Protein sequences bound to mineral surfaces persist into deep time
Figures
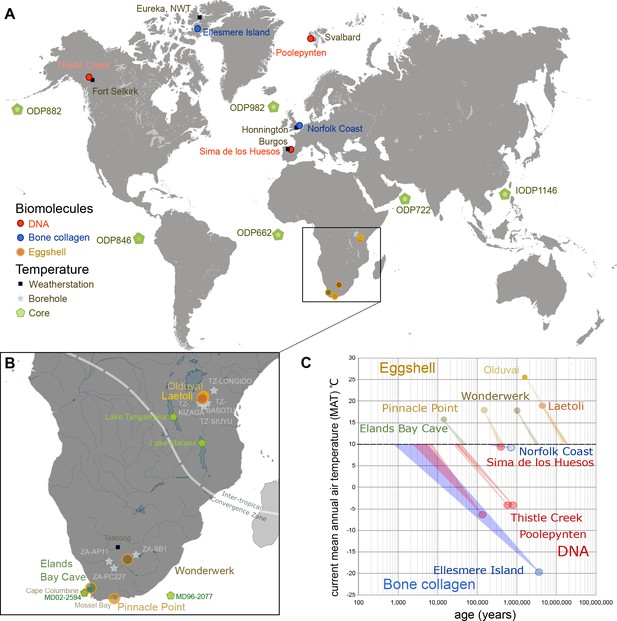
Eggshell peptide sequences from Africa have thermal ages two orders of magnitude older than those reported for DNA or bone collagen.
(A) Sites reporting the oldest DNA and collagen sequences are from high latitude sites compared to ostrich eggshell samples from sites in Africa illustrated in (B) for which the current mean annual air temperatures are much higher. (C) Kinetic estimates of rates of decay for DNA (Lindahl and Nyberg, 1972), collagen (Buckley and Collins, 2011) and ostrich eggshell proteins (Crisp et al., 2013) were used to estimate thermal age assuming a constant 10°C (Figure 1—source data 1; Appendix 1. For Elands Bay Cave and Pinnacle Point the oldest samples are shown). Note log scale on the z-axis: struthiocalcin-1 peptide survival is two orders of magnitude greater than any previously reported sequence, offering scope for the survival of peptide sequences into deep time.
-
Figure 1—source data 1
Data and calculations for thermal ages reported in Figure 1 and in Supplementary file 1.
- https://doi.org/10.7554/eLife.17092.004
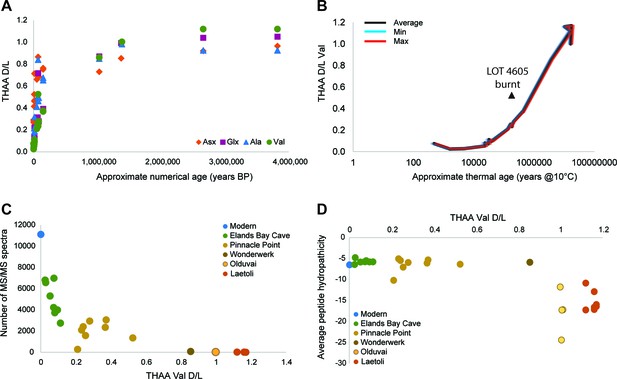
Proteome persistence and patterns of degradation.
(A) Amino acid racemization in ostrich eggshell up to 3.8 Ma old from sites in South Africa and Tanzania. (B) Linear increase of THAA Val D/L values with the log of thermal age. (C) Exponential decrease of the number of identified MS/MS spectra with age (THAA Val D/L). (D) The average hydropathicity of the peptides identified remains stable up to Val D/Ls ~1. Note that Val D/Ls > 1.0 are unexpected and may be due to decomposition processes occurring in the closed system. The intracrystalline proteome composition in modern eggshell does not vary across microstructural layers (Figure 2—figure supplement 1), but patterns of degradation are different between fossil samples and purified proteins degraded at high temperature in the absence of the mineral (Figure 2—figure supplement 2).

Structure and composition of OES.
(A) modern (left) and fossil (LOT 13898; right) OES: crystalline (1), prismatic (2), cone (3) and organic (4) layers. (B) comparison of total THAA yields in each layer before and after bleaching. (C) comparison between the composition of bleached eggshell powder from the cone, palisade and crystalline layers.
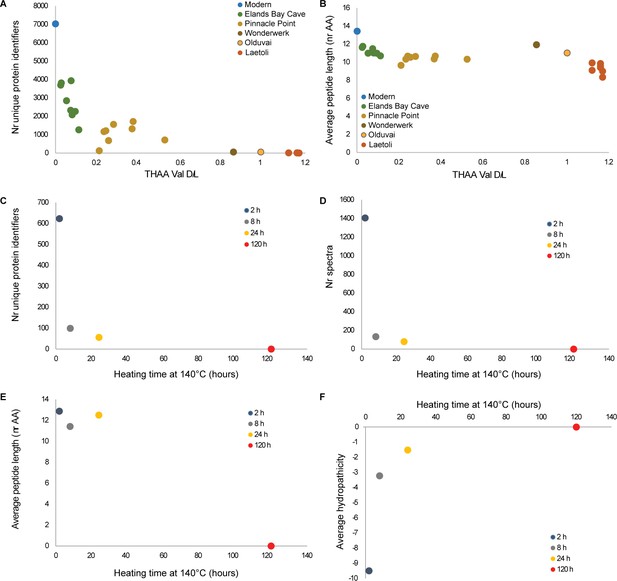
Proteome degradation.
(A–B) fossil OES: (A) number of unique proteins; (B) mean peptide length (excluding contaminants). (C–E) degradation of purified proteins in water: (C) number of unique proteins identified; (D) number of identified product ion spectra; (E) mean peptide length; (F) average hydropathicity. No peptides were detected in the 120 hr heated sample.
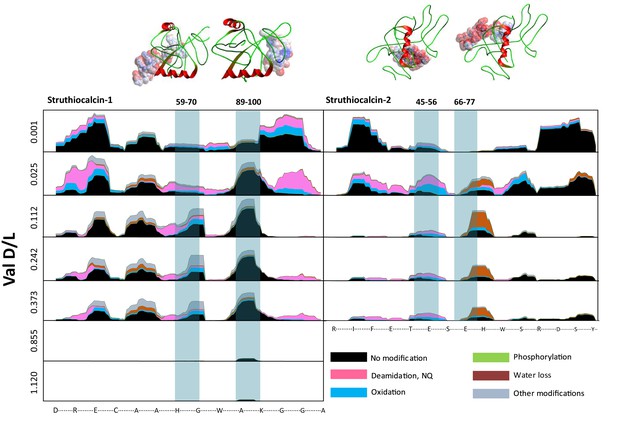
Survival of struthiocalcin 1 and struthiocalcin 2 peptides.
