The exon junction complex regulates the splicing of cell polarity gene dlg1 to control Wingless signaling in development
Figures
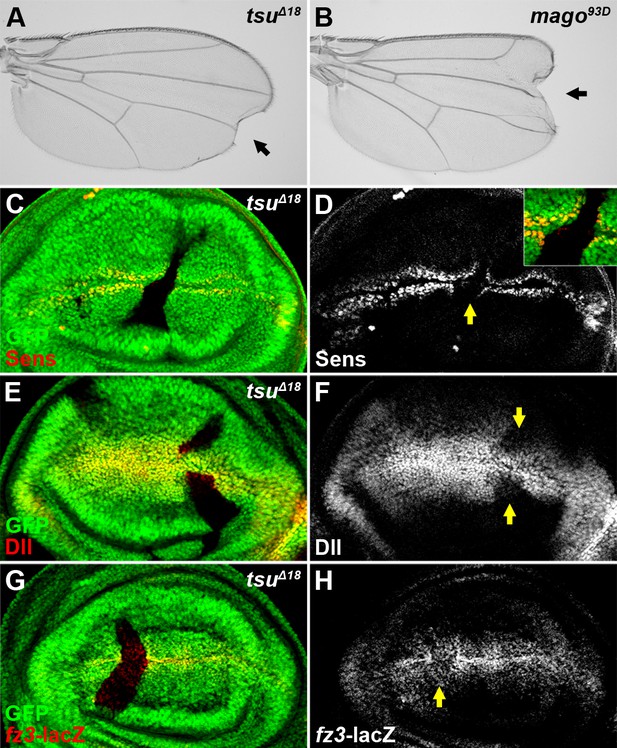
The pre-EJC positively regulates Wg signaling.
(A,B) A typical loss of Wgsignaling wing margin phenotype was observed when tsuΔ18 or mago93D mutant somatic clones were generated in adult wings. Arrows indicate serrated wing margin. (C–H) The production of Wg signaling targets Sens (C,D), Dll (E,F) and fz3-lacZ (G,H) was reduced in tsuΔ18 clones (marked by the absence of GFP and hereafter in subsequent figures). The positions of clones are indicated by arrows.
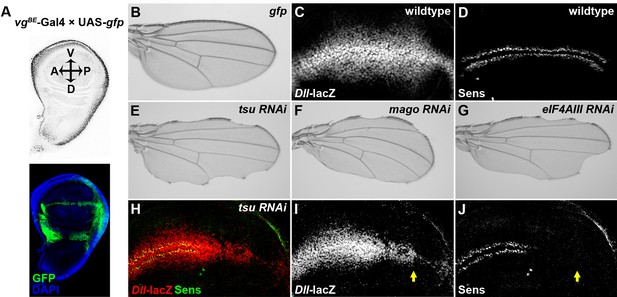
Knocking down individual components of the pre-EJC reduces Wg signaling in the developing wing.
(A) The vgBE-Gal4 driver used in the screen confers expression of RNAi transgenes along the dorsal-ventral (D-V) boundary in the third instar wing imaginal disc as marked by the expression of UAS-gfp transgene (bottom panel). DAPI labeling marks the nuclei. (B) Expression of gfp alone by vgBE-Gal4 did not produce any defect in the adult wing. (C,D) Shown are differential activation of Wg signaling target genes along the D-V boundary in wildtype wing discs. Moderate Wg signaling induces the expression of Dll-lacZ in a broad region across the wing pouch (C). High level of Wg activity results in activation of Sens immediately adjacent to the D-V boundary (D). (E–G) Reduced expression of either component of the pre-EJC, tsu (E), mago (F) or eIF4AIII (G) by RNAi resulted in a typical loss of wing margin phenotype. (H–J) The activity of Dll-lacZ was reduced when tsu RNAi was expressed by hh-Gal4 in the posterior compartment of the wing imaginal disc (I). Similarly, the expression of a high threshold Wg signaling target Sens was abolished (J). Arrows mark the posterior part of the wing disc.
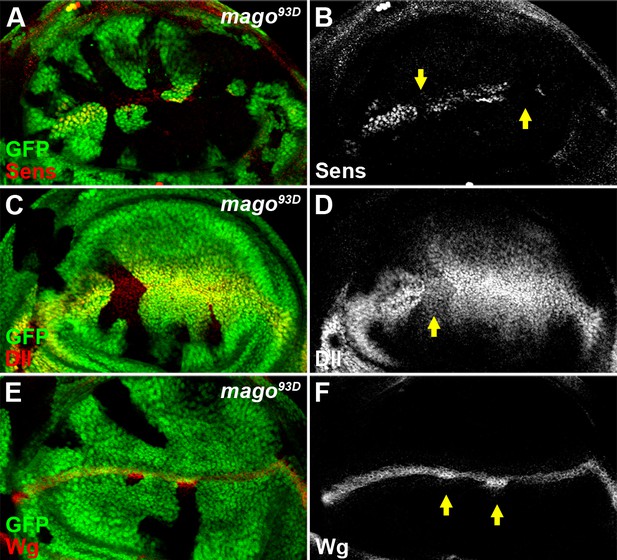
The pre-EJC component Mago positively regulates Wg signaling.
(A–D) The expression of Wg signaling targets Sens (A,B) and Dll (C,D) was reduced in loss of function mago93D somatic clones (marked by the absence of GFP). (E–F) Wg protein stained with the conventional method was accumulated in mago93D clones.
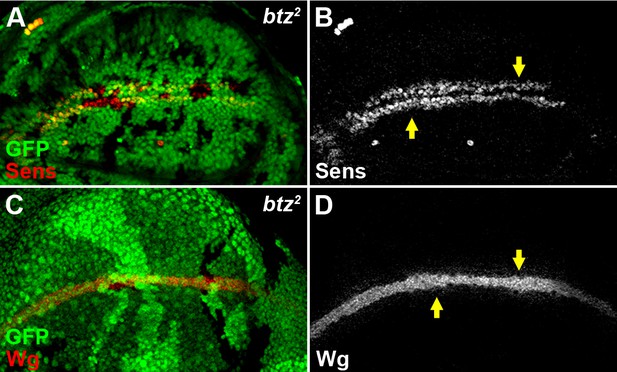
The EJC cytoplasmic component Btz does not regulate Wg signaling.
