Presence versus absence of CYP734A50 underlies the style-length dimorphism in primroses
Figures
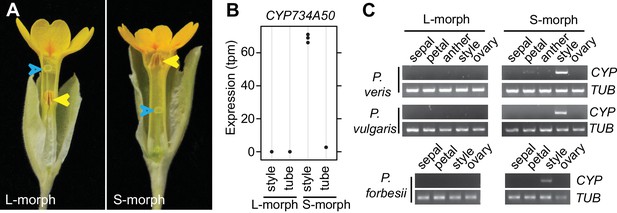
Identification of CYP734A50 as an S-morph and style-specific gene.
(A) Dissected P. veris L- (left) and S-morph flowers (right), showing reciprocal differences in style length and anther position (yellow arrowheads). Blue arrowheads indicate stigmas on top of styles. (B) Expression levels of CYP734A50 in P. veris based on RNA-seq (n = 3 biological replicates each). See also Figure 1—source data 1. (C) Expression of CYP734A50 and TUBULIN in dissected floral organs. Full gel images and qRT-PCR quantification based on three biological replicates are shown in Figure 1—figure supplement 2 and 3.
-
Figure 1—source data 1
Candidates for S-morph style-specific genes.
TPM values and annotations are indicated for candidate transcripts showing significantly (p<0.05) higher expression in S-morph styles than in S-morph corolla tubes and in either L-morph styles or corolla tubes.
- https://doi.org/10.7554/eLife.17956.004
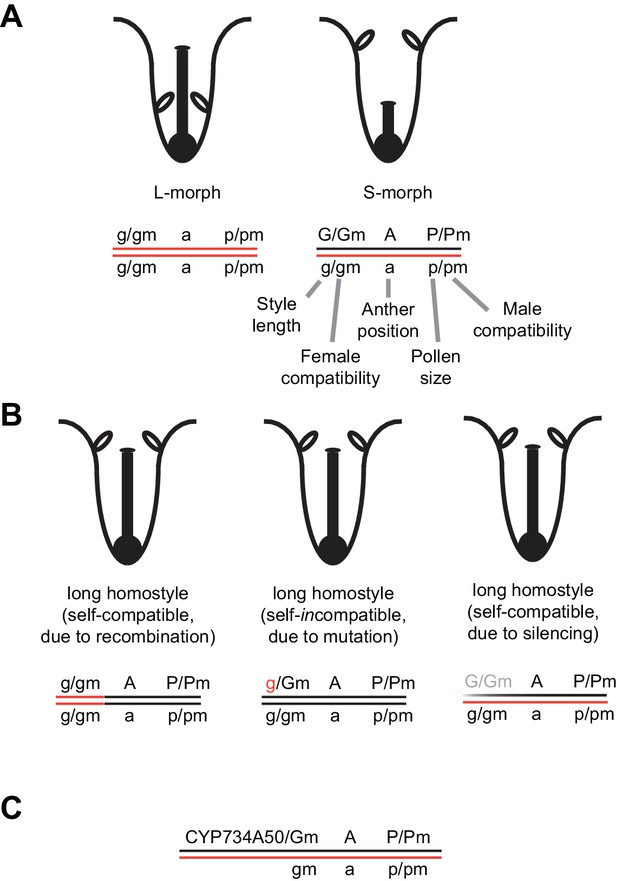
Schematic of the genetic control of heterostyly in Primula.
(A) Schematic representation of L-morph (left) and S-morph flowers (right) and the classic model of corresponding S-locus genotypes, with the functions of individual genes indicated. Open ellipses represent anthers. The order of G, A and P at the S-locus is that suggested by Kurian and Richards (1997) for Primula sect. Primula; in Primula subgenus Auriculastrum the order appears to be GPA (Lewis and Jones, 1992). Also note that the positions of Gm and Pm relative to G and to P are unknown, respectively (indicated by a slash). (B) Schematic representation of three types of long homostyles and their S-locus genotypes arising from recombination within the S-locus (left), from mutation of the dominant G allele (middle), and from silencing the G and Gm genes (right). (C) Proposed model of the S-locus indicating the identity of CYP734A50 to the dominant G allele and its absence from the recessive haplotype.
