Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome
Figures
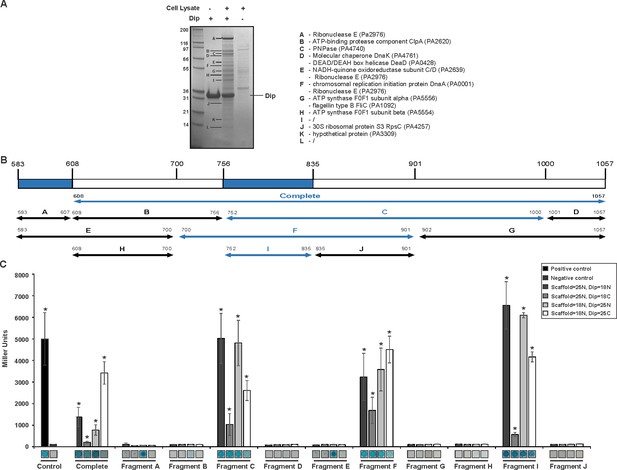
Interaction analyses of Dip and the P. aeruginosa RNA degradosome.
(A) In vitro pull down of P. aeruginosa cell lysate, using his-tagged Dip as a bait. Eluted samples were loaded on a 12% SDS-PAGE gel. The letters indicate bands submitted for identification by mass spectrometry analysis (“/” represents bands which could not be confidently identified). (B) Fragments of the scaffold domain of RNase E used in the bacterial two-hybrid assay. Blue and black arrows indicate fragments with a positive and no signal, respectively. The numbers indicate the number of the residue of RNase E. (C) Bacterial two-hybrid assay in which the T25 (25) or T18 (18) domain of CyaA is fused to the N-terminal (N) or C-terminal (C) side of a target protein. Non-fused T25 or T18 domains were used as negative control (No insert). The leucine zipper of GCN4 was used as a positive control. Interactions were visualized by a drop test on selective medium (shown below the graphs) and β-galactosidase activity was measured quantitatively in Miller units. Error bars represent SD and P‐values were calculated using Student's t‐test (n = 3), *p<0.05.
-
Figure 1—source data 1
Data bacterial two-hybrid.
- https://doi.org/10.7554/eLife.16413.003
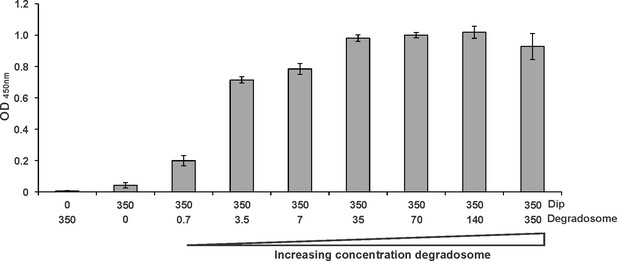
ELISA using 350 nM of Dip (fused to a C-terminal his-tag) as a bait protein and increasing amounts of P. aeruginosa RNA degradosome (carrying a C-terminal Strep-tag II on RNase E) as a prey (numbers in nM).
As negative control, 350 nM of only Dip or RNA degradosome was used. Error bars represent standard deviation (n = 3). (Note: the amount of RNA degradosome used in this assay is a crude estimate, assuming the overall mass of the degradosome assembly is 1 Mega Dalton).
-
Figure 1—figure supplement 1—source data 1
Data ELISA.
- https://doi.org/10.7554/eLife.16413.005
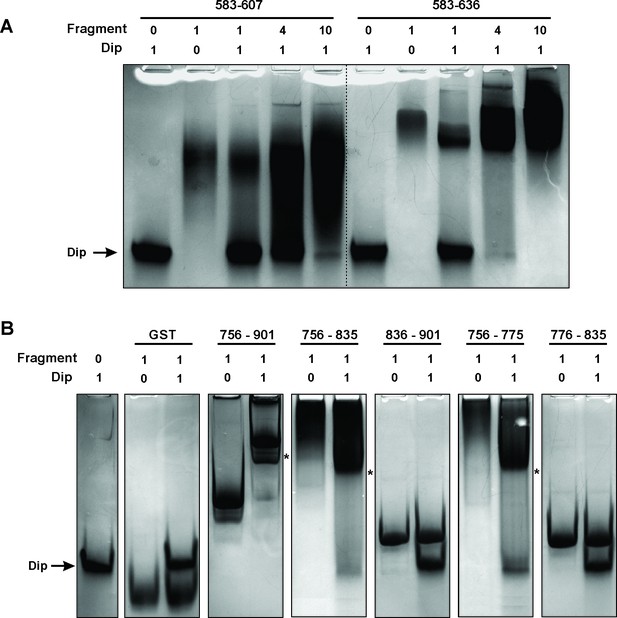
Electrophoretic Mobility shift assays using Dip.