The expression of Sens (A,B) and the accumulation of Wg (C,D) were not altered in loss-of-function btz2 somatic clones (marked by the absence of GFP).
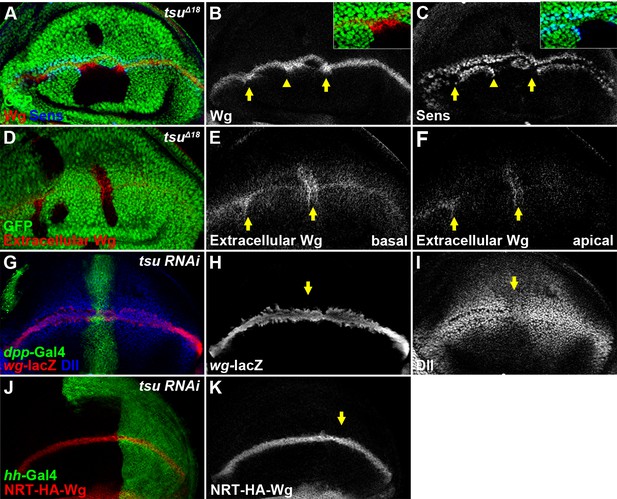
The pre-EJC is required for Wg morphogen reception.
(A–C) Wg protein stained with the conventional method (B) was accumulated in tsuΔ18 clones where the expression of Sens was reduced (arrows). The regions marked by arrowheads are shown in insets. Note that Sens was activated in cells outside the mutant clone (C). (D–F) Extracellular Wg was accumulated at the basal (E) and apical extracellular spaces of the tsuΔ18 clones (F). (G–I) The activity of wg-lacZ (H) did not change when tsu RNAi was expressed by ptc-Gal4 (marked by GFP and arrows). Note that Dll expression was reduced in tsu RNAi-expressing cells (I). (J,K) The expression of plasma membrane bound NRT-HA-Wg did not change when tsu RNAi was expressed by hh-Gal4.
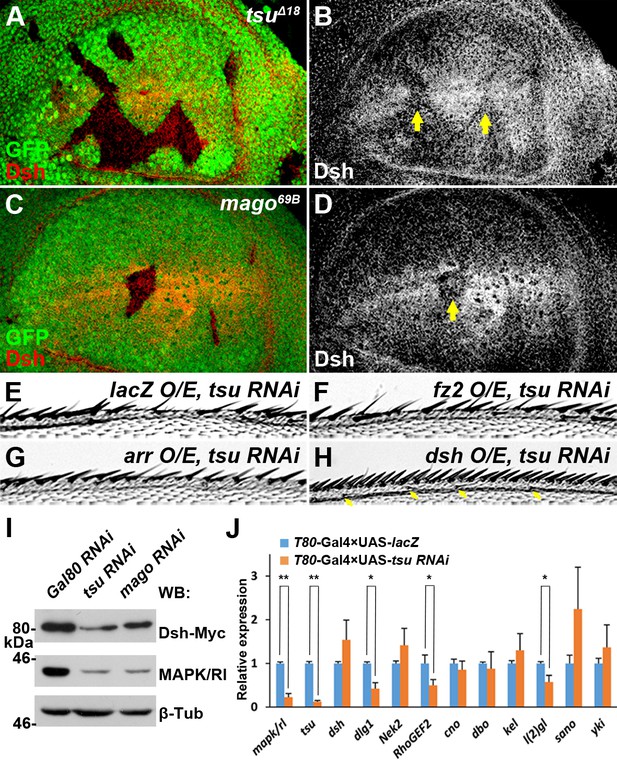
The pre-EJC regulates Wg signaling through Dsh.
(A–D) The amount of Dsh protein was reduced in tsuΔ18 and mago69B clones in the wing disc. (E–H) Overexpressing dsh (H), but not fz2 (F) or arr (G), rescued the loss of Wg signaling wing margin phenotype caused by tsu knockdown (E). Arrows indicate sensory bristles along the wing margin. (I) The production of Myc-tagged Dsh was reduced when tsu or mago dsRNA was expressed in S2 cells. Yeast Gal80 dsRNA served as a negative control for RNAi treatment. MAPK/Rl, a known pre-EJC target served as a positive control for defective pre-EJC. β-Tubulin was used as a loading control for all experiments. (J) Real time RT-PCR revealed that the abundance of dlg1, RhoGEF2 and l(2)gl, but not dsh mRNA, was reduced when tsu dsRNA was expressed in wing discs. mapk/rl served as a positive control. α-Tubulin 84B was used to normalize the amount of cDNA template. The experiments were performed in triplicates, and data were represented as the mean+S.D. (*p<0.05; **p<0.01; Student’s t-test).
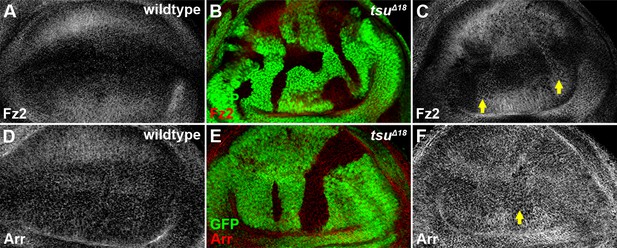
The pre-EJC does not obviously regulate the expression of Fz2 and Arr.
Shown are expression patterns of Fz2 (A) and Arr (D), two co-receptors for Wg morphogen reception, in wildtype wing discs. The expression of Fz2 (B,C) was slightly upregulated in some tsuΔ18 clones, while the expression of Arr (E,F) remained largely unchanged.