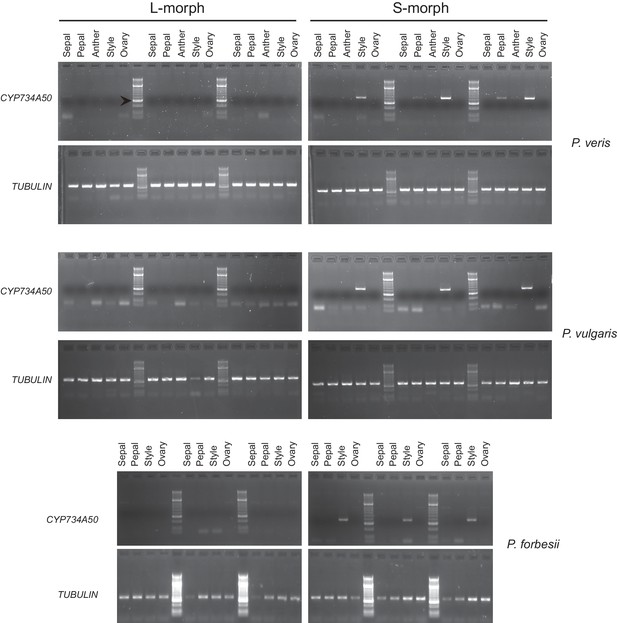
Style-specific expression of CYP734A50.
Expression of CYP734A50 and TUBULIN in dissected floral organs of L- and S-morph plants of the indicated species as determined by RT-PCR. Three biological replicate sets of dissected flowers were analysed. Arrowhead marks 300 bp band in DNA-size standard.
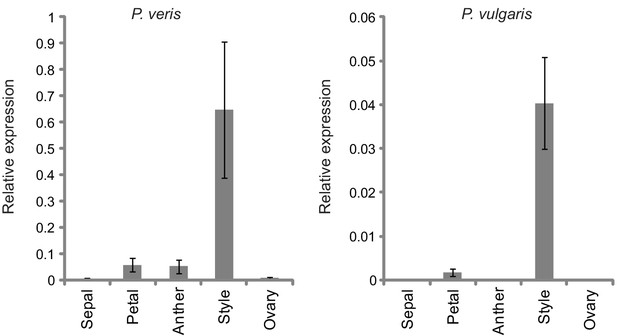
Quantification of style-specific expression of CYP734A50.
Expression of CYP734A50 in dissected floral organs of L- and S-morph plants of P. veris (left) and P. vulgaris (right) as determined by qRT-PCR, normalized to α-TUBULIN. Values are mean ± SD from three biological replicates.
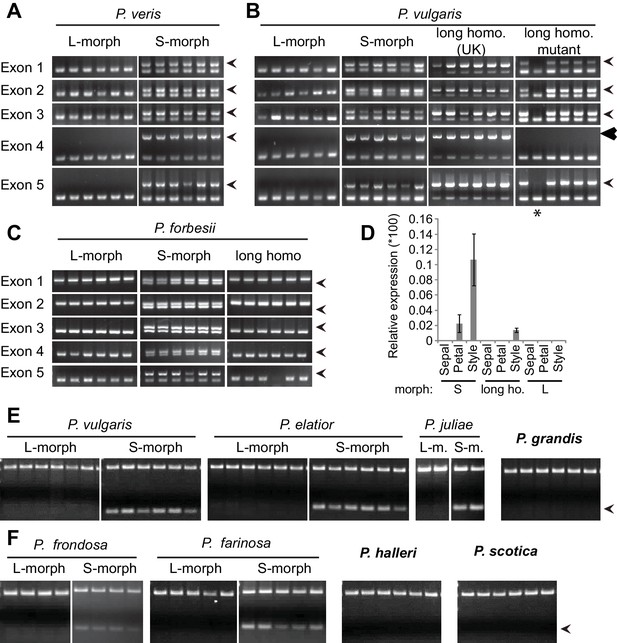
Co-segregation of CYP734A50 with the style phenotype.
