Cyclin Kinase-independent role of p21CDKN1A in the promotion of nascent DNA elongation in unstressed cells
Figures
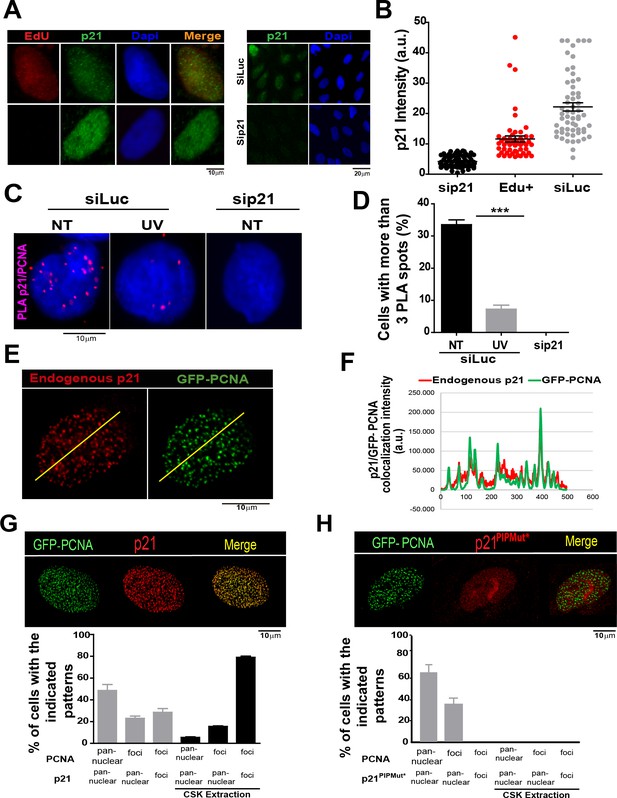
The PCNA interacting region of p21 facilitates the recruitment of p21 to replication factories in cycling cells.
(A) Representative images of p21 in EdU positive and negative cells (left panel) and from U2OS cells transfected with control siRNA (siLuc) or sip21 (right panel). (B) p21 intensity in the indicated samples. Nuclei were counterstained with DAPI. 70 nuclei/sample; two independent experiments were performed. (C) Representative images from Proximity ligation assay (PLA) performed after mild extraction on the indicated samples. (D) Quantification of PLA experiments described in C. 100 nuclei/sample; two independent experiments were performed. (E) Colocalization of p21 foci with GFP-PCNA which reveal replication factories (mild extraction was applied). (F) Profiles of signal intensity along an arbitrary line (showed in E) drawn across the nuclei. (G and H) Samples were transfected with the indicated p21 mutants. Representative images are shown. Samples treated or not with CSK buffer were classified into the indicated categories.100 nuclei/ sample; two independent experiments were analysed. For all figures in this manuscript: significance of the differences are: *p<0.1; **p<0.01; ***p<0.001. When the difference is not statistically significant, the p value is not shown. Error bars represent SEM (standard error of the mean).
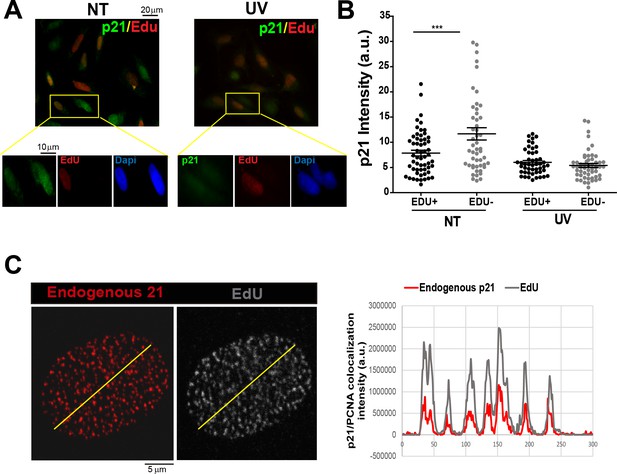
Endogenous p21 localizes to replication factories.