(A) EMSA of fragments of RNase E corresponding to a first interaction site for Dip, incubated with or without Dip. The numbers above the horizontal line indicate the residues of RNase E corresponding to the two tested fragments. The numbers below indicate the relative amount of the fragments and Dip. (B) EMSA using fragments of the RNase E belonging to the second site of interaction for Dip. The numbers indicate the residues of RNase E that encompass the fragments. A shift in migration is indicated with an asterisk.
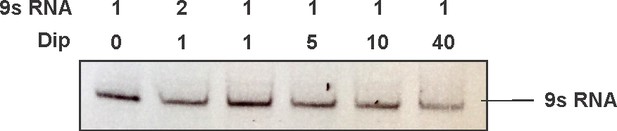
EMSA of increasing amounts of Dip (0.5 pmol, 1 pmol, 5 pmol, 10 pmol and 40 pmol) and 1 pmol of 9S RNA of E. coli.
All samples were run on a 10% native acrylamide gel.
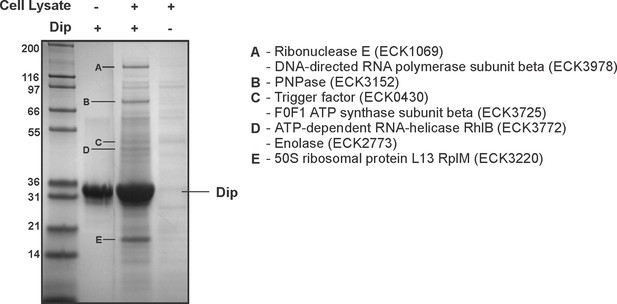
Interaction analyses of Dip and the E. coli RNA degradosome.
In vitro pull down of E. coli cell lysate, using his-tagged Dip as a bait. Eluted samples were loaded on a 12% SDS-PAGE gel. The letters indicate proteins identified by mass spectrometry analysis.
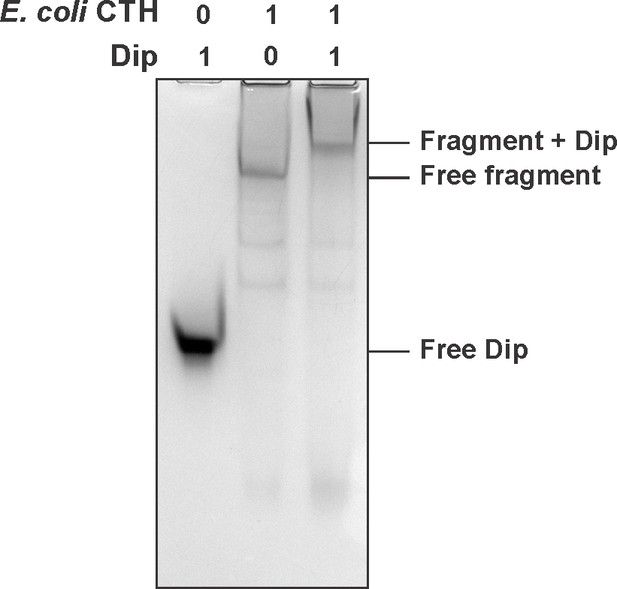
EMSAs of Dip and the CTH (catalytic half (1–26/498–1061) of the E. coli RNase E. Sample were run on an 8% native acrylamide gel.
