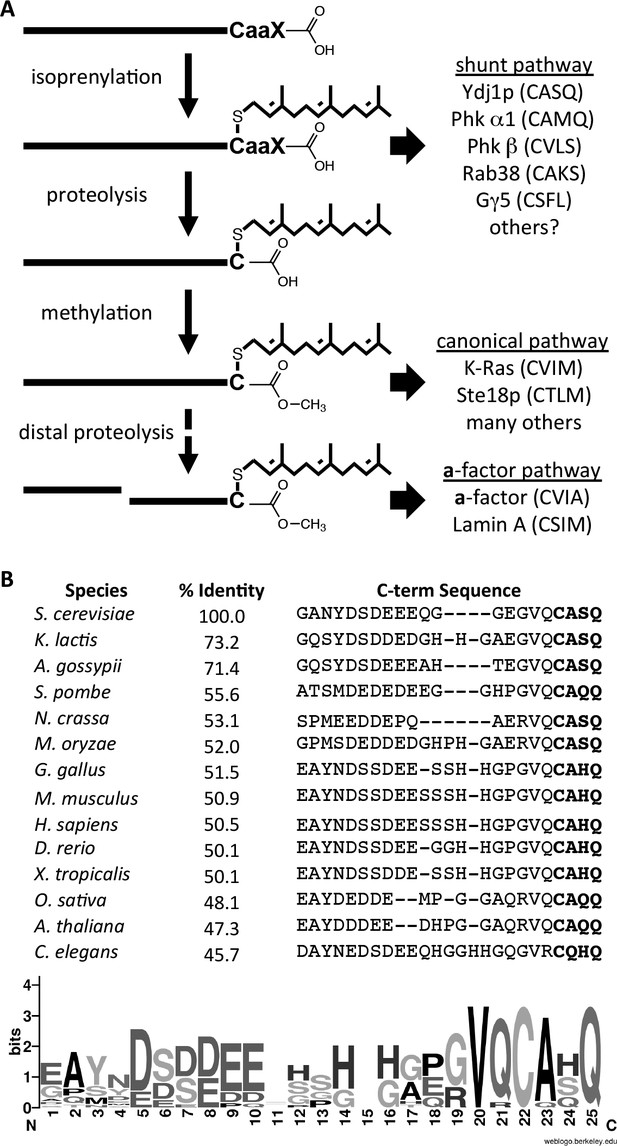
Model for post-translational modification of Ydj1p.
(A) The CaaX motif directs isoprenylation of the protein. The isoprenylated species is either the endpoint modification (e.g. Ydj1p) or an intermediate for further modification (e.g. Ras, a-factor, lamin A). (B) Alignment of the COOH-termini of Ydj1p and its homologs along with percent identity scores for entire sequences relative to S. cerevisiae Ydj1p. The sequences and alignment scores were retrieved from the Homologene database (http://www.ncbi.nlm.nih.gov/homologene). A WebLogo representation of amino acid frequency within the COOH-terminal region is shown.
Tables
Strains used in this study.
| Strain Identifier | Genotype | Reference |
|---|---|---|
| IH1783; ATCC#204278 | MATa trp1 leu2 ura3 his4 can1 | (Michaelis and Herskowitz, 1988) |
| IH1793; ATCC#204279 | MATα lys1 | (Michaelis and Herskowitz, 1988) |
| SM1188; ATCC# 204273 | MATa trp1 leu2 ura3 his4 can1 ste14-3::TRP1 | (Hrycyna et al., 1991) |
| SM2331 | MATa trp1 leu2 ura3 his4 can1 mfa1-∆1 mfa2-∆1 | (Chen et al., 1997) |
| SM3689 | MATa trp1 leu2 ura3 his4 can1 mfa1-∆1 mfa2-∆1 rce1::TRP1 | (Tam et al., 1998) |
| yWS42; BY4741 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 | (Shoemaker et al., 1996) |
| yWS164 | MATa trp1 leu2 ura3 his4 can1 mfa1-∆1 mfa2-∆1 rce1::TRP1 ste24::KANR | (Cadiñanos et al., 2003) |
| yWS304 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ydj1::KANR | (Shoemaker et al., 1996) |
| yWS1577 | MATα his3∆1 leu2∆0 met15∆0 ura3∆0 ydj1::KANR | This study |
| yWS1626 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ste14::KANR | (Shoemaker et al., 1996) |
| yWS1629 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 rce1:: KANR | (Shoemaker et al., 1996) |
| yWS1632 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ram1::KANR | (Shoemaker et al., 1996) |
| yWS1635 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ste14::KANR ydj1::KANR | This study |
| yWS1682 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ste24:: KANR | (Shoemaker et al., 1996) |
| yWS1685 | MATα his3∆1 leu2∆0 met15∆0 ura3∆0 ste24:: KANR | This study |
| yWS1686 | MATα his3∆1 leu2∆0 met15∆0 ura3∆0 rce1:: KANR | This study |
| yWS1689 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 rce1::KANR ydj1::KANR | This study |
| yWS1693 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ste24::KANR ydj1::KANR | This study |
| yWS2078 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ydj1::KANR [RNQ+] | (Summers et al., 2009) |
| yWS2109 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 YDJ1 | This study |
| yWS2110 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 YDJ1 (SASQ) | This study |
| yWS2111 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 YDJ1 (CTLM) | This study |
| yWS2112 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 YDJ1 (CVIA) | This study |
| yWS2117 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ste14:: KANR YDJ1 | This study |
| yWS2118 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ste14:: KANR YDJ1 (SASQ) | This study |
| yWS2119 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ste14:: KANRYDJ1 (CTLM) | This study |
| yWS2120 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ste14:: KANRYDJ1 (CVIA) | This study |
Plasmids used in this study.