Over time (and increasing THAA Val D/L values) the spectral count decreases as degradation progresses. Blue bars highlight the peptides investigated computationally (represented by the filled atoms in the models). Highly degraded samples (Val D/L ~0.8–1.1) preserve the DDDD-containing peptide. The full time series is shown for SCA-1 in Figure 3—figure supplement 1 and for SCA-2 in Figure 3—figure supplement 2.
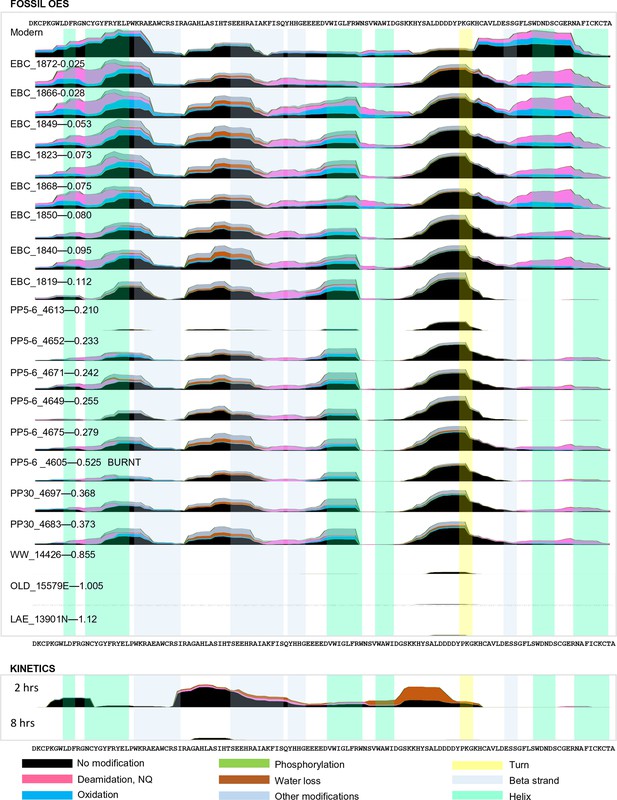
Frequency of identified spectra of SCA-1 in bleached OES (fossils) and purified proteins (kinetics).
Spectral count scale: 0–400 for fossil OES; 0–200 for kinetics. Sample 4605 has been recognized as burnt (Crisp, 2013) but excellent sequence preservation is observed. Low spectral counts for sample 4613 are likely due to sample preparation, as AAR did not identify this sample as problematic. Coloured bars show protein structure.
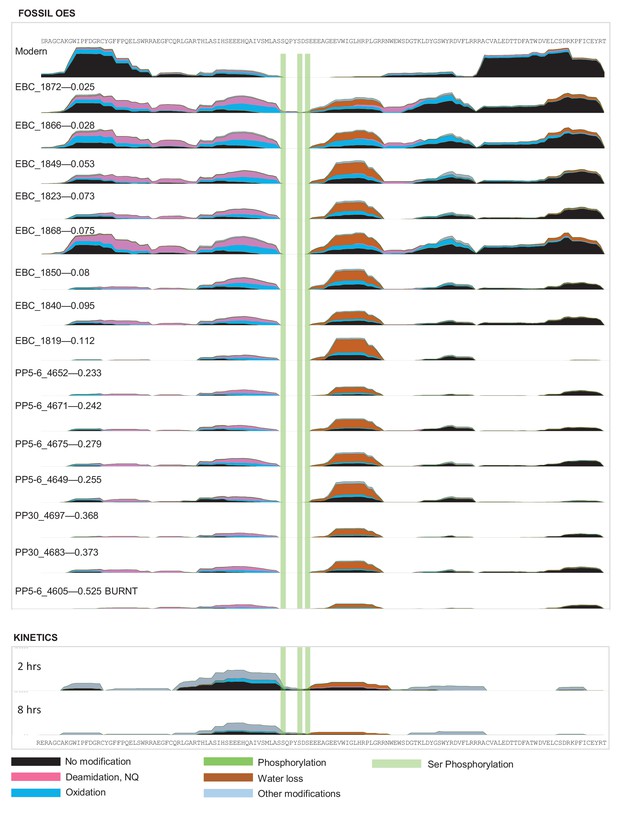
Frequency of identified spectra of SCA-2 in bleached OES (fossils) and purified proteins (kinetics).
Spectral count scale: 0–400 for fossil OES; 0–200 for kinetics. Sample 4605 has been recognized as burnt (Crisp 2013) but excellent sequence preservation is observed. No SCA-2 peptides were found for sample 4613; this is likely due to sample preparation. Wonderwerk and Laetoli samples yielded some peptide sequence, but not consistently.
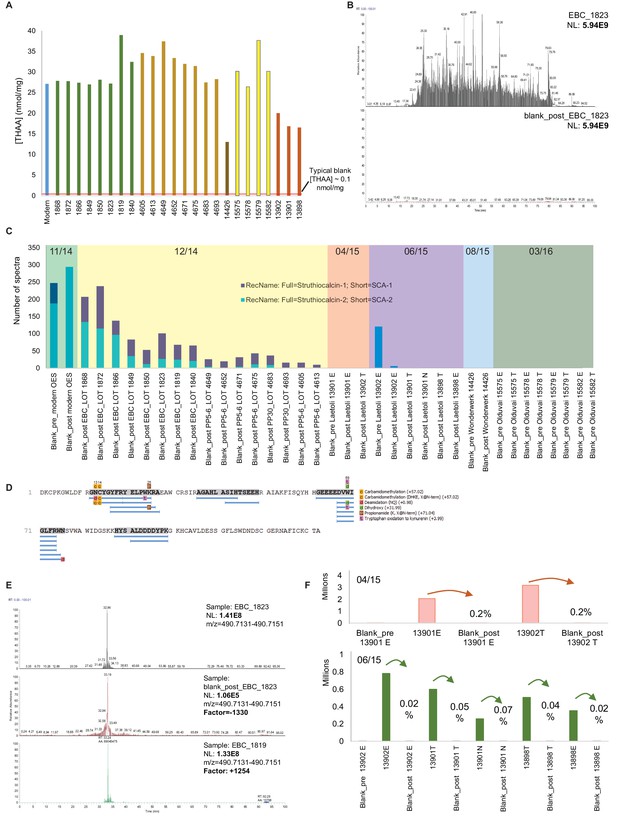
Authenticity of the ancient sequences.
Amino acid analyses (A): Total concentrations in all eggshell samples (sum of Asx, Glx, Gly, Ala, Val and Ile). Carry-over: (B) Total ion chromatogram for one eggshell sample (EBC_1823) and the blank analysed immediately after (blank_post_EBC1823). (C) Spectral abundance of SCA-1 and SCA-2 in LC-MS/MS blanks. (D) SCA-1 coverage in the blank analysed after a Pinnacle Point eggshell sample PP_4652. Note 'DDDD-' and 'EEEED-' peptides and Asn deamidation. (E) Extracted ion chromatogram for LDDDDYPK in EBC_1823, blank_post_EBC1823 and EBC_1819. (F) Absolute and relative total abundance of 'DDDD' peptides in Laetoli samples/blanks. Signal reduction is at least 100-fold (more often 1000- or 10,000-fold). Independent replication and manual de novo sequencing of the peptides from Laetoli (Appendix 5, section A; Supplementary file 2), consistency of diagenesis-induced modifications (Appendix 5, section D; Supplementary file 3) and volatile organic compound analyses (Appendix 5, section E) were also used to validate the results obtained.