https://doi.org/10.7554/eLife.16413.009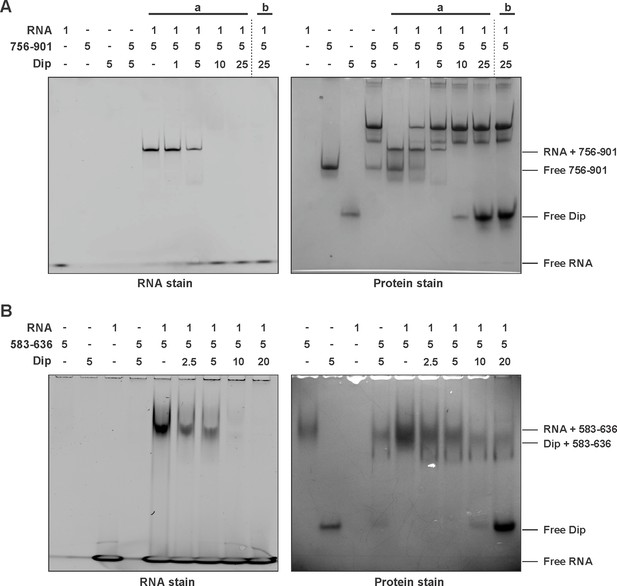
Competitive binding of RNA and Dip to RNase E.
(A) EMSA of RNA (27mer), the 756–901 fragment of RNase E (fused to a GST-tag) and Dip. (a) indicates that RNA was incubated with the fragment prior to the addition of Dip. (b) indicates that Dip was incubated with the peptide before adding the RNA. (B) EMSA of 9S RNA, the 583–636 fragment of RNase E (fused to a GST-tag) and Dip. The samples were run on an 8% native acrylamide gel. Concentrations are presented in µM.
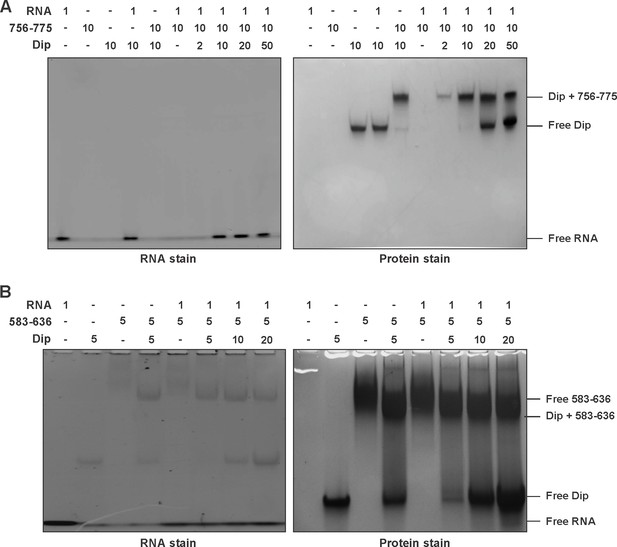
Electrophoretic mobility shift assay of RNase E peptides, 27mer RNA and Dip.
(A) EMSA of 27mer RNA, the untagged 756–775 fragment of RNase E and Dip. (B). EMSA of RNA (27mer), the 583–636 fragment of RNase E (fused to a gst-tag) and Dip. The samples were run on an 8% native acrylamide gel.
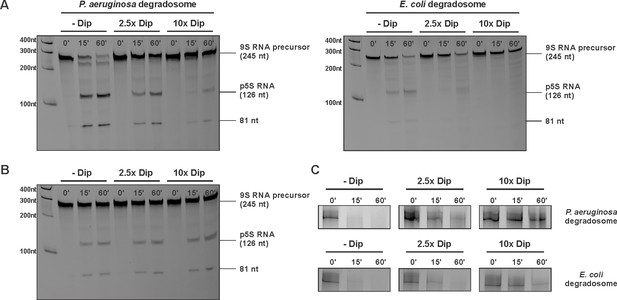
Degradation and processing assays in the presence of Dip.