(A) U2OS cells were not treated (NT) or UV irradiated with 20 J/m2 and 2 hr later EdU was incorporated for 10 min. EdU and p21 were revealed by Click-IT technology and specific antibodies respectively. Representative panels are showed. Zoomed images correspond to the indicated yellow boxes. (B) Intensity of p21 in the experiment shown in zA. 50 nuclei were counted and two independent experiments were performed. (C) EdU was incorporated for 10 min to U2OS cells. A CSK extraction buffer was used to retain only chromatin bound proteins. Endogenous p21 was revealed by immunofluorescence and the colocalization between p21 and EdU was determined generating profiles of signal intensity along an arbitrary line drawn across the nuclei. For all supplemental figures in this manuscript: significance of the differences are: *p<0.1; **p<0.01; ***p<0.001. When the difference is not statistically significant, the p value is not shown. Error bars represent SEM (standard error of the mean).
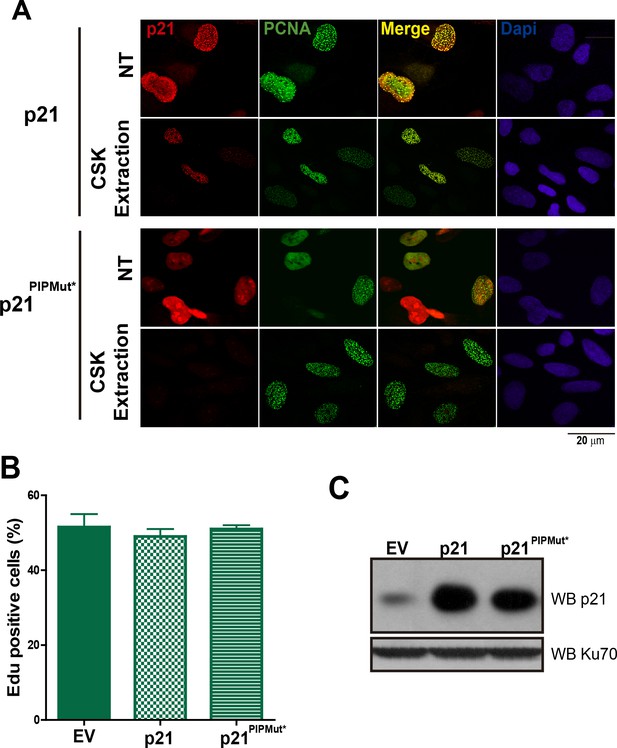
Chromatin bound p21 is localized at replication factories.
(A) The colocalization of p21 and p21PIPMut* to replication factories was evaluated before and after CSK extraction. While the localization of p21 to replication factories is evident in both conditions, p21PIPMut* does not localize to replication factories before extraction and is removed from the nuclei by CSK extraction. (B) The mutation of the CDK-binding domain of p21 allows unperturbed cell cycle progression in the presence of high levels of p21. U2OS cells were transfected with the indicated expression vectors and the percentage of EdU positive cells was determined by analyzing 200 nuclei in each of the three independent experiments performed. (C) Western blot of p21 and p21PIPMut* transfection in U2OS cells.
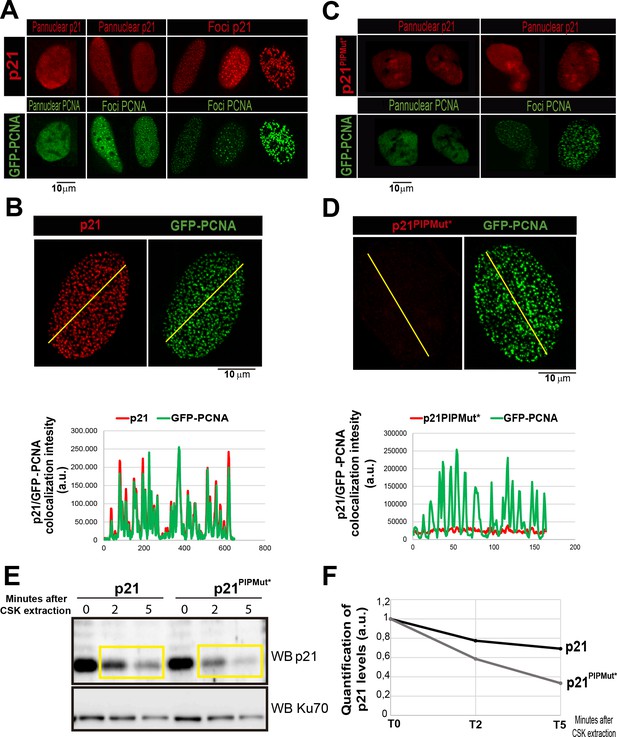
The PIR domain is required for p21 recruitment to replication factoriesU2OS cells were transfected with GFP-PCNA and p21 or p21PIPMut* respectively.