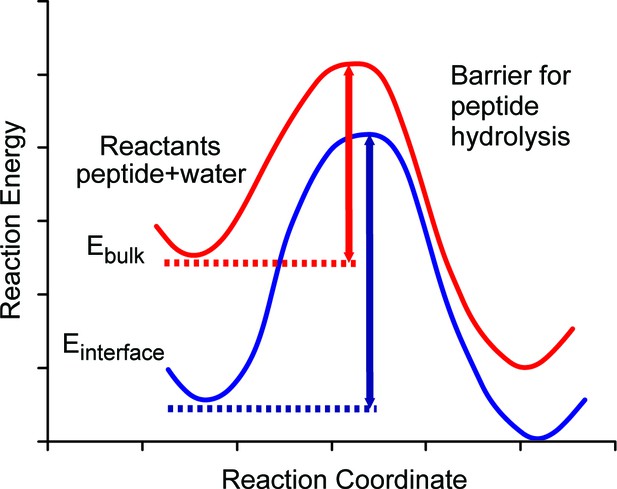
Schematic diagram of energy barriers for peptide hydrolysis.
A pictorial representation of the energy barriers associated with the lysis of the peptide. The process in bulk water is depicted in red and the process at the surface is depicted in blue. The surface process shows a larger barrier due to the stabilization of the reactants at the surface.
Tables
Summary of archaeological and paleontological eggshell samples analysed in this study. See also Tables 3–7.
| Site | Approximate age range (ka) | Approximate thermal age range (ka@10°C) | Number of specimens |
|---|---|---|---|
| Elands Bay Cave | 0.3–16 | 0.5–45 | 8 |
| Pinnacle Point 5/6 | 50–150 | 120–470 | 8 |
| Wonderwerk | ~1000 | ~3600 | 1 |
| Olduvai | ~1340 | ~16300 | 4 |
| Laetoli | 2600–4300 | 8900–20400 | 3 |
Binding of proteins to the (10.4) calcite surface. The binding energies calculated as (a) mean for the full protein (by minimization, see Appendix 3); (b) for four individual domains within the proteins (by molecular dynamics [ovocleidin]; by minimization [struthiocalcin]); (c) for the four domains treated as peptides (by molecular dynamics, see Appendix 3).
| OC-17 | SCA-1 | SCA-2 | |
|---|---|---|---|
| Charge on the protein | +7 (balanced by Cl-) | −11 (balanced by Na+, Ca2+) | −10 (balanced byCa2+) |
| Binding energy (mean): kJ mol−1 | −197 ± 22 | −142 ± 33 | |
| Binding energy (domains): kJ mol−1 | −422 ± 43 | −423 ± 42 (YHHGEEEEDVWI) −611 ± 44 (YSALDDDDYPKG) | −255 ± 72 (SDSEEEAGEEVW) −231 ± 68 (ASIHSEEEHQAIV) |
| Binding energy (peptides only) kJ mol−1 | −142 ± 19 (YHHGEEEEDVWI) −219 ± 24 (YSALDDDDYPKG) | −131 ± 32 (SDSEEEAGEEVW) −122 ± 41 (ASIHSEEEHQAIV) | |
| Water molecules displaced | 21.3 | 20.2 | 23.1 |
| Estimated binding free energy: kJ mol−1 | −188 ± 37 | −159 ± 24 | −99 ± 39 |
| Residence times for water (ps) [average surface bound water molecule = 120 ps] | 130 ± 3 (YHHGEEEEDVWI) 135 ± 3 (YSALDDDDYPKG) | 124 ± 4 (SDSEEEAGEEVW) 123 ± 5 (ASIHSEEEHQAIV) |
Summary of samples from Elands Bay Cave, South Africa. The stratigraphic layers have been independently dated by radiocarbon. Unpublished uncalibrated dates provided by J. Parkington. Date calibration was performed with OxCal v.4.2 (Ramsey, 2009. Calibration curves: IntCal13 for dates obtained on charcoal; Marine13 for dates obtained on shells/crayfish, DeltaR = 93 ± 28 [Dewar et al., 2012]). Age estimates for undated layers based on estimating the median (mid-point) of two dates obtained on layers bracketing the layer with OES samples.
| LOT | NEaar | Layer | Age (cal BP) 95.4% | Material used for 14C dating/notes |
|---|---|---|---|---|
| 1868 | 6887 | Kaunda | <323 (estimate) | Layer above dates on layer NKOM |
| 1872 | 6888 | George Best | 322–1008 | Layer between dates on layers NKOM and EDDI |
| 1866 | 6889 | D. Lamour | 906–2282 | Layer between dates on layers EDDI and LARM |
| 1849 | 6891 | Maroon Robson | 8773 ± 125 | Charcoal |
| 1850 | 6893 | Nero | 8748–10096 | Layer between dates on layers Maroon Robson / Burnt Robeson |
| 1823 | 6896 | Crayfish | 11545 ± 441 | Crayfish |
| 1819 | 6899 | Smoke | 12589 ± 104 | Charcoal |
| 1840 | 6907 | OBS 1 | 15208–15940 | Layer between dates on layers Smoke and SOSE |
Sample details for sub-fossil OES analysed for LC-MS/MS; from Pinnacle Point, South Africa. Stratigraphic information and weighted mean OSL age estimates (ka) for PP 5–6 (Karkanas et al., 2015) and PP 30 (Rector and Reed, 2010).
| Site | LOT | NEaar | Archaeological sample information | Stratigraphic aggregate | Age (ka) |
|---|---|---|---|---|---|
| PP5-6 | 4613 | 7676 | Plotted Find 102627, Lot 3151 | RBSR | 51 ± 2 |
| PP5-6 | 4649 | 7283 | Plotted Find 165702, Lot 8038 | SGS | 64 ± 3 |
| PP5-6 | 4671 | 7316 | Specimen 273467, Lot 3255 | SADBS | 71 ± 3 |
| PP5-6 | 4605 | 7198 | Specimen 273489, Lot 3277 | SADBS | 71 ± 3 |
| PP5-6 | 4652 | 7286 | Plotted Find 178331, Lot 8172 | ALBS | 72 ± 3 |
| PP5-6 | 4675 | 7320 | Specimen 273514, Lot 7980 | LBSR | 81 ± 4 |
| PP 30 | 4683 | 7328 | Specimen 66008, Lot 1795 | Single horizon | ~151 |
| PP 30 | 4697 | 7342 | Specimen 65168, Lot 1750 | Single horizon | ~151 |
Sample details for sub-fossil OES samples from Wonderwerk Cave, South Africa. Ages based on cosmogenic isotope burial dating and magnetostratigraphy, from Matmon et al. (2012).