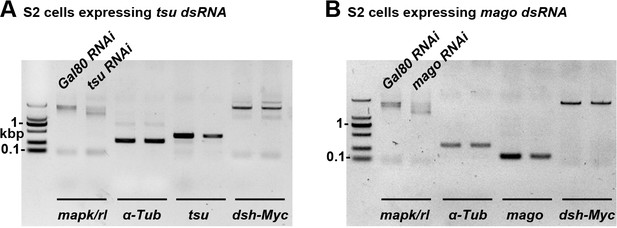
The mRNA abundance of cDNA-derived dsh is not altered when the pre-EJC activity is knocked down in S2 cells.
The mRNA expression of cDNA-derived dsh was not altered when tsu (A) or mago (B) dsRNA was expressed in S2 cells. Yeast Gal80 dsRNA served as a negative control for RNAi treatment. mapk/rl served as a positive control. α-Tubulin 84B served as a loading control.
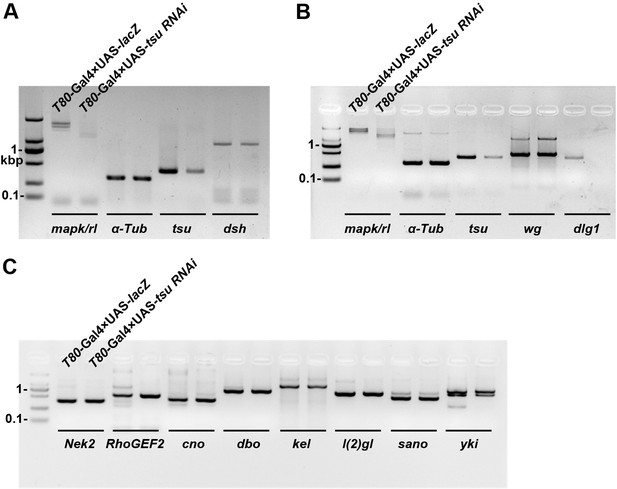
The mRNA abundance of dlg1 but not those encoding other Dsh-interacting proteins is reduced when the pre-EJC is knocked down in wing discs.
The mRNA expression of dlg1 (B) but not dsh (A) nor genes that encode for other Dsh-interacting proteins (C) was obviously reduced when tsu RNAi was expressed by T80-Gal4 in wing discs. mapk/rl served as a positive control. α-Tubulin 84B served as a loading control.
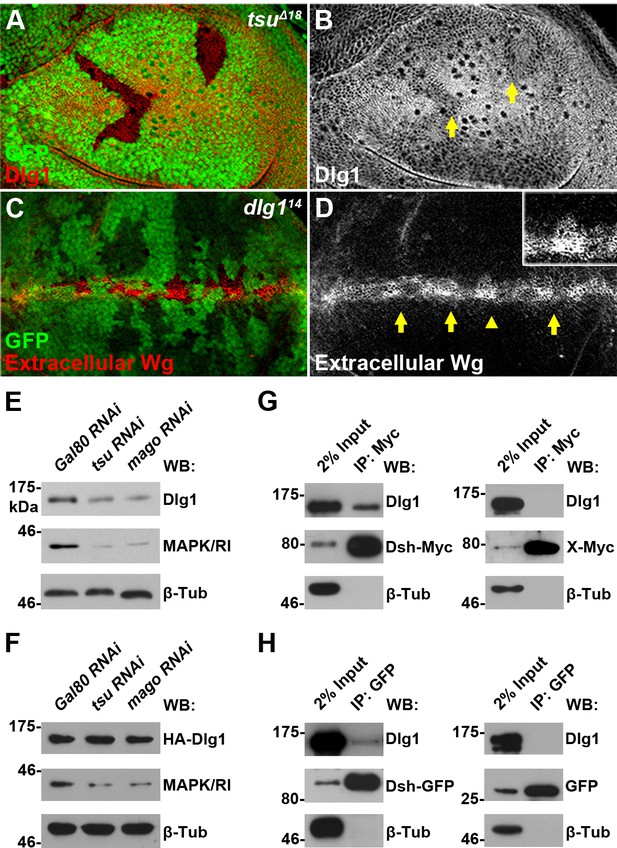
The pre-EJC regulates Dsh-interacting protein Dlg1.
(A,B) The expression of endogenous Dlg1 protein was reduced in tsuΔ18 clones (indicated by an arrow). (C,D) Extracellular Wg was accumulated in dlg114 clones. Shown in the inset is an enlarged clone with accumulated extracellular Wg. (E,F) The amount of endogenous (E) but not a cDNA-derived Dlg1 protein (F) was reduced when tsu or mago dsRNA was expressed in S2 cells. MAPK/Rl served as a positive control for dysfunctional pre-EJC activity. (G,H) Endogenous Dlg1 interacted with Myc-tagged Dsh in S2 cells (G) and GFP-tagged Dsh in wing discs expressing the dsh-gfp under the control of the dsh promoter (H). A Myc-tagged irrelevant protein X or GFP alone served as negative controls, respectively.
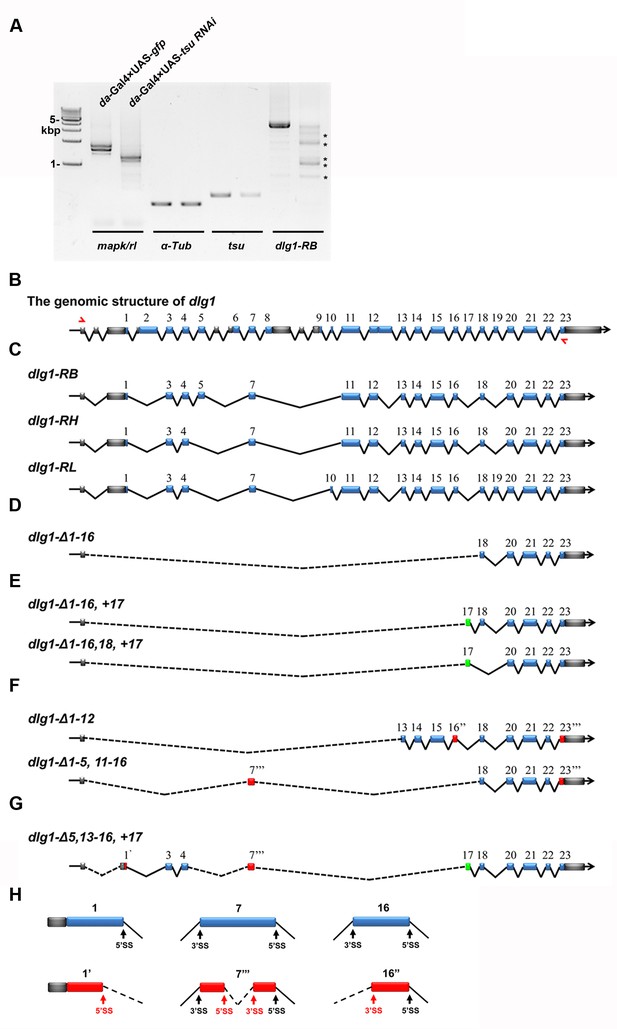
The pre-EJC regulates the precise splicing of dlg1.