(A–C) PCR-genotyping of individuals with the indicated phenotypes of P. veris (A), P. vulgaris (B) and P. forbesii (C). ‘Long homo (UK)’ are naturally occurring long homostyles from England; ‘long homo mutant’ are long homostyles from the commercially grown population. ‘Long homo’ P. forbesii are long-homostylous putative recombinants found in our experimental population. Multiplex PCR amplified CYP734A50 exons (arrowheads) and ITS as an internal control. Enlarged arrowhead marks absence of exon 4 in the mutant. Asterisk in (B) marks a segregating L-morph individual. (D) Expression of CYP734A50 in dissected floral organs of naturally occurring long homostyles, S- and L-morphs of P. vulgaris, normalized to α-TUBULIN. Values are mean ± SD from three biological replicates. (E,F) PCR-genotyping of individuals with the indicated phenotypes from species in Primula sect. Primula (E) and Primula sect. Aleuritia (F). Long homostylous species are indicated in bold. Multiplex PCR was performed using primers to amplify exon 3 of CYP734A50 (arrowhead) and ITS as an internal control. Full gel images are shown in Figure 2—figure supplement 2. Specificity of PCR amplification for CYP734A50 is shown in Figure 2—figure supplement 3. Each set of plants was genotyped at least twice independently.
-
Figure 2—source data 1
Read counts from Illumina whole-genome sequencing of S- and L-morph plants of P. vulgaris and P. forbesii.
Reads were mapped to the exons of CYP734A50 and CYP734A51 including the 200 surrounding intronic nucleotides (± 100 bp). Absolute read counts are given. Due to the very similar target lengths for mapping against both genes, absolute read counts can be directly compared. All P. vulgaris samples are from the SRA project deposited under http://www.ncbi.nlm.nih.gov/bioproject/PRJEB9683.
- https://doi.org/10.7554/eLife.17956.009
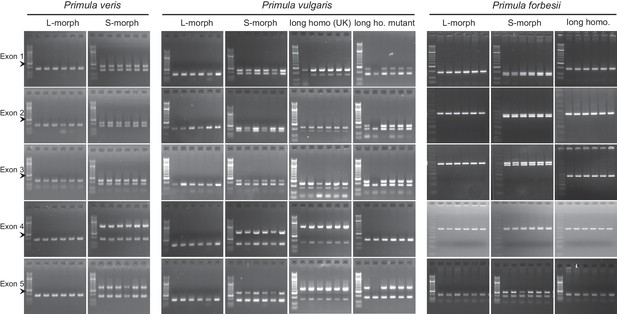
Co-segregation of CYP734A50 with the style phenotype.
PCR-genotyping of individuals with the indicated phenotypes of P. veris (left), P. vulgaris (middle) and P. forbesii (right). Multiplex PCR amplified CYP734A50 and ITS as an internal control. Arrowhead marks 300 bp band in DNA-size standard.
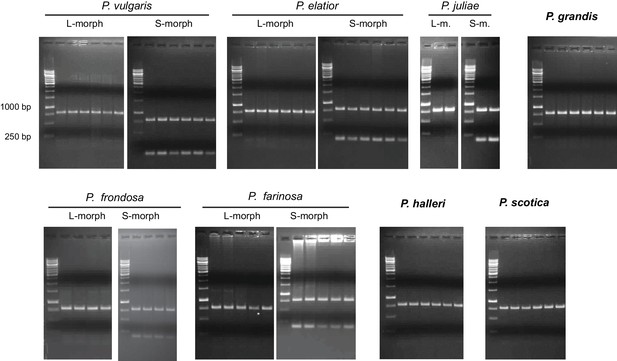
Loss of CYP734A50 from natural long homostylous species.
PCR-genotyping of individuals with the indicated phenotypes from species in Primula sect. Primula (top) and Primula sect. Aleuritia (bottom). Long homostylous species are indicated in bold. Multiplex PCR was performed using primers to amplify exon 3 of CYP734A50 and ITS as an internal control.