(A) The 9S RNA precursor of 5S rRNA of E. coli was incubatedin vitro with RNA degradosome in the absence or presence of Dip. (B) E. coli 9S RNA was incubated with the catalytic domain of the E. coli RNA degradosome (1–529) in the absence or presence of Dip. (C) A late φKZ RNA transcript was incubated with the degradosome in the absence or presence of Dip. All samples were loaded on a 5-8-20% (according to the size) denaturing polyacrylamide gel and visualized with SYBR gold stain.

Influence of Dip on the RNA degradosome activity.
(A) The 9S RNA of E. coli was incubated in the absence or presence of Dip. (B) The RNA fragment fdhE of E. coli was incubated with the RNA degradosome of P. aeruginosa and E. coli in the absence or presence of Dip. The samples were loaded on an 8% denaturing polyacrylamide gel.
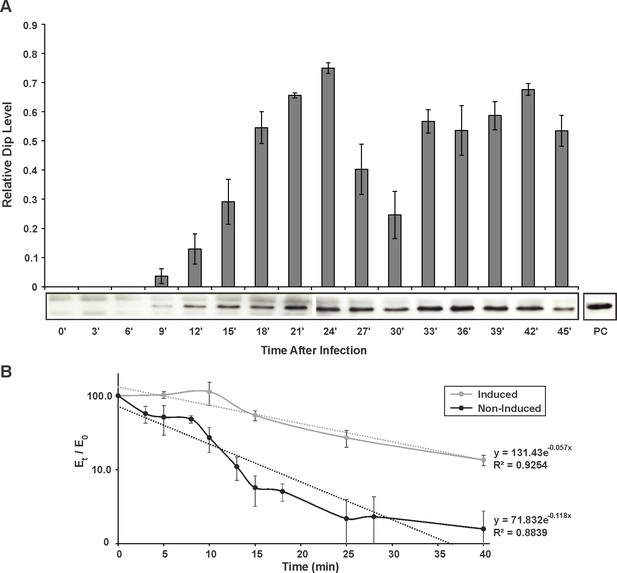
In vivo detection and influence of Dip.
(A) Western blot for the in vivo detection of Dip during φKZ infection using anti-Dip antibodies. Pixel number and intensity were quantified and normalized against the positive control (PC) in imageJ. Error bars represent standard deviation (n = 3). (B) In vivo RNA decay of the household gene OprL. The quantity of OprL transcripts from Rif200-treated P. aeruginosa cells (wild type cells (Non-Induced) or cells expressing Dip) was determined by qRT-PCR and normalized to the total RNA content. The amount of RNA (Et) was compared to the amount of RNA at time point 0 (E0) and plotted in a semi-logarithmic plot as a function of time. Error bars represent standard deviation (n = 3). Dotted lines represent a trend line of a data set.
-
Figure 6—source data 1
Data Western blot.
- https://doi.org/10.7554/eLife.16413.015
-
Figure 6—source data 2
Data qPCR.
- https://doi.org/10.7554/eLife.16413.016
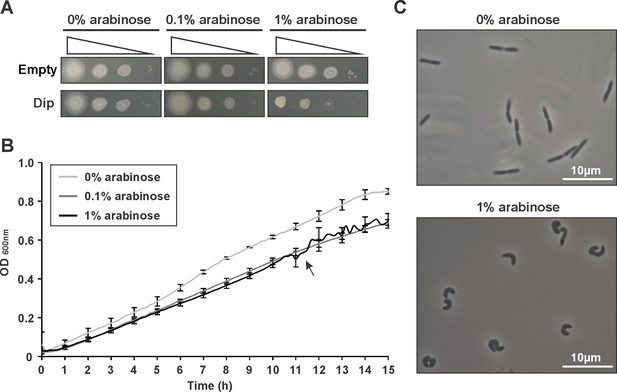
Dip was expressed in P. aeruginosa cells from a pHERD20T vector with an arabinose-inducible PBAD promoter.
(A) Dilution series (100-10−2-10−4-10−6) were spotted on solid medium. As a control P. aeruginosa cells with an empty pHERD20T vector were used. (B) A bioscreen following the OD600 nm in liquid medium. The arrow indicates the time point were cells were sampled in C. Error bars represent SD. (n = 3) (C) Microscopic view of the cells at single cell level at OD600 nm 0.5 with and without the induction of Dip expression.