(A) Aberrant dlg1 transcripts (indicated by asterisks) were detected by RT-PCR in RNA samples prepared from fly larvae expressing tsu RNAi driven by da-Gal4. (B) Shown is the genomic structure of dlg1 that contains 23 coding exons (blue boxes). The positions of primer pairs used to amplify RB, RH and RL splicing isoforms are indicated by red arrows. (C) Shown are RB, RH, RL splicing isoforms that are amplified by the above indicated primer pair. (D–H) Aberrant isoforms indicated by asterisks in panel A. were excised and purified from the agarose gel, cloned into a pGEM-T vector and subsequently subject to sequencing. After comparing the mRNA sequences deduced from sequencing results with wildype RB, RH and RL isoforms, three classes of aberrant splicing events were detected. The first class represents a previously reported exon skipping event, which was observed in all splicing forms (D–G). The second class represents exon inclusion, which contains the exons that are normally efficiently spliced in wildtype isoforms (green boxes). The third class includes exons that are generated by previously unidentified splice sites (SS; red boxes). These three classes are not mutually exclusive as classes 1 and 2 were found in aberrant isoforms shown in panel E, classes 1 and 3 in panel F, and classes 1–3 in panel G. Δ indicates missing exons. 1’ indicates a 5’ SS in exon 1. 16’’ indicates a 3’SS sites in exon 16. 7’’’ and 23’’’ indicate 5’ and 3’ SS present in the exon 7 and 23, respectively. Dashed lines indicate novel ligation of exons. Details on usage of previously unidentified splice sites are shown (H).
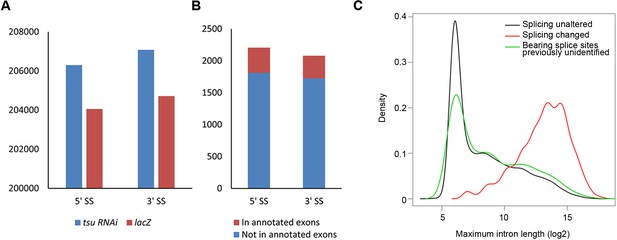
The pre-EJC regulates the alternative splicing.
(A) The previously identified 5’ splice sites (SS) and 3’ SS extracted from reference (BDGP v5.78) were used 206,299 and 207,082 times in the tsu sample, respectively, while those used in the lacZ sample were 204,053 and 204,713 times. Thus, compared with the lacZ sample, the tsu sample has a 1.4% (2246/204,053) higher usage for previously identified 5’SS and a 1.5% (2269/204,713) for 3’SS, respectively. p-value for 5' splice site usage is 0.079, and 0.057 for 3’ splice sites (Student’s t-test). (B) Previously unidentified splice sites were detected in RNA-seq when the pre-EJC was knocked down. In total, 2,207 of 5’ SS and 2,081 of 3’ SS were identified, among which 394 of 5’ SS and 395 of 3’ SS were located in annotated exons in reference (BDGP v5.78), respectively. (C) Distribution plot highlights the correlation between maximum intron length and genes whose splicing were subject to differential regulation by the pre-EJC. Shown are distribution plots of genes whose splicing pattern was neither affected (black line) nor changed (red line) by reduced pre-EJC activity. The genes with altered splicing have an overall larger maximum intron length (average length>1000 nt). This result is consistent with previous reports (Ashton-Beaucage et al., 2010; Roignant and Treisman, 2010). In addition, a new class of genes was identified in which previously unidentified splice sites were utilized for generating novel transcripts (green line). Interestingly, no correlation with overall larger maximum intron length was detected in this new class of genes (green lines), implying an unknown mechanism for the pre-EJC to recognize splice sites.
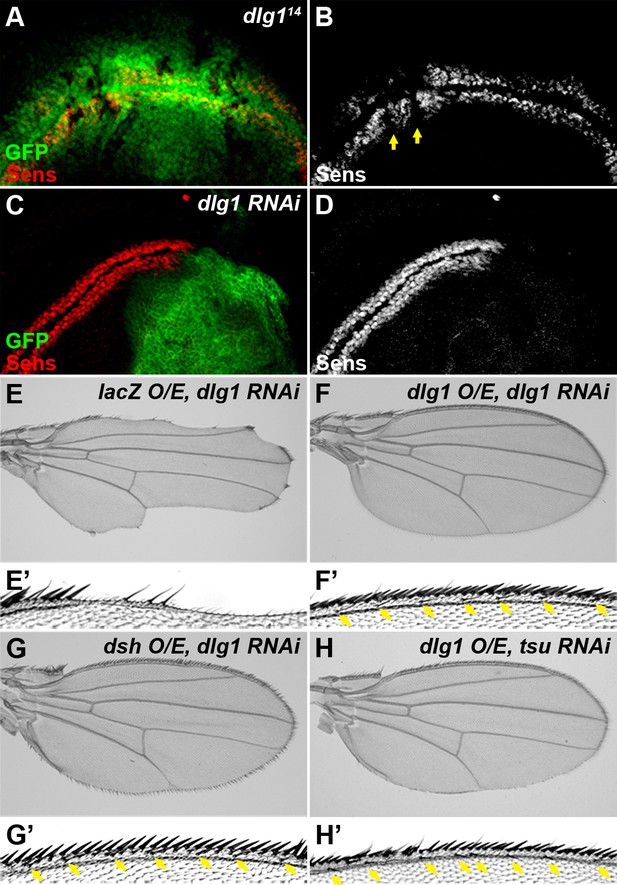
Dlg1 is a positive regulator of Wg signaling.