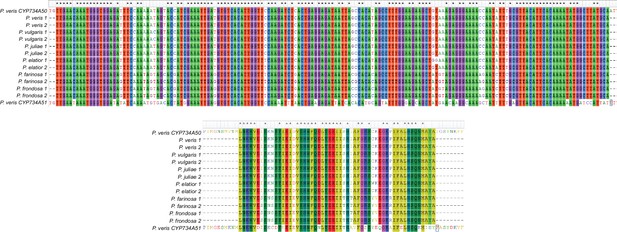
Specificity of PCR-genotyping in different Primula species.
Multiple-sequence alignments at the DNA (top) or protein (bottom) levels of the CYP734A50-specific fragments (see Figure 2E,F) amplified from the indicated species. PCR-products from two S-morph individuals were sequenced from each species. The corresponding region of CYP734A51 is included for comparison to show specificity of amplification of the CYP734A50 orthologues.
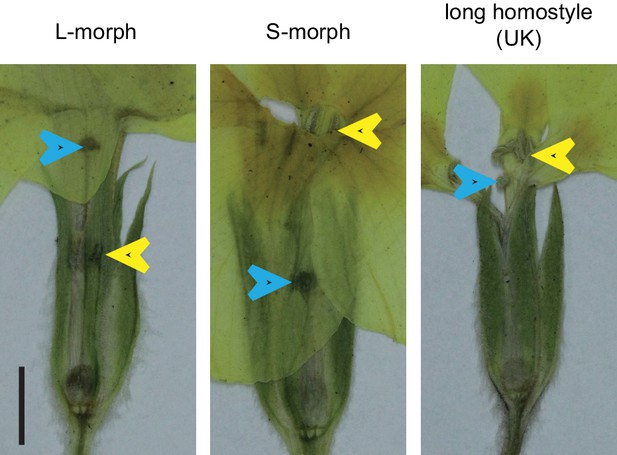
Naturally occurring long homostyles of P. vulgaris.
Phenotypes of naturally occurring long homostyles, L- and S-morph plants of P. vulgaris collected near Wanstrow (UK). Flowers were pressed before photographing. Blue arrowheads indicate the position of the stigma, yellow arrowheads indicate the position of anthers. Scale bar is 5 mm.
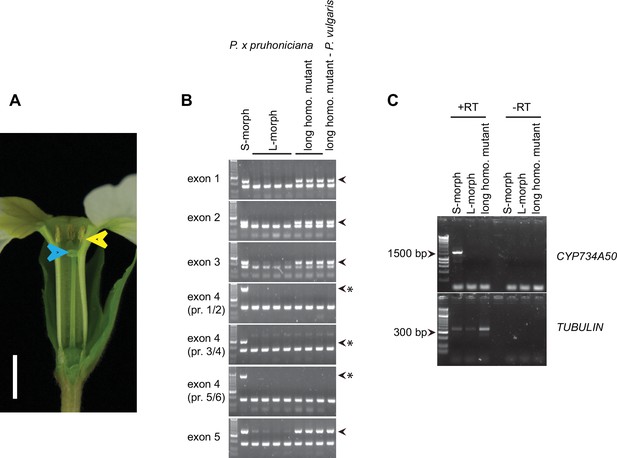
Characterization of long-homostylous mutants of P. vulgaris and P. x pruhoniciana.
(A) Phenotype of long homostyle mutant of P. x pruhoniciana. Blue arrowheads indicate the position of the stigma, yellow arrowheads indicate the position of anthers. Scale bar is 5 mm. (B) PCR-genotyping of S- and L-morph P. x pruhoniciana individuals, three long homostyle mutant P. x pruhoniciana plants and one long homostyle mutant P. vulgaris individual. Multiplex PCR was performed using primers to amplify the indicated CYP734A50 exons and ITS as an internal control. Arrowheads mark CYP734A50 specific bands; asterisks indicate exon-4 deletion in the mutants. Three different primer pairs were used to test for the presence of exon 4. Each genotyping PCR was performed twice independently. (C) Expression of full-length CYP734A50 transcript and of TUBULIN control in S- and L-morph plants, as well as the long homostyle exon-4 deletion mutant from the commercially grown P. vulgaris population. Fragment lengths of DNA-size standard are indicated. RT: reverse transcription.