-
Figure 6—figure supplement 1—source data 1
Data Bioscreen.
- https://doi.org/10.7554/eLife.16413.018
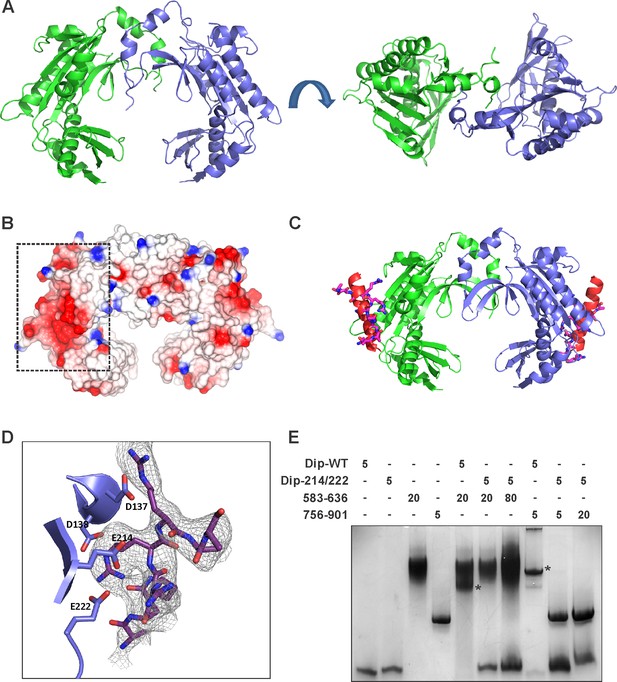
The crystal structure of Dip.
(A) Two views of the dimeric Dip structure. Protomers are coloured in green and blue. (B) Electrostatic surface representation (red = negative, blue = positive) of the Dip dimer. The negatively charged, patch/pocket on the outer surface of the Dip-dimer is indicated with the dashed box. (C) The structural interaction between peptide 756–775 (of RNase E) and Dip. The in-silico docked peptide is shown as a red helix, and the peptide modelled from X-ray crystal data is shown as purple and blue sticks. (D) Experimental structure of the complex, showing a close view of the interacting amino acids of Dip (D137, D138, E214 and E222) and RNase E peptide 756–775. The electron density map is shown for the RNase E peptide only for clarity. (E) EMSA of wild type Dip and the mutant Dip-E214/E222 (substituted to Ala) and the RNase E peptides 583–636 and 725–901. The samples were run on an 8% native acrylamide gel. Concentrations are presented in µM.
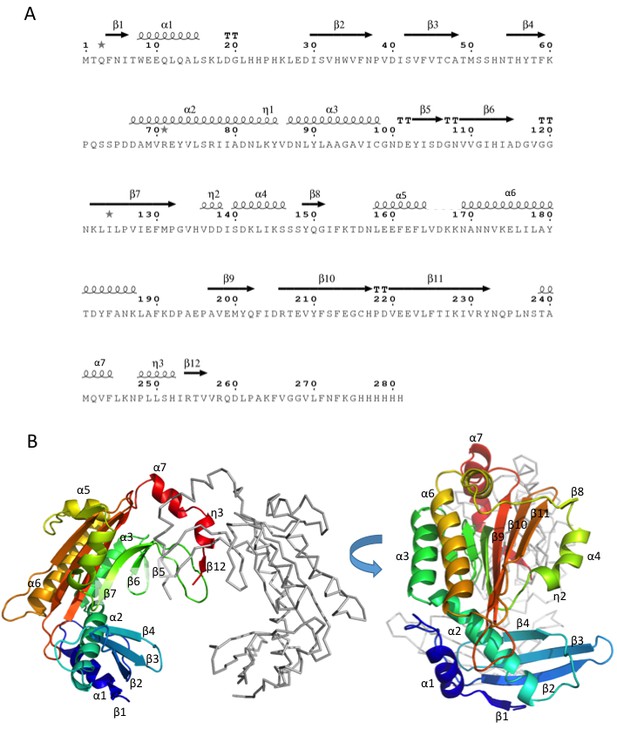
Structural analysis of Dip.