(A–D) The expression of Sens was reduced in dlg114 mutant clones (A,B, marked by the absence of GFP) or when dlg1 RNAi was expressed by the hh-Gal4 in the posterior compartment of the wing disc (C, D, marked by GFP). The expression pattern of Sens in a wildtype wing disc is shown in Figure 1—figure supplement 1D. (E–H) Overexpressing dlg1 (F,H) or dsh (G) largely rescued the loss of Wg signaling phenotype along the wing margin caused by knockdown of dlg1 (E) or pre-EJC (H, cf. Figure 3E). Enlarged images of the wing margin from panels E–H are shown as E’–H’. Arrows indicate sensory bristles along the wing margin.
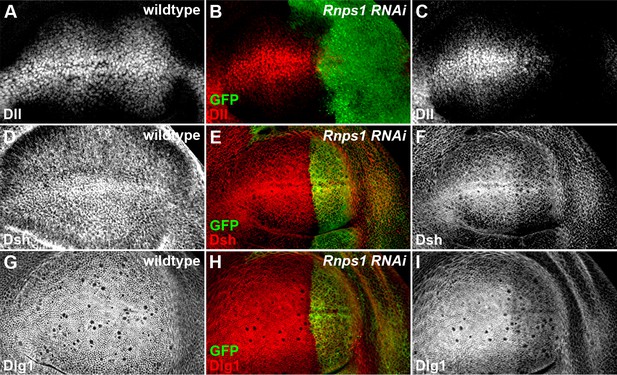
Splicing factor Rnps1 regulates Wg signaling.
(A,D,G) Shown are the expression patterns of Dll (A), Dsh (D) and Dlg1 (G) in wildtype wing discs. (B,C,E,F,H,I) The production of Dll (C), Dsh (F) and Dlg1 (I) protein was reduced when Rnps1 RNAi was expressed by hh-Gal4 in the posterior compartment of the wing disc (marked by GFP).
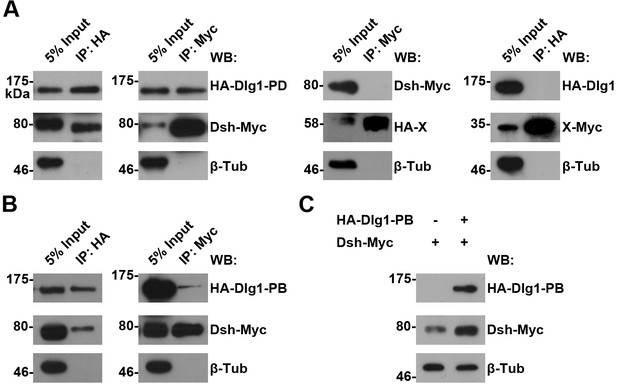
The interaction between Dlg1 and Dsh in S2 cells.
(A,B) The interaction between Dlg1-PD (A) or Dlg1-PB (B) and Dsh was detected in S2 cells transiently transfected with HA-dlg1 and dsh-Myc by co-immunoprecipitation (co-IP). Irrelevant proteins tagged with either HA or Myc served as negative controls for co-IPs, respectively. β-Tubulin served as a loading control. (C) Overexpressing dlg1-RB increased the abundance of Dsh protein in S2 cells.
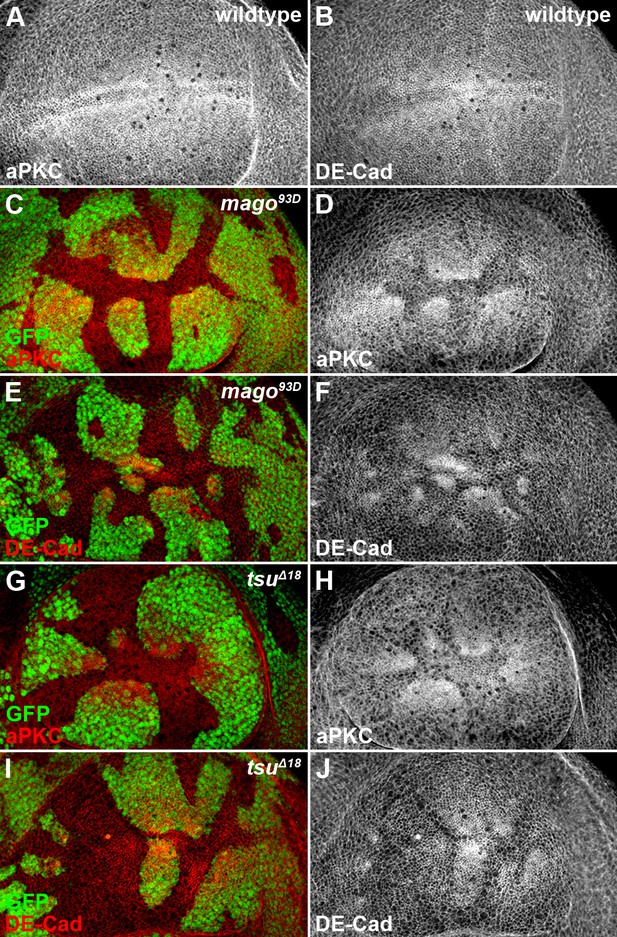
The pre-EJC components Mago and Tsu regulate cell polarity in the wing disc.
(A,B) Shown are the expression patterns of cell polarity proteins aPKC (A) and DE-Cad (B) in wildtype wing discs. (C–J) The expression patterns of aPKC (C,D,G,H) and DE-Cad (E,F,I,J) were altered in loss-of-function mago93D (C–F) or tsuΔ18 (G–J) somatic clones (marked by the absence of GFP).