| LOT | NEaar | Stratum | Independent age (Ma) |
|---|---|---|---|
| 14426 | 10581 | ME46, SPF#4390, Exc. 1, stratum 10, square Q33, depth 15–20 cm | 1.07–0.99 |
Sample details for fossil OES samples from Olduvai, Tanzania.
| LOT | NEaar | Locality/Stratum | Independent age (Ma) |
|---|---|---|---|
| 15575 | 10955 | Sample BK09-3150 | 1.338 ± 0.024 |
| 15578 | 10958 | Sample BK10-5309 | 1.338 ± 0.024 |
| 15579 | 10959 | Sample BK09-2627 | 1.338 ± 0.024 |
| 15582 | 10962 | Sample BK09-2706 | 1.338 ± 0.024 |
Sample details for fossil OES samples from Laetoli, Tanzania. Ages of the strata and localities (40Ar/39Ar) from Deino (2011). LOT 13901 is attributed to Struthio camelus. LOTs 13902 and 13898 are attributed to Struthio kakesiensis (Harrison and Msuya, 2005).
| LOT | NEaar | Locality/Stratum | Independent age (Ma) |
|---|---|---|---|
| 13901 | 10574 | Loc 15, Upper Ndolanya Beds | ~2.66 |
| 13902 | 10573 | Loc 10 West, Upper Laetolil Beds | ~3.8–3.85 |
| 13898 | 10575 | Kakesio 1−6, Lower Laetolil Beds | ~3.85 -> 4.3 |
Historical temperature data. Weather station temperature data derived from the NOAA Baseline Climatological Dataset - Eischeid et al. (1995): The quality control of long-term climatological data using objective data analysis. Preprints of AMS Ninth Conference on Applied Climatology, Dallas, TX., January 15–20, 1995.
| WMO ID | Wmo station name | latitude | longitude | Alt. (m) | MAT °C |
|---|---|---|---|---|---|
| 7191700 | Eureka,N.W.T. | 79.98 | −85.93 | 10 | −19.6 |
| 100800 | Svalbard Lufthavn | 78.25 | 15.47 | 27 | −6.3 |
| 420201 | Dundas Radio Greenland | 76.6 | −68.8 | 20 | −10.5 |
| 420200 | Thule A.B. | 76.52 | −68.5 | 77 | −12.1 |
| 7195702 | Fort Mcpherson | 67.4 | −134.9 | 30 | −9.3 |
| 7196501 | Fort Selkirk | 62.8 | −137.4 | 454 | −3.9 |
| 358600 | Honington | 52.33 | 0.77 | 54 | 9.6 |
| 3605801 | Ust-Kan | 50.92 | 84.75 | 1037 | 0.6 |
| 807500 | Burgos/Villafria | 42.37 | 3.63 | 894 | 10.2 |
| 6832800 | Tsabong | −26.05 | 22.45 | 960 | 20.14 |
| 6871200 | Cape Columbine | −32.83 | 17.85 | 60 | 15.6 |
| 892800 | Mossel Bay (Cape St.) | −34.18 | 22.15 | 59 | 17.7 |
| Borehole data: data from NOAA Paleoclimatology Borehole Datasets http://www.ncdc.noaa.gov/paleo/borehole | |||||
|---|---|---|---|---|---|
| CA-289-2 | 60.99 | −134 | 1524 | 0 | |
| UK-STOWLANGTOFT | 52.28 | 0.85 | 47 | 9.1 | |
| ES-ROMANERA | 37.69 | −7.33 | 166 | 20.2 | |
| ES-AC-1BILLITON | 37.6 | −6.83 | 110 | 18.7 | |
| ES-PB1ADAROVALVERDE | 37.56 | −6.78 | 237 | 18.6 | |
| TZ-LONGIDO | −2.61 | 36.47 | 1316 | 24.3 | |
| TZ-BASOTU | −4.38 | 35.17 | 1736 | 23.9 | |
| TZ-KIZAGA | −4.42 | 34.37 | 1472 | 23.2 | |
| TZ-SIUYU | −4.9 | 34.88 | 1678 | 23.1 | |
| ZA-SB1 | −27.28 | 25.5 | 1357 | 19.2 | |
| ZA-AP11 | −28.3 | 21.05 | 928 | 23.7 | |
| ZA-PC227 | −29.33 | 21.78 | 1052 | 21.5 | |
Long term climate records: cores used in this study to assess the extent of depression at LGM (used to scale).
| Record | latitude | longitude | Method | SST (°C) during LGM | Ref |
|---|---|---|---|---|---|
| ODP982 | 58 | −16 | UK37 | 13 | (Lawrence et al., 2009) |
| ODP882 | 50.35 | −167.58 | UK37 | 9.1 | (Martínez-Garcia et al., 2010) |
| MD02-2594 | −33.30 | 17.30 | Mg/Ca | ~16 | doi.pangaea.de/10.1594/PANGAEA.810663 |
| MD962077 | −33.17 | 31.25 | Mg/Ca | ~16 | doi.pangaea.de/10.1594/PANGAEA.810716 |
| ODP722A | 16.62 | 59.80 | UK37 | 20.2 | (Herbert et al., 2010) |
| ODP662 | −1.39 | −11.74 | UK37 | 18.7 | (Herbert et al., 2010) |
| ODP846 | −3.10 | −90.82 | Mg/Ca | 18.6 | (Herbert et al., 2010) |
| IODP1146 | 19.46 | 116.27 | UK37 | 24.3 | (Herbert et al., 2010) |
Kinetic parameters used for estimating thermal age.
| Target | Chemical reaction | Activation energy (kJ mol−1) | Reference |
|---|---|---|---|
| Eggshell proteins | Valine racemization | 117 | (Crisp et al., 2013) |
| DNA | Depurination | 123 | (Lindahl and Nyberg, 1972) |
| Collagen | Gelatinization | 173 | (Holmes et al., 2005) |
Thermal age calculations.