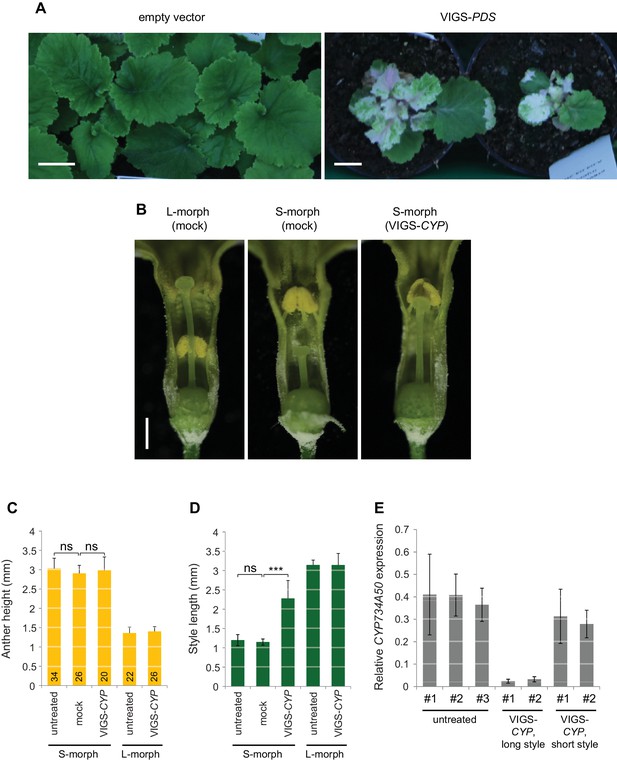
Virus-induced gene silencing in P. forbesii.
(A) Phenotypes of P. forbesii seedlings injected with empty TRV-vectors as negative control (left) and with PDS-containing TRV vectors (right) showing bleaching due to silencing of phytoene desaturase (PDS) expression. No phenotypic alteration is seen in the negative control seedlings compared to untreated plants. Scale bar is 1 cm. (B) Flower phenotypes from the indicated treatments. Mock indicates injection of the inflorescence stems with water. (C,D) Anther height (C) and style length (D) of plants from the indicated treatments. Numbers of flowers for stamen and style-length measurements are indicated. Values are mean ± SD. Means of indicated samples were compared using a two-sided t-test assuming unequal variance, followed by Bonferroni correction. ***p<0.001. (E) Expression of CYP734A50 in dissected styles of plants from the indicated treatments and phenotypes as determined by qRT-PCR, normalized to α-TUBULIN. Each sample (#1, #2, etc) is derived from pooling styles of one individual plant. Values are mean ± SD from three technical replicates per sample.
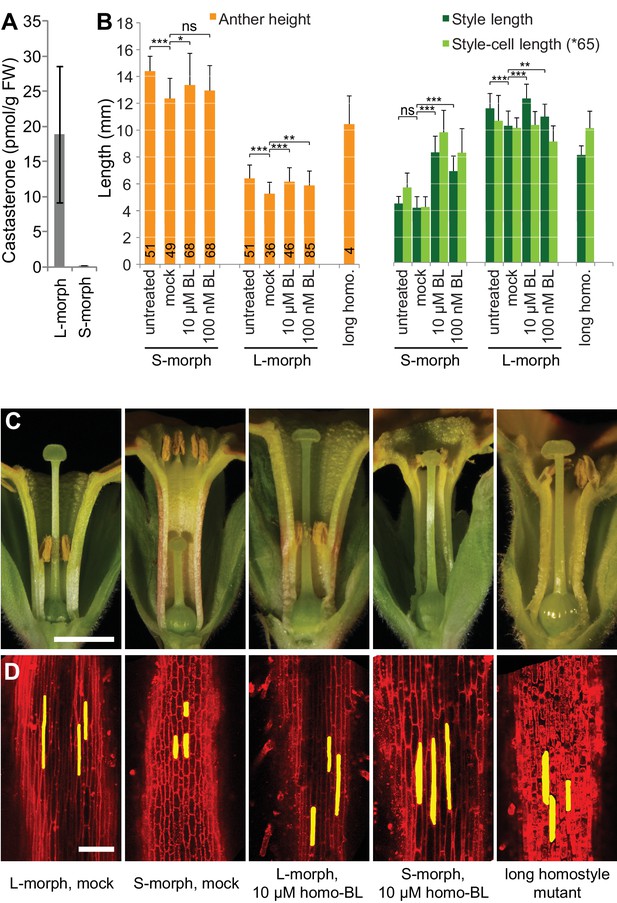
Endogenous brassinosteroid content in styles and response to exogenous brassinosteroid treatment in P. vulgaris.