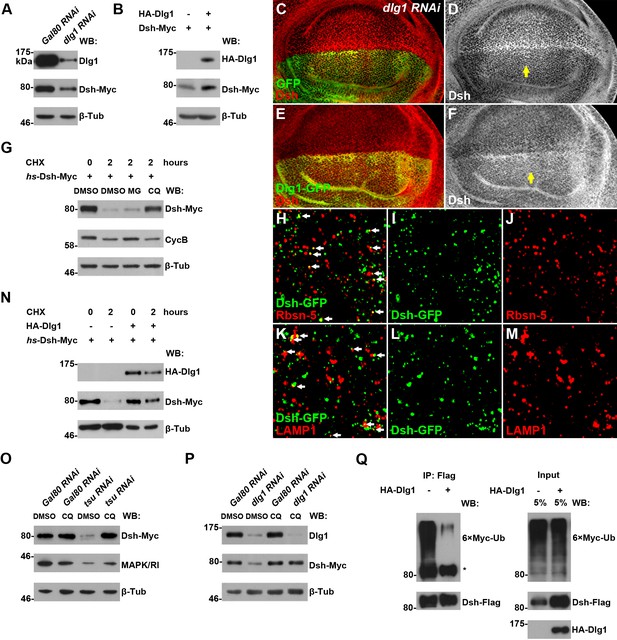
Dlg1 regulates Dsh protein turnover.
(A) Knocking down dlg1 by RNAi in S2 cells reduced the amount of Dsh protein. (B) Overexpressing dlg1 increased the abundance of Dsh protein in S2 cells. Note that Dlg1-PD form was used in all experiments unless mentioned otherwise. (C–F) The amount of Dsh was altered respectively when dlg1 RNAi (D) or dlg1-gfp (F) was expressed in wing discs by ap-Gal4. (G) CHX treatment-induced Dsh protein degradation was inhibited by lysosome inhibitor chloroquine (CQ) but not by proteasome inhibitor MG132 (MG). Cyclin B is known to be degraded in the proteasome, which served as a positive control for MG treatment (Zhang et al., 2014). DMSO served as a negative control. (H–M) Dsh protein was detected in endocytic compartments in wing discs expressing Ubpy RNAi to prevent lysosome function. Over 10% of GFP-tagged Dsh (dsh-gfp under the control of the dsh promoter) colocalized with early endosome protein Rbsn-5 [H–J; n (field of view) = 8] and late endosome/lysosome protein marker LAMP1 in wing discs [K–M; n (field of view) = 10]. (N) CHX treatment-induced Dsh degradation was inhibited when dlg1 was overexpressed in S2 cells. (O,P) Dsh degradation resulted from tsu RNAi (O) or dlg1 RNAi (P) treatment was inhibited by lysosome inhibitor CQ. (Q) Overproduced Dlg1 reduced the level of ubiquitination of Dsh. Asterisk indicated non-specific signal of Myc antibody reactivity.
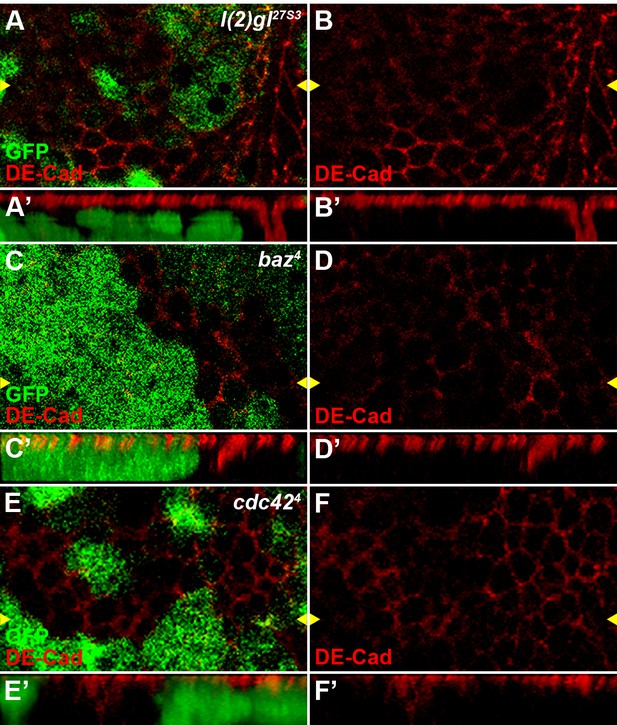
Reduced activity of cell polarity determinants l(2)gl, baz or cdc42 result in polarity defects in wing disc cells.
The localization of adherens junction, marked by DE-cadherin, was obviously changed in l(2)gl27S3 (A–B), baz4 (C–D) or cdc424 (E–F) loss-of-function somatic clones (marked by the absence of GFP) in wing discs. Reconstituted optical cross sections along the Z-axis (indicated by the yellow arrowheads) are shown in A’–F’.