| Site | Lat | Long | Alti (m) | MAT (°C) | Scale | ΔT LGM (°C) | Ref | Age (ka) | Rate | Ea(kJ) | ln A | Mean thermal age (Ma@10°C) | Min thermal age (Ma@10°C) | Max thermal age (Ma@10°C) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ellesmere Island | 78.55 | −82.5 | 292 | −19.7 | 5.0 | −21 | (Ballantyne et al., 2010) | 3600 | Collagen | 173 | 61 | 0.003 | 0.001 | 0.007 |
| Ellesmere Island | 78.55 | −82.5 | 292 | −19.7 | 5.0 | −21 | (Ballantyne et al., 2010) | 3800 | Collagen | 173 | 61 | 0.004 | 0.001 | 0.011 |
| Poolepynten | 79 | 11.43 | 4 | −6.3 | 4.0 | −16 | (Hubberten, 2004) | 130 | DNA | 127 | 41.2 | 0.002 | 0.002 | 0.002 |
| Thistle Creek | 63.11 | −139.54 | 483 | −4.1 | 1.0 | −4 | (Elias, 2001) | 560 | DNA | 127 | 41.2 | 0.02 | 0.02 | 0.02 |
| Thistle Creek | 63.11 | −139.54 | 483 | −4.1 | 1.0 | −4 | (Elias, 2001) | 700 | DNA | 127 | 41.2 | 0.03 | 0.03 | 0.03 |
| Happisburgh | 52.82 | 1.53 | 2 | 9.1 | 2.0 | −9 | (Harrison et al., 2015) | 700 | Collagen | 173 | 61 | 0.22 | 0.19 | 0.25 |
| Sima de los Huesos | 42.35 | −3.52 | 1004 | 9.5 | 1.0 | −4 | (Moreno et al., 2012) | 400 | DNA | 127 | 41.2 | 0.25 | 0.22 | 0.28 |
| Elands Bay Cave | −32.32 | 18.32 | 10 | 15.7 | 1.0 | −4 | (Martínez-Méndez, 2010) | 16 | Valine | 118 | 38 | 0.04 | 0.03 | 0.05 |
| Pinnacle Point | −34.2 | 22.09 | 20 | 18 | 1.0 | −4 | (Bar-Matthews et al., 2010) | 150 | Valine | 118 | 38 | 0.42 | 0.38 | 0.47 |
| Wonderwerk | 27.8 | 23.55 | 1645 | 18.6 | 0.6 | −2.5 | (Ecker et al., 2015) | 1000 | Valine | 118 | 38 | 3.6 | 3.1 | 4.1 |
| Olduvai | −2.99 | 35.35 | 576 | 26 | 0.6 | −2.5 | (Tierney et al., 2010) (Berke et al., 2012) | 1338 | Valine | 118 | 38 | 16.3 | 14.4 | 18.5 |
| Laetoli | −3.22 | 35.19 | 1700 | 18.7 | 0.6 | −2.5 | (Tierney et al., 2010) (Berke et al., 2012) | 3800 | Valine | 118 | 38 | 16.0 | 13.6 | 18.7 |
THAA concentrations (pmol/mg) detected in OES microstructural layers; bleached and unbleached powders were analysed. Analytical errors measured on replicate analyses are < 5% (Asx = 3.80%; Glx = 3.81%; Gly = 4.89%; Ala = 3.67%; Val = 3.98%; Ile = 3.63%).
| [Asx] | [Glx] | [Ser] | [L-Thr] | [L-His] | [Gly] | [L-Arg] | [Ala] | [Tyr] | [Val] | [Phe] | [Leu] | [Ile] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cone- bleached | 4925 | 5409 | 3992 | 2589 | 1520 | 6923 | 2655 | 4314 | 1579 | 2815 | 1983 | 4264 | 2280 |
| Cone- unbleached | 14452 | 17835 | 13455 | 9140 | 4894 | 19136 | 8108 | 10234 | 4342 | 9734 | 4334 | 10500 | 7676 |
| Palisade- bleached | 5990 | 6434 | 4479 | 2347 | 2079 | 6641 | 3427 | 5183 | 1525 | 2768 | 2659 | 5032 | 3052 |
| Palisade- unbleached | 10018 | 11545 | 7967 | 4507 | 3667 | 12691 | 6247 | 9085 | 3052 | 4843 | 4505 | 8446 | 5102 |
| Whole- bleached | 6323 | 6533 | 4662 | 2445 | 2032 | 6976 | 3322 | 5314 | 1263 | 2943 | 2732 | 5289 | 3177 |
| Whole- unbleached | 12412 | 13774 | 9927 | 5420 | 4216 | 14951 | 7222 | 10670 | 4364 | 6003 | 5342 | 10152 | 6342 |
| Crystalline- bleached | 4518 | 4181 | 3085 | 1301 | 1399 | 4097 | 1981 | 3528 | 49 | 1704 | 1838 | 3299 | 2225 |
| Crystalline- unbleached | 24422 | 24212 | 17990 | 8100 | 6748 | 23365 | 12371 | 20055 | 6445 | 9912 | 10755 | 19079 | 12535 |
30 major protein groups identified in bleached modern OES (>20 unique peptides only).
| Protein group | Accession | -lgP | Coverage (%) | #Unique | Description |
|---|---|---|---|---|---|
| 1 | gi|46396750 | 344 | 100 | 346 | RecName: Full = Struthiocalcin-1; Short = SCA-1 |
| 2 | gi|46396751 | 293 | 98 | 222 | RecName: Full = Struthiocalcin-2; Short = SCA-2 |
| 7 | gi|697501075, gi|678217626 | 281 | 39 | 208 | Aggrecan core protein [Struthio camelus australis] |
| 8 | gi|697508924 | 268 | 88 | 126 | Vitelline membrane outer layer protein 1-like [Struthio camelus australis] |
| 10 | gi|697509029 | 268 | 80 | 113 | Serum albumin-like [Struthio camelus australis] |
| 13 | gi|697455783 | 246 | 40 | 89 | Tenascin isoform X3 [Struthio camelus australis] |
| 11 | gi|697477202 | 234 | 75 | 84 | Carbonic anhydrase 4 [Struthio camelus australis] |
| 14 | gi|697481828 | 237 | 15 | 82 | Mucin-5AC [Struthio camelus australis] |
| 9 | gi|697523391, gi|678221588 | 228 | 69 | 77 | Apolipoprotein D [Struthio camelus australis] |
| 12 | gi|375162648 | 231 | 75 | 76 | Immunoglobulin A heavy chain constant region secretory form partial [Struthio camelus] |
| 15 | gi|697470179, gi|697470177 | 213 | 51 | 63 | Mesothelin isoform X1 [Struthio camelus australis] |
| 22 | gi|697433909, gi|678206587 | 195 | 40 | 45 | Golgi apparatus protein 1 [Struthio camelus australis] |
| 17 | gi|375162644 | 192 | 63 | 42 | immunoglobulin M heavy chain constant region secretory form partial [Struthio camelus] |
| 19 | gi|678209093, gi|697441180 | 197 | 66 | 41 | Stanniocalcin-1 partial [Struthio camelus australis] |
| 18 | gi|697514088, gi|678219803 | 197 | 86 | 39 | Ovomucoid [Struthio camelus australis] |
| 20 | gi|697488611 | 191 | 89 | 39 | Serotriflin-like [Struthio camelus australis] |
| 21 | gi|678214778 | 192 | 42 | 38 | Delta and Notch-like epidermal growth factor-related receptor partial [Struthio camelus australis] |
| 23 | gi|697475278, gi|697475274, gi|697475280 | 192 | 41 | 37 | Polymeric immunoglobulin receptor [Struthio camelus australis] |
| 26 | gi|697430975 | 181 | 33 | 33 | BPI fold-containing family B member 4-like [Struthio camelus australis] |
| 25 | gi|697430936 | 161 | 42 | 32 | Uncharacterized protein LOC104140623 [Struthio camelus australis] |
| 31 | gi|697485873, gi|678214846 | 172 | 68 | 28 | Pigment epithelium-derived factor [Struthio camelus australis] |
| 28 | gi|697430934, gi|678205748 | 153 | 34 | 26 | BPI fold-containing family B member 4-like [Struthio camelus australis] |
| 29 | gi|697432918 | 172 | 41 | 26 | Prosaposin [Struthio camelus australis] |
| 16 | gi|375162654 | 178 | 96 | 24 | immunoglobulin lambda constant region partial [Struthio camelus] |
| 30 | gi|697522911, gi|697522913, gi|697522916 | 172 | 50 | 22 | Pantetheinase-like isoform X1 [Struthio camelus australis] |
| 24 | gi|697446419 | 165 | 71 | 22 | Beta-2-microglobulin [Struthio camelus australis] |
| 27 | gi|678210026,gi|678210025 | 154 | 63 | 22 | Cygnin [Struthio camelus australis] |
| 34 | gi|678216365 | 173 | 31 | 21 | Signal peptide CUB and EGF-like domain-containing protein 1 partial [Struthio camelus australis] |
| 32 | gi|697492053 | 143 | 42 | 21 | Ovalbumin {ECO:0000303|PubMed:21058653} [Struthio camelus australis] |
| 37 | gi|697505689 | 147 | 13 | 20 | Ovostatin-like [Struthio camelus australis] |
List of contaminant proteins detected in the 66 analyses, with total number of spectra identified per protein.