(A) Castasterone levels in styles of P. vulgaris. Values are mean ± SD from four biological replicates. (B) Anther height, style and style-cell lengths of plants with the indicated phenotypes after brassinolide (BL) treatment. Numbers of flowers used for stamen and style-length measurements are indicated in the bars. Six cells from 10 styles each were measured, except for long homostyle mutant (four styles). Values are mean ± SD. Samples were compared using a two-sided t-test assuming unequal variance, followed by Bonferroni correction. *p<0.05; **p<0.01; ***p<0.001. (C,D) Flower phenotypes (C) and style cells (D) from the indicated treatments. Representative cells are marked in yellow. Scale bars: 5 mm (C), 100 μm (D). A replicate experiment using P. forbesii is shown in Figure 3—figure supplement 1.
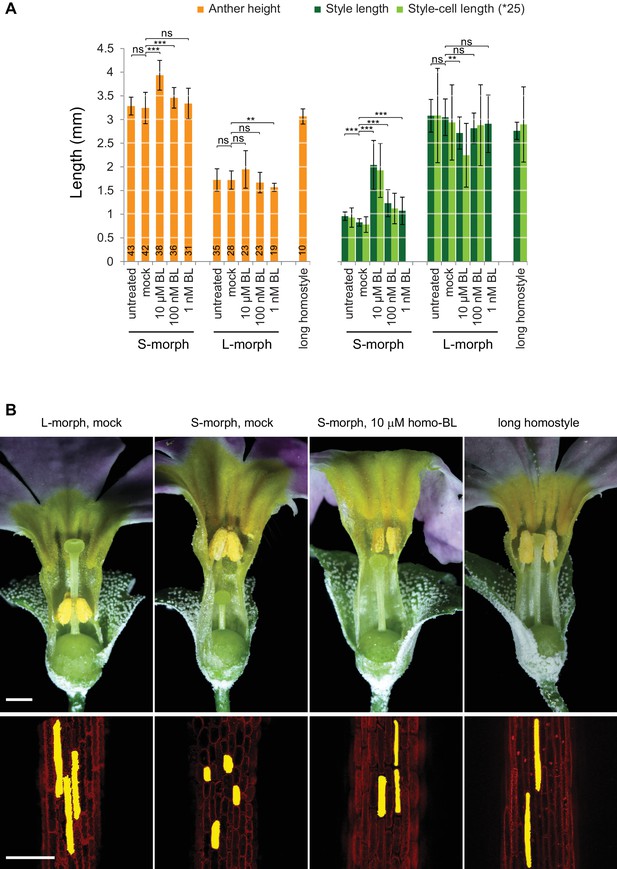
Response of P. forbesii flowers to exogenous brassinosteroid treatment.
(A) Anther height, style and style-cell lengths of plants with the indicated phenotypes after brassinolide (BL) treatment. Numbers of flowers for stamen and style-length measurements are indicated in the bars. Six cells from at least 10 styles each were measured. Values are mean ± SD. Means of indicated samples were compared using a two-sided t-test assuming unequal variance, followed by Bonferroni correction. *p<0.05; **p<0.01; ***p<0.001. (B) Flower phenotypes (top) and style cells (bottom) from the indicated treatments. Representative cells are filled for clarity. Scale bars are 1 mm (top) and 100 μm (bottom).