(A) Amino acid sequence of Dip with secondary structural elements indicated above (α = alpha-helix, β = beta-strand, η = 3–10 helix, TT = beta-turn. (B) Two views of the structure of dimeric Dip, with one protomer shown as cartoon coloured blue to red rainbow from N to C terminus, and the second protomer shown as grey ribbon. Secondary structure elements as in (A) are labelled.
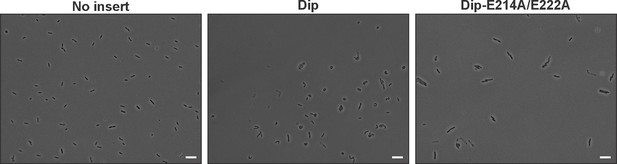
Microscopic view of P. aeruginosa cells induced (1% arabinose at OD600 nm 0.07) for the in vivo expression of No insert (empty vector), Dip and Dip-E214A/E222A using the pHERD20T vector.
Bars represent 10 µm.
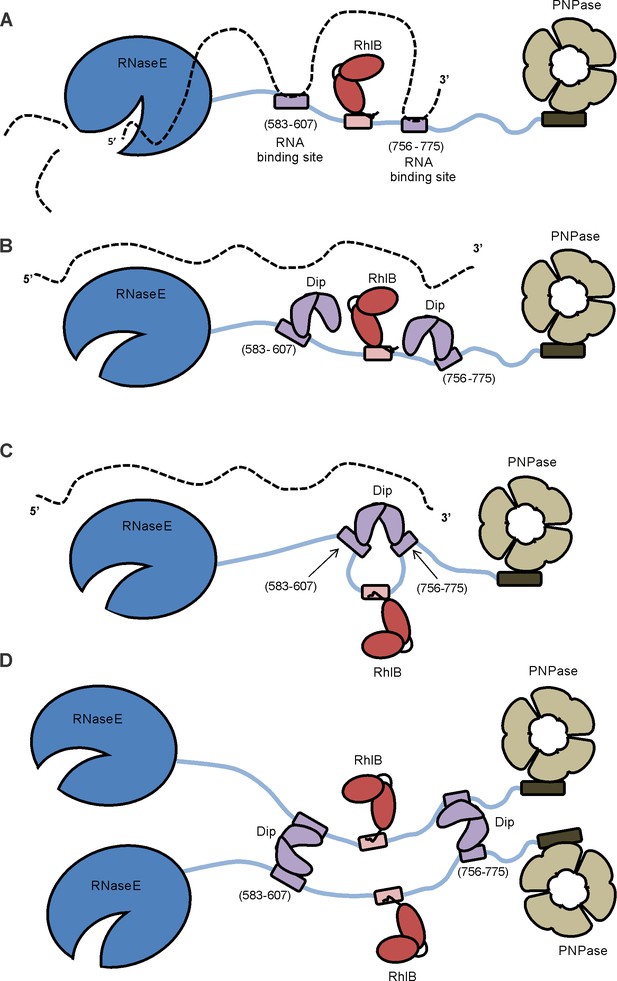
Model of the working mechanism of Dip.
(A) The wild type P. aeruginosa degradsome consisting of the RNase E subunit (blue), the ATP-dependent RNA helicase RhlB (red) and the PNPase (beige). The RNA molecule (dotted line) binds to the RNA-binding sites of the scaffold domain of RNase E (purple) and is cut by the catalytic domain of RNase E. (B) A putative model hypothesizing that one Dip-dimer binds to each RNA-binding site during the infection by phage φKZ. (C) A second model hypothesizing that one Dip-dimer binds to both RNA-binding sites at the same time, yielding a looping of the scaffold domain of RNase E. (D) A model in which the Dip-dimers form a link between two RNA degradosome protomers.
Additional files
-
Supplementary file 1
Mass spectrometry results for the Rne::StrepII affinity purification.
Table 1. MS results of the affinity purifications on Rne::StrepII, infected with one of seven Pseudomonas phages. Table 2. Diffraction statistics and refinement statistics of the crystals of Dip.
- https://doi.org/10.7554/eLife.16413.023