| Row labels | Count of #Spec |
|---|---|
| sp|TRYP_PIG| | 771 |
| sp|K2C1_HUMAN| | 493 |
| Chymotrypsin-like elastase family member 1 OS = Sus scrofa GN = CELA1 PE = 1 SV = 1 | 422 |
| sp|K1C9_HUMAN| | 301 |
| sp|K1C10_HUMAN| | 216 |
| sp|K22E_HUMAN| | 213 |
| sp|TRFE_HUMAN| | 98 |
| sp|ALBU_HUMAN| | 94 |
| PREDICTED: keratin type II cytoskeletal cochleal isoform X1 [Struthio camelus australis] | 80 |
| PREDICTED: keratin type II cytoskeletal cochleal isoform X2 [Struthio camelus australis] | 32 |
| PREDICTED: keratin type II cytoskeletal 5-like [Struthio camelus australis] | 25 |
| sp|RS27A_HUMAN| | 23 |
| Keratin type II cytoskeletal 75 partial [Struthio camelus australis] | 20 |
| sp|GFP_AEQVI| | 19 |
| sp|TRY1_BOVIN| | 19 |
| Keratin type II cytoskeletal cochleal partial [Struthio camelus australis] | 16 |
| PREDICTED: keratin type II cytoskeletal cochleal isoform X3 [Struthio camelus australis] | 16 |
| sp|ANT3_HUMAN| | 8 |
| sp|HBB_HUMAN| | 8 |
| Keratin type II cytoskeletal 75 [Struthio camelus australis] | 5 |
| sp|TRFL_HUMAN| | 5 |
| Keratin type I cytoskeletal 14 partial [Struthio camelus australis] | 3 |
| PREDICTED: keratin type II cytoskeletal 4-like [Struthio camelus australis] | 3 |
| sp|HBA_HUMAN| | 3 |
| sp|K1C15_SHEEP| | 2 |
| sp|GSTP1_HUMAN| | 1 |
D/L values of archaeological OES samples analysed in this study. b = bleached; H* = THAA obtained by 24-hr acid hydrolysis; F = FAA. Ile A/I values could not be calculated for Laetoli samples due to the presence of a compound co-eluting with D-alloisoleucine. Analytical errors measured on replicate analyses are <5% (D/Ls: Asx = 0.54%; Glx = 1.18%; Ala = 3.94%; Val = 2.09%; Ile = 3.77%. Concentrations: Asx = 3.80%; Glx = 3.81%; Gly = 4.89%; Ala = 3.67%; Val = 3.98%; Ile = 3.63%). (★) Sample 4605 yielded high D/L values because this sample had been exposed to high temperatures in the burial environment (burning [Crisp, 2013]). Note: thermal age calculations were performed on the basis of the Hansen model (Hansen et al., 2013); due to the absence of more continuous record for younger (last 2000 years) samples in the Hansen record, the Elands Bay Cave time points <1600 years refined using the SST record from core MD02-2594 - Dyez14 (Dyez et al. 2014).
| LOT | NEaar | Asx D/L | Glx D/L | Ala D/L | Val D/L | Ile A/I | Thermal age (years) |
|---|---|---|---|---|---|---|---|
| 1868 | 6887bH* | 0.291 | 0.061 | 0.066 | 0.075 | 0.052 | 401−564 |
| 1868 | 6887bF | 0.346 | 0.074 | 0.119 | 0.098 | 0.076 | |
| 1872 | 6888bH* | 0.268 | 0.056 | 0.061 | 0.025 | 0.032 | 1313–1932 |
| 1872 | 6888bF | 0.212 | 0.079 | 0.100 | 0.000 | 0.000 | |
| 1866 | 6889bH* | 0.278 | 0.057 | 0.066 | 0.028 | 0.052 | 3962−5704 |
| 1866 | 6889bF | 0.331 | 0.062 | 0.116 | 0.000 | 0.076 | |
| 1849 | 6891bH* | 0.414 | 0.095 | 0.142 | 0.053 | 0.081 | 19,759–27,951 |
| 1849 | 6891bF | 0.555 | 0.136 | 0.226 | 0.138 | 0.169 | |
| 1850 | 6893bH* | 0.524 | 0.141 | 0.215 | 0.080 | 0.133 | 19,759–27,951 |
| 1850 | 6893bF | 0.698 | 0.177 | 0.368 | 0.166 | 0.254 | |
| 1823 | 6896bH* | 0.462 | 0.117 | 0.185 | 0.073 | 0.091 | 25,657–36,249 |
| 1823 | 6896bF | 0.621 | 0.161 | 0.301 | 0.158 | 0.208 | |
| 1819 | 6899bH* | 0.714 | 0.222 | 0.317 | 0.112 | 0.184 | 26,878–37,877 |
| 1819 | 6899bF | 0.806 | 0.294 | 0.442 | 0.217 | 0.332 | |
| 1840 | 6907bH* | 0.469 | 0.123 | 0.187 | 0.095 | 0.122 | 32,379–44,863 |
| 1840 | 6907bF | 0.665 | 0.171 | 0.327 | 0.168 | 0.241 | |
| 4605(★) | 7198bH* | 0.866 | 0.717 | 0.840 | 0.525 | 0.708 | 166,094–198,795 |
| 4605(★) | 7198bF | 0.931 | 0.782 | 0.931 | 0.692 | 0.955 | |
| 4613 | 7676bH* | 0.661 | 0.226 | 0.411 | 0.210 | 0.269 | 122,139–148,223 |
| 4613 | 7676bF | 0.800 | 0.345 | 0.570 | 0.349 | 0.472 | |
| 4649 | 7283bH* | 0.712 | 0.314 | 0.500 | 0.255 | 0.320 | 151,169–181,807 |