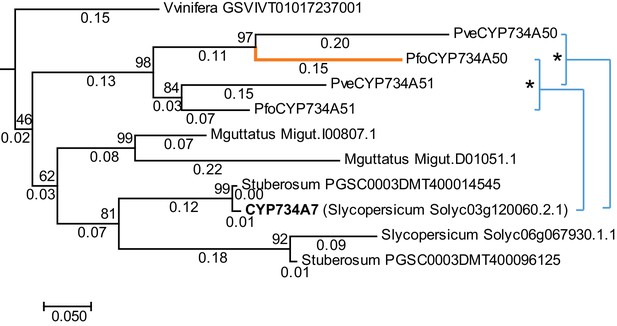
Phylogenetic analysis of CYP734A-like proteins.
Molecular phylogeny of CYP734A50-related proteins based on Neighbour Joining. The tree is drawn to scale, with branch lengths (below branches) measured as number of substitutions per site. Numbers next to nodes indicate percent support from bootstrap analysis (1000 replicates). Cyan brackets with asterisks show significant differences in evolutionary rates compared to CYP734A7 (bold) as outgroup as assessed by Tajima’s relative rate test (p<0.01). CYP734A7 causes BR-deficient phenotypes when overexpressed (Ohnishi et al., 2006). The orange branch shows evidence for relaxed constraint, but not for positive selection based on branch-site tests 1 and 2. The full tree is shown in Figure 4—figure supplement 1. Full results of branch-site tests 1 and 2 are shown in Figure 4—source data 2.
-
Figure 4—source data 1
Comparison of synonymous and non-synonymous substitution rates between CYP734A50 and CYP734A51 genes.
Results of Ka and Ks calculations for CYP734A50 and CYP734A51 genes using the Nei-Gojobori method with complete deletion of missing data. Ka and Ks rates were compared using Fisher’s exact test.
- https://doi.org/10.7554/eLife.17956.019
-
Figure 4—source data 2
Full results of branch-site tests 1 and 2 for relaxed constraints or positive selection.
(A) Subtree as in Figure 4 with numbered branches for which branch-site tests were performed. The orange branch shows evidence of relaxed selection; the green branch shows evidence of positive selection. (B) Results of branch-site tests 1 and 2 for the indicated branches labelled in (A).
- https://doi.org/10.7554/eLife.17956.020
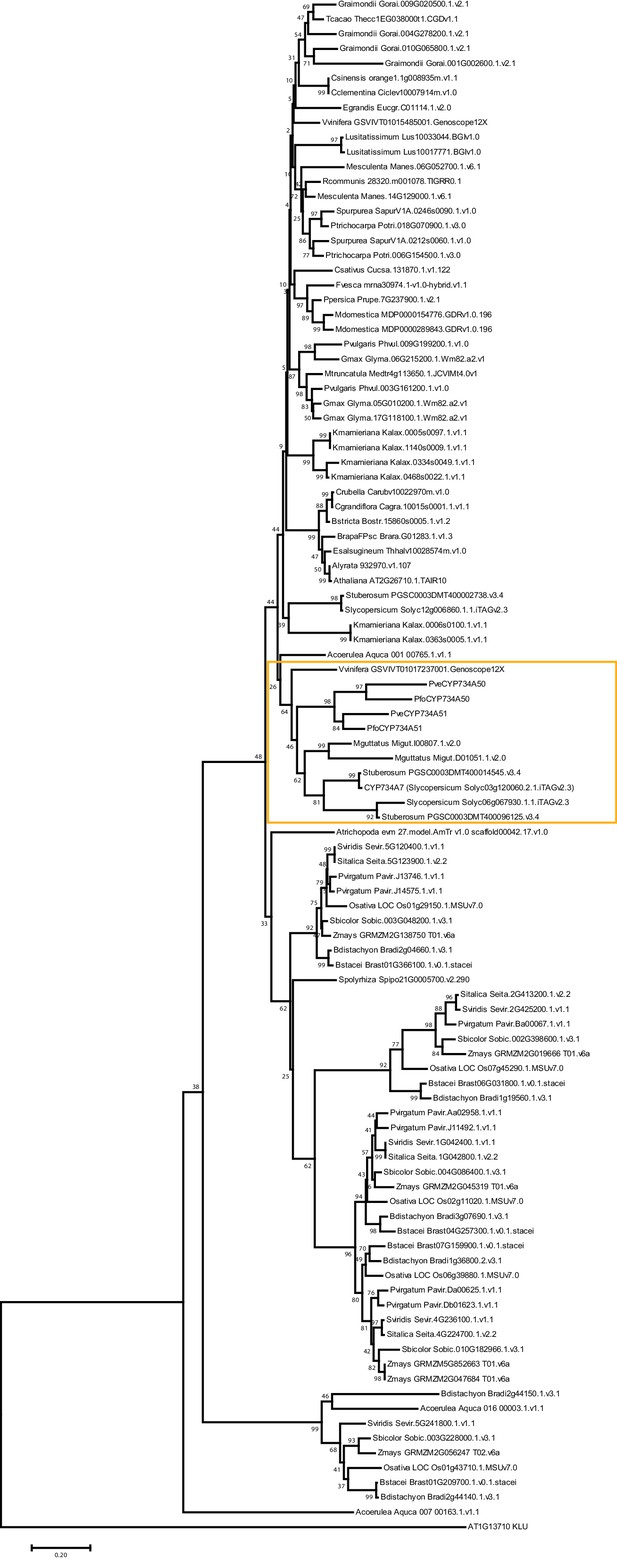