| Plasmid Identifier | Genotype | Reference |
|---|---|---|
| pGAL-HO | CEN URA3 PGAL-HO | (Herskowitz and Jensen, 1991) |
| pRS315 | CEN LEU2 | (Sikorski and Hieter, 1989) |
| pRS316 | CEN URA3 | (Sikorski and Hieter, 1989) |
| pRS415 | CEN LEU2 | (Sikorski and Hieter, 1989) |
| pRS416 | CEN URA3 | (Sikorski and Hieter, 1989) |
| pSM703 | 2µ URA3 PPGK | (Zhang et al., 2001) |
| pSM1093 | CEN URA3 STE24 | (Fujimura-Kamada et al., 1997) |
| pSM1194 | 2µ URA3 STE24 | (Fujimura-Kamada et al., 1997) |
| pSM1275 | CEN URA3 RCE1 | (Schmidt et al., 1998) |
| pSM1316 | CEN LEU2 STE14 | (Romano and Michaelis, 2001) |
| pWS270 | CEN URA3 PGAL-GFP-RAS2 | (Manandhar et al., 2007) |
| pWS523 | CEN URA3 PGAL-GST-YDJ1 | (Zhu et al., 2001) |
| pWS546 | CEN URA3 PGAL-GFP-RAS2-CASQ | This study |
| pWS610 | CEN LEU2 MFA1 | (Krishnankutty et al., 2009) |
| pWS612 | CEN LEU2 MFA1-CASQ | (Krishnankutty et al., 2009) |
| pWS613 | CEN LEU2 MFA1-CTLM | This study |
| pWS882 | CEN URA3 PGAL-GFP-YDJ1 | This study |
| pWS942 | CEN URA3 YDJ1 | This study |
| pWS948 | 2µ URA3 PPGK-YDJ1 | This study |
| pWS972 | 2µ URA3 PPGK-YDJ1-SASQ | This study |
| pWS1132 | CEN URA3 YDJ1-SASQ | This study |
| pWS1246 | CEN URA3 YDJ1-CTLM | This study |
| pWS1247 | 2µ URA3 PPGK-YDJ1-CTLM | This study |
| pWS1286 | CEN URA3 YDJ1-CVIA | This study |
| pWS1291 | 2µ URA3 PPGK-YDJ1-CVIA | This study |
| pWS1298 | 2µ URA3 PPGK-GST-YDJ1 | This study |
| pWS1307 | 2µ URA3 PPGK-His6-YDJ1 | This study |
| pWS1326 | CEN LEU2 YDJ1 | This study |
| pWS1327 | CEN LEU2 YDJ1-SASQ | This study |
| pWS1328 | CEN LEU2 YDJ1-CTLM | This study |
| pWS1329 | CEN LEU2 YDJ1-CVIA | This study |
| pWS1389 | CEN URA3 GFP-YDJ1 | This study |
| pWS1390 | CEN URA3 GFP-YDJ1-SASQ | This study |
| pWS1391 | CEN URA3 GFP-YDJ1-CTLM | This study |
| pWS1392 | CEN URA3 GFP-YDJ1-CVIA | This study |
| pWS1430 | CEN URA3 PGal1-PrD | (Summers et al., 2009) |
| pWS1431 | CEN URA3 PCUP1-PrD-GFP | (Summers et al., 2009) |