| 4649 | 7283bF | 0.846 | 0.380 | 0.729 | 0.428 | 0.590 | |
| 4652 | 7286bH* | 0.678 | 0.277 | 0.463 | 0.233 | 0.302 | 168,899–202,088 |
| 4652 | 7286bF | 0.822 | 0.355 | 0.698 | 0.391 | 0.528 | |
| 4671 | 7316bH* | 0.681 | 0.270 | 0.468 | 0.242 | 0.299 | 166,094–198,795 |
| 4671 | 7316bF | 0.863 | 0.339 | 0.687 | 0.384 | 0.577 | |
| 4675 | 7320bH* | 0.687 | 0.279 | 0.494 | 0.279 | 0.330 | 192,147–230,607 |
| 4675 | 7320bF | 0.832 | 0.321 | 0.688 | 0.423 | 0.590 | |
| 4683 | 7328bH* | 0.764 | 0.392 | 0.676 | 0.373 | 0.505 | 378,398–467,602 |
| 4683 | 7328bF | 0.879 | 0.425 | 0.821 | 0.523 | 0.737 | |
| 4697 | 7342bH* | 0.752 | 0.388 | 0.653 | 0.368 | 0.497 | 378,398–467,602 |
| 4697 | 7342bF | 0.904 | 0.659 | 0.920 | 0.777 | 0.973 | |
| 14426 | 10581bH* | 0.730 | 0.870 | 0.855 | 0.855 | 0.985 | 3,238,624–4,188,207 |
| 14426 | 10581bF | 0.790 | 0.930 | 0.960 | 1.010 | 1.280 | |
| 15575 | 10955bH* | 0.860 | 1.035 | 0.981 | 1.001 | >1.2 | 14,387,543–18,460,416 |
| 15575 | 10955bF | 0.924 | 1.001 | 0.961 | 1.005 | >1.2 | |
| 15578 | 10958bH* | 0.891 | 1.040 | 0.984 | 1.007 | >1.2 | 14,387,543–18,460,416 |
| 15578 | 10958bF | 0.916 | 1.009 | 0.960 | 0.999 | >1.2 | |
| 15579 | 10959bH* | 0.811 | 1.021 | 0.965 | 1.007 | >1.2 | 14,387,543–18,460,416 |
| 15579 | 10959bF | 0.926 | 1.006 | 0.969 | 1.005 | >1.2 | |
| 15582 | 10962bH* | 0.881 | 1.033 | 0.976 | 1.012 | >1.2 | 14,387,543–18,460,416 |
| 15582 | 10962bF | 0.915 | 1.002 | 0.947 | 1.016 | >1.2 | |
| 13902 | 10573bH* | 0.965 | 1.050 | 0.925 | 1.12 | >1.2 | 13,764,246–18,893,425 |
| 13902 | 10573bF | 0.935 | 1.030 | 0.925 | 1.065 | >1.2 | |
| 13901 | 10574bH* | 0.920 | 1.040 | 0.945 | 1.16 | >1.2 | 8,943,148–11,841,107 |
| 13901 | 10574bF | 0.935 | 1.015 | 0.930 | 1.08 | >1.2 | |
| 13898 | 10575bH* | 0.945 | 1.050 | 0.930 | 1.17 | >1.2 | 14,746,875–20,367,942 |
| 13898 | 10575bF | 0.935 | 1.010 | 0.930 | 1.095 | >1.2 |
%FAA values (%FAA = [FAA]/[THAA] * 100). b = bleached. Total% FAA for Laetoli are calculated on the basis of Asx, Gly, Ala, Val only. * [Ala] and [Gly] > 100% are likely due to the effect of decomposition of other amino acids to FAA Gly and FAA Ala (e.g. Ser) (Walton, 1998). (★) Sample 4605 had been exposed to high temperatures in the burial environment (burning) (Crisp 2013).
| LOT | NEaar | Asx | Gly | Ala | Val | Ile | Average |
|---|---|---|---|---|---|---|---|
| 1868 | 6887 | 7 | 16 | 18 | 19 | 6 | 13 |
| 1872 | 6888 | 4 | 11 | 10 | 3 | 2 | 6 |
| 1866 | 6889 | 18 | 35 | 39 | 15 | 13 | 24 |
| 1849 | 6891 | 26 | 41 | 41 | 21 | 18 | 30 |
| 1850 | 6893 | 17 | 14 | 57 | 34 | 30 | 30 |
| 1823 | 6896 | 36 | 41 | 47 | 23 | 21 | 34 |
| 1819 | 6899 | 22 | 45 | 56 | 31 | 28 | 36 |
| 1840 | 6907 | 18 | 40 | 51 | 26 | 23 | 32 |
| 4605(★) | 7198 | 70 | 74 | 83 | 61 | 56 | 69 |
| 4613 | 7676 | 53 | 57 | 66 | 44 | 41 | 52 |
| 4649 | 7283 | 49 | 49 | 63 | 40 | 39 | 48 |
| 4652 | 7286 | 48 | 47 | 61 | 42 | 40 | 48 |
| 4671 | 7316 | 50 | 61 | 72 | 47 | 45 | 57 |
| 4675 | 7320 | 62 | 59 | 74 | 48 | 47 | 58 |
| 4683 | 7328 | 90 | 71 | 91 | 64 | 66 | 76 |
| 4697 | 7342 | 55 | 83 | 96 | 64 | 58 | 71 |
| 14426 | 10581 | 50 | 122* | 138* | 90 | 84 | 97 |
| 15575 | 10955 | 53 | 47 | 55 | 46 | n/a | 50 |
| 15578 | 10958 | 70 | 62 | 72 | 62 | n/a | 67 |
| 15579 | 10959 | 29 | 27 | 31 | 26 | n/a | 28 |
| 15582 | 10962 | 78 | 62 | 74 | 64 | n/a | 69 |
| 13902 | 10573 | 74 | 64 | 82 | 65 | n/a | 71 |
| 13901 | 10574 | 88 | 78 | 92 | 74 | n/a | 83 |
| 13898 | 10575 | 87 | 76 | 93 | 73 | n/a | 82 |
: Proteins identified in the purified extract from bleached OES, heated at 140°C in ultrapure water for 2, 8, 24 and 120 hr (identified by at least 2 unique peptide sequences). Values are number of peptide sequences identified.