Phylogenetic analysis of CYP734A-like proteins.
Molecular phylogeny of CYP734A50-related proteins based on Neighbour Joining. The tree is drawn to scale, with branch lengths proportional to number of substitutions per site. Numbers next to nodes indicate percent support from bootstrap analysis (1000 replicates). A. thaliana KLUH/CYP78A5 was used as outgroup. The subtree shown in Figure 4 is indicated by the orange box.
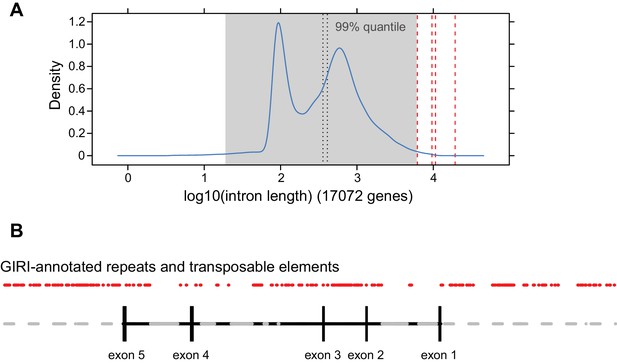
Analysis of CYP734A50 and CYP734A51 introns.
(A) Density distribution of intron lengths in the P. veris genome. The four introns of the CYP734A50 locus are indicated by dashed red lines, as is the 99% quantile. CYP734A50 introns range from 6.2 to 19.3 kb, while the mean and median of P. veris intron sizes are 668 and 410 bp, respectively. This difference is significant at p<10–4 based on Wilcoxon rank sum test. (B) Annotation of transposable elements and repeats in CYP734A50 introns from P. veris according to GIRI. Grey areas in introns indicate stretches of unresolved sequences.
Additional files
-
Supplementary file 1
Oligonucleotide sequences used.
- https://doi.org/10.7554/eLife.17956.023
-
Supplementary file 2
Coding sequences of CYP734A family members from sequenced genomes in fasta format.
- https://doi.org/10.7554/eLife.17956.024
-
Supplementary file 3
Multiple-sequence alignment of coding sequences of CYP734A family members in MEGA format.
- https://doi.org/10.7554/eLife.17956.025
-
Supplementary file 4
Protein sequences of CYP734A family members from sequenced genomes in fasta format.
- https://doi.org/10.7554/eLife.17956.026
-
Supplementary file 5
Multiple-sequence alignment of protein sequences of CYP734A family members in MEGA format.
- https://doi.org/10.7554/eLife.17956.027