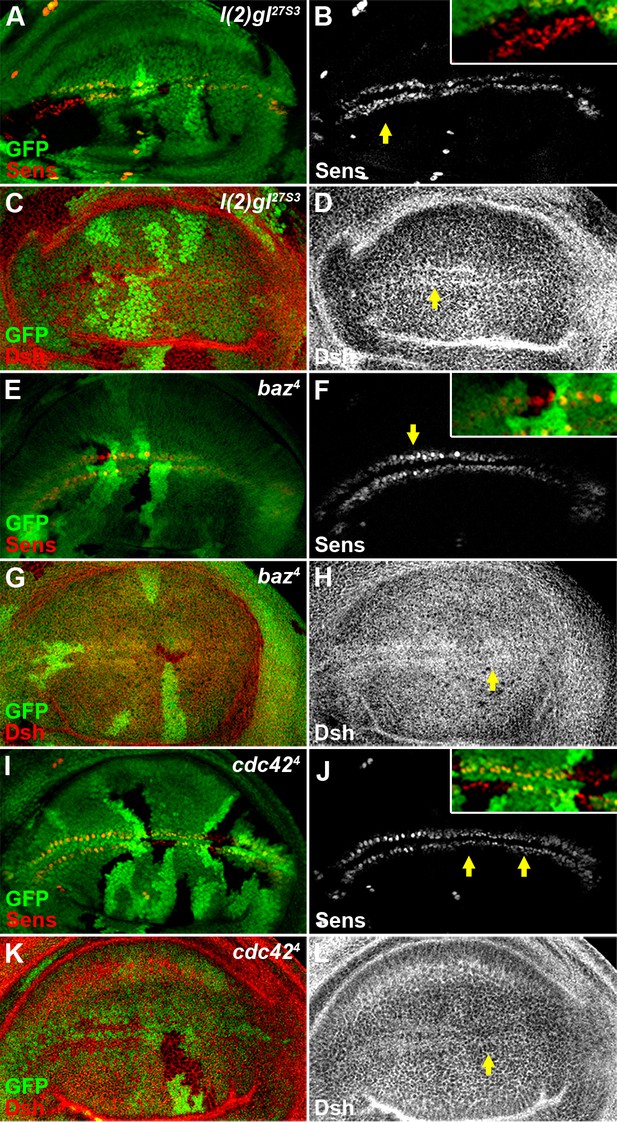
Reduced activity of cell polarity determinants l(2)gl, baz or cdc42 does not result in an obvious Wg signaling defect.
The expression pattern of Sens (B,F,J) or Dsh (D,H,L) was not obviously changed in l(2)gl27S3 (A–D), baz4 (E–H) or cdc424 (I–L) loss-of-function somatic clones (marked by the absence of GFP) induced in wing discs. Shown in insets (B,F,J) are enlarged images of regions marked by arrows.
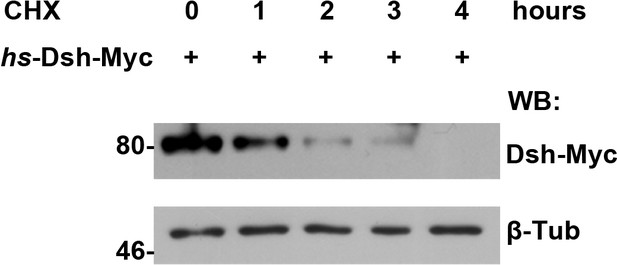
The turnover of fly Dsh protein was measured in S2 cells treated with CHX followed by a four-hour chase.
https://doi.org/10.7554/eLife.17200.022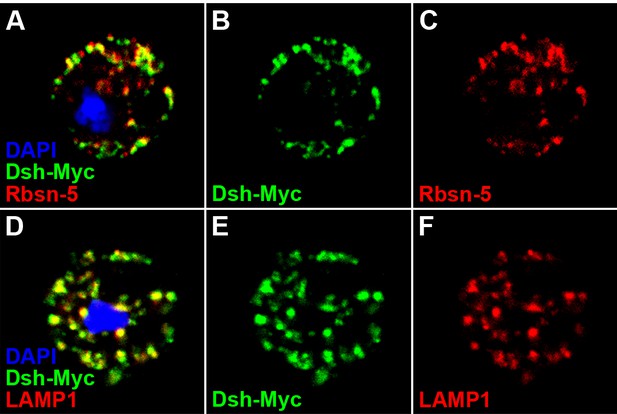
Dsh colocalizes with endosome and lysosome markers in S2 cells.
S2 cells expressing dsh-Myc were treated with CQ for 8 hr to disrupt lysosome function. Over 60% of Dsh-Myc protein colocalized with early endosome protein Rbsn-5 [A–C, n (number of cells) = 10] and late endosome/lysosome marker LAMP1 [D–F, n (number of cells) = 10].
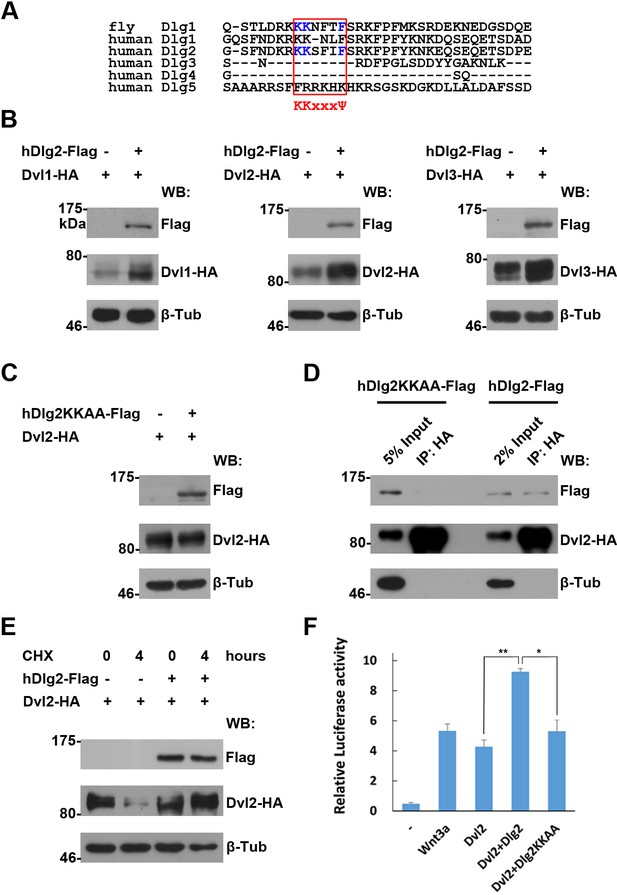
The regulation of Dvl proteins by Dlg is conserved in vertebrates.