| Protein description | 2 hr | 8 hr | 24 hr | 120 hr | Total |
|---|---|---|---|---|---|
| RecName: Full = Struthiocalcin-1; Short = SCA-1 | 295 | 21 | 316 | ||
| RecName: Full = Struthiocalcin-2; Short = SCA-2 | 124 | 8 | 132 | ||
| Vitelline membrane outer layer protein 1-like [Struthio camelus australis] | 22 | 2 | 24 | ||
| von Willebrand factor partial [Struthio camelus australis] | 20 | 20 | |||
| immunonoglobulin heavy chain variable region partial [Struthio camelus] | 18 | 18 | |||
| Apolipoprotein D [Struthio camelus australis] | 14 | 2 | 16 | ||
| Mitogen-activated protein kinase MLT [Struthio camelus australis] | 7 | 9 | 16 | ||
| Aggrecan core protein [Struthio camelus australis] | 13 | 13 | |||
| iron binding protein [Struthio camelus] | 9 | 9 | |||
| Histone H4 [Struthio camelus australis] | 4 | 4 | 8 | ||
| Cyclin-K [Struthio camelus australis] | 7 | 7 | |||
| BPI fold-containing family B member 4 partial [Struthio camelus australis] | 4 | 2 | 6 | ||
| Histone H2B 1/2/3/4/6 [Struthio camelus australis] | 3 | 3 | 6 | ||
| Complement C3 [Struthio camelus australis] | 2 | 3 | 5 | ||
| PREDICTED: histone H2B 1/2/3/4/6 [Struthio camelus australis] | 2 | 2 | 4 | ||
| PREDICTED: polymeric immunoglobulin receptor [Struthio camelus australis] | 3 | 3 | |||
| Carbonic anhydrase 4 partial [Struthio camelus australis] | 2 | 2 |
Peptides identified in all Laetoli OES samples, prepared in York (analysed in Oxford) and Copenhagen. (*Gla) = this residue was found to be in the decarboxylated form, i.e. glutamate residues that have been post-translationally modified by vitamin K-dependent carboxylation to form gamma-carboxyglutamic acid (Gla), which binds calcium. We conducted manual de novo analyses of all product ion spectra and determined either complete or partial sequences for all of them, independently identifying sequences assigned (assisted de novo) by the software (PEAKS Studio).
| Sequence | Protein | Laboratory | Score | MS/MS count |
|---|---|---|---|---|
| AGAHLASIHTSEEHR | SCA-1 | Copenhagen | 47.14 | 4 |
| HYSALDDDDYPKGK | SCA-1 | Copenhagen | 35.69 | 2 |
| AGAHLASIH | SCA-1 | Copenhagen | 31.45 | 3 |
| ERNAFICK | SCA-1 | Copenhagen | 28.12 | 1 |
| GNCYGYFR | SCA-1 | Copenhagen | 28.1 | 1 |
| DVWIGLFR | SCA-1 | Copenhagen | 26.38 | 5 |
| ALDDDDYPK | SCA-1 | York | 39.39 | 28 |
| ALDDDDYPKG | SCA-1 | York | 41.34 | 14 |
| DDDDYPKGK | SCA-1 | York | 40.79 | 3 |
| DDDYPKGK | SCA-1 | York | 32.89 | 1 |
| HYSALDDDDYPK | SCA-1 | York | 51.09 | 1 |
| KHYSALDDDDYPK | SCA-1 | York | 34.86 | 2 |
| LDDDDYPK | SCA-1 | York | 34.35 | 12 |
| LDDDDYPKG | SCA-1 | York | 35.3 | 6 |
| LDDDDYPKGK | SCA-1 | York | 35.66 | 3 |
| SALDDDDYPK | SCA-1 | York | 41.04 | 10 |
| SALDDDDYPKG | SCA-1 | York | 39.15 | 5 |
| YSALDDDDYPK | SCA-1 | York | 34.46 | 3 |
| YSALDDDDYPKG | SCA-1 | York | 31.9 | 3 |
| RAEAWCR | SCA-1 | York | 30.65 | 1 |
| CYGFFPQELSWR | SCA-2 | Copenhagen | 30.98 | 1 |
| KPFICEYRT | SCA-2 | Copenhagen | 25.03 | 1 |
| GE(*Gla)EVWIGLHRPLGR | SCA-2 | York | 37.33 | 2 |
| LDYGSWYR | SCA-2 | York | 35.1 | 1 |
| AGE(*Gla)EVWIGLHRPLGR | SCA-2 | York | 34.64 | 2 |
Crushed eggshell sulfur-containing VOC emissions from 2.7 Ma OES (Laetoli LOT 13901). NA = no structural isomer can be determined.
| Compound | Retention time (min) | Measured m/z | Chemical formula | NIST MS similarity | Reverse fit | |
|---|---|---|---|---|---|---|
| Ethane thiol/dimethylsulfide | 3.42 | 62.0196 | C2H6S | NA | NA |   |
| Methylthioethane | 5.59 | 76.0354 | C3H8S | 726 | 821 |  |
| Diethylsulfide | 8.24 | 90.0511 | C4H10S | 725 | 834 |  |
| 1-methylthiopropane | 8.63 | 90.0509 | C4H10S | 646 | 721 |  |
| Unknown isomer | 8.86 | 104.0663 | C5H12S | NA | NA | NA |
| 2-ethylthio-propane | 9.66 | 104.0664 | C5H12S | 535 | 723 | 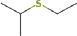 |
| 2-methyl-1-(methylthio)-propane | 10.26 | 104.0670 | C5H12S | 636 | 798 | 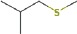 |
| ethylpropylsulfide | 10.85 | 104.0671 | C5H12S | 628 | 801 |  |
| 2-methyl-2-(methylthio)-butane | 11.89 | 118.0823 | C6H14S | 567 | 777 | 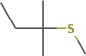 |
| 1-(ethylthio)-2-methyl-propane | 12.18 | 118.0824 | C6H14S | 817 | 856 | 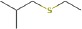 |
| 2-methyl-3-(methylthio)-butane | 12.286 | 118.0823 | C6H14S | 600 | 820 | 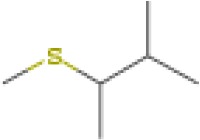 |
| Unknown isomer | 12.791 | 118.0826 | C6H14S | NA | NA | |
| 3-methyl-1-(methylthio)-butane | 12.821 | 118.0827 | C6H14S | 779 | 872 | 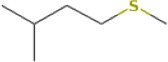 |
| Tetrahydro-2,5-dimethylthiophene | 13.06 | 116.0670 | C6H12S | 657 | 741 | 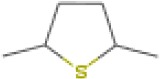 |
| Tetrahydro-2,5-dimethylthiophene | 13.14 | 116.0670 | C6H12S | 622 | 707 | 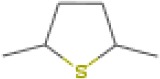 |
Additional files
-
Supplementary file 1
Survival of ostrich eggshell proteins in time.
The proteins identified in each ostrich eggshell sample are reported, together with the number of identified peptides and the percentage coverage, Val D/L value and hydropathicity.
- https://doi.org/10.7554/eLife.17092.020
-
Supplementary file 2
Product ion spectra.
Raw spectra (manually annotated on the basis of PEAKS assignments) of all the identified sequences identified in panel 1. 2–9: Copenhagen dataset; 10–26: York/Oxford dataset.
- https://doi.org/10.7554/eLife.17092.021
-
Supplementary file 3
Diagenesis-induced modifications.
Modifications detected in SCA-1 and SCA-2 sequences in all OES samples analysed.
- https://doi.org/10.7554/eLife.17092.022
-
Supplementary file 4
Full proteomics dataset.
This Excel file reports all the peptide and protein data for ostrich eggshell samples and the blanks.
- https://doi.org/10.7554/eLife.17092.023