(A) Sequence alignment of the I3-insert of Hook domains presented in Dlg proteins reveals sequence conservation between fly Dlg1 and human Dlg orthologs. The K-K-x-x-x-Ψ motif is highlighted in a red box. Amino acids essential for Dsh interaction are shown in blue. (B,C) Overproduced wildtype (B), but not the KKAA mutant form of hDlg2 (C), increased the amount of Dvl proteins in HEK293T cells. (D) Dvl protein interacted with wildtype, but not the KKAA mutant form of hDlg2. (E) CHX treatment-induced Dvl protein degradation was prevented by overproduced hDlg2. (F) Overexpressing wildtype, but not the KKAA mutant form of hDlg2, further increased Dvl2-induced TOPFlash Wnt signaling reporter activity. Ectopic Wnt3a served as a positive control. Experiments were repeated thrice, and data were represented as the mean+S.D. after normalized to Renilla activity (**p<0.01; *p<0.05; Student’s t-test).
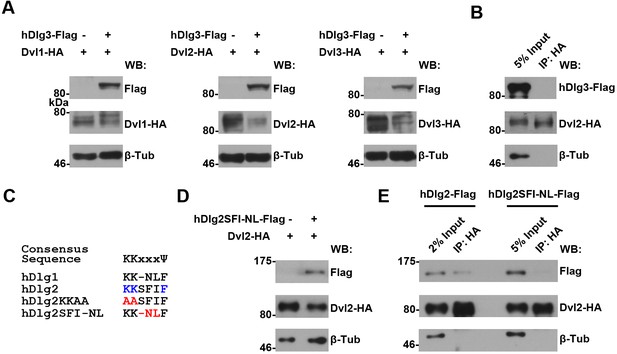
An intact K-K-x-x-x-Ψ motif is required for human Dlg proteins to stabilize Dvl2 in HEK293T cells.
(A) Overexpressing hdlg3 in HEK293T cells did not increase the amount of hDvl2 proteins. (B) No interaction between hDlg3 and hDvl2 was detected by co-IP in HEK293T cells. (C) Shown is the alignment of the K-K-x-x-x-Ψ motives in human Dlg 1 and 2. Two hDlg2 mutant forms, KKAA and SFI-NL, are also compared. (D) Overexpressing the SFI-NL mutant form of hdlg2 (B) did not result in increased hDvl2 production. (E) The SFI-NL mutant form of hDlg2 displayed reduced interaction with hDvl2 in HEK293T cells when compared with that of wildtype hDlg2.
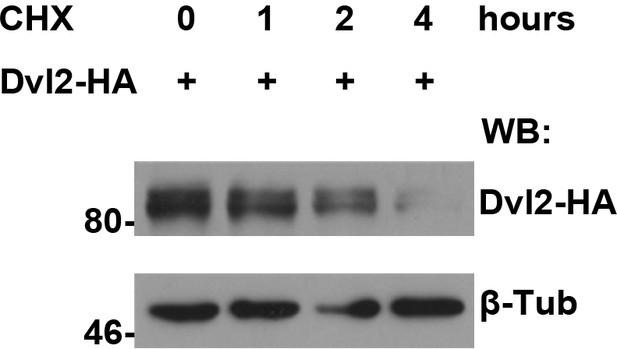
Protein turnover of human Dvl2 protein was measured in HEK293T cells treated with CHX followed by a four-hour chase.
https://doi.org/10.7554/eLife.17200.026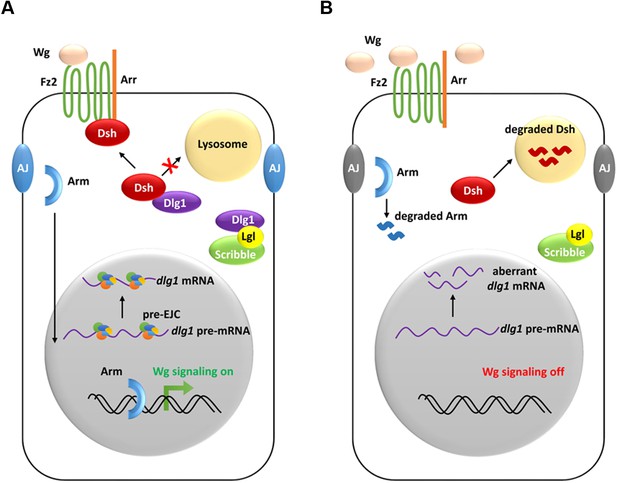
A model showing how the pre-EJC functions in the Wg signaling.
(A) In wildtype wing disc epithelial cells, the pre-EJC activity is required for precise splicing of target transcripts, including dlg1. Dlg1 protein functions independently from its role in cell polarity determination to prevent Dsh protein from degradation in the lysosome. Stabilized Dsh works together with Fz2 and Arr, two Wg co-receptors, to facilitate Wg signal reception. Consequently, through a series of cytoplasmic events, Arm protein becomes stabilized, which translocates to the nucleus to activate Wg target gene transcription. (B) When the pre-EJC activity was defective in wing disc epithelium, precise splicing of dlg1 gene is not maintained. Decreased Dlg1 production is not sufficient to stabilize Dsh, resulting in reduced Wg morphogen reception. As a result, Wg is accumulated in the extracellular space of the epithelium, which ultimately fails to protect Arm from degradation, hence reduced Wg signaling.
Additional files
-
Supplementary file 1
Fly genetics information and Primers.
- https://doi.org/10.7554/eLife.17200.028
-
Supplementary file 2
RNA-seq analyses identify candidate genes whose mRNA expression are subject to the pre-EJC regulation.
Two criteria were used to determine candidate genes that are subject to the regulation by the pre-EJC: 1) the mRNA abundance values (measured by FPKM method) are above 10 in either tsu or lacZ samples (except for the value of sens that is listed at the bottom of the table), and 2) the expression levels are changed by more than 25% in the tsu sample. In total, 1447 genes were identified, in which the expression of 818 genes was downregulated.
- https://doi.org/10.7554/eLife.17200.029
-
Supplementary file 3
Bioinformatic analyses deduce novel transcripts from the pre-EJC regulated candidate genes.
408 novel transcripts in 227 genes were identified by Cufflinks (v2.2.1).
- https://doi.org/10.7554/eLife.17200.030