(A and C) Representative images of panuclear and foci distribution of GFP-PCNA and p21 is shown for each non-extracted condition. (B and D) After pre-extraction with CSK buffer, the colocalization of p21 and PCNA was analyzed by confocal microscopy, generating profiles of signal intensity along an arbitrary line drawn across the nuclei. (E) p21 and p21PIPMut*expressing cells were subjected to CSK extraction for the indicated times. (F) The p21 relative retention into the insoluble fraction was quantified by densitometric analysis. The values on the plot were normalized to the KU70 intensity for each sample.
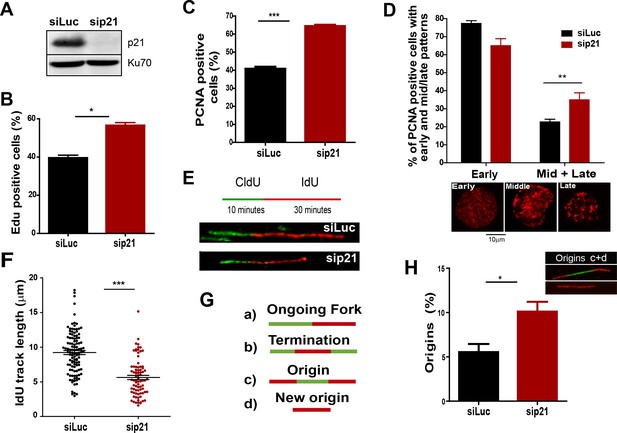
The depletion of endogenous p21 impairs the choreography of unperturbed DNA replication.
(A) Western blot (WB) analysis showing p21 levels in U2OS cells transfected with the indicated siRNAs. (B) EdU positive cells. 200 nuclei/sample were analysed in three independent experiments. (C) The percentage of cells with CSK-resistant PCNA nuclear retention. 300 nuclei/sample were analysed in three independent experiments. (D) Relative amount of cells with early or mid/late PCNA distribution. 100 nuclei/sample were examined in three independent experiments. (E) Representative fibers from control (siLuc) or sip21 transfected cells. (F) IdU track length. 100 fibers/samples were analysed in three independent experiments. (G) Schematic representation of the different structures that can be measured in the fiber analysis. (H) Samples in F were used to analyse the frequency of origin firing as the relative number of origins [(red-green-red + red only fibers)/total fibers]. 200 fibers/samples were analysed in three independent experiments.
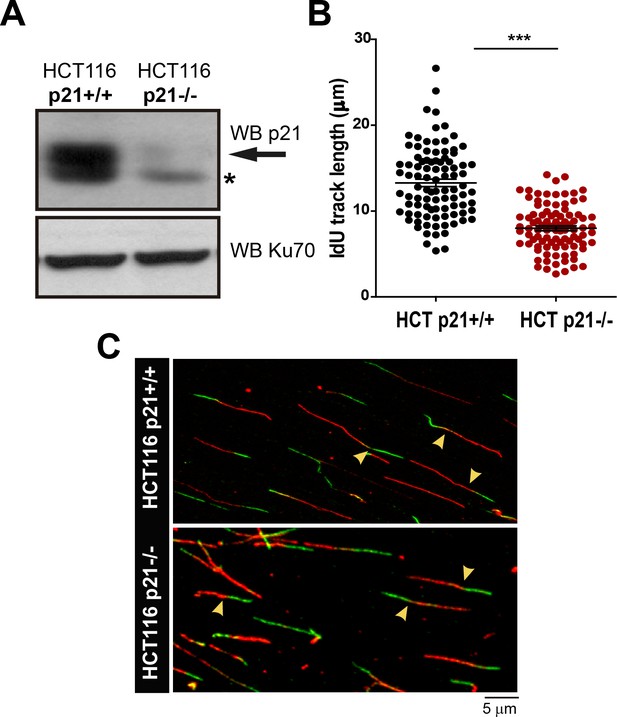
Stable p21 depletion cause alterations in the DNA replication choreography of HCT116 cells.
(A) Whole cell extracts from isogenic HCT116 p21+/+ and HCT116 p21−/− were subjected to Western Blot analysis to verify p21 elimination. (*) represents unspecific band. (B) IdU track length was measured after examining 100 fibers in three independent experiments (C) Representative field showing DNA fibers in HCT116 p21+/+ and HCT116 p21−/−. Yellow arrows indicate representative of bicolor fibers in each condition.
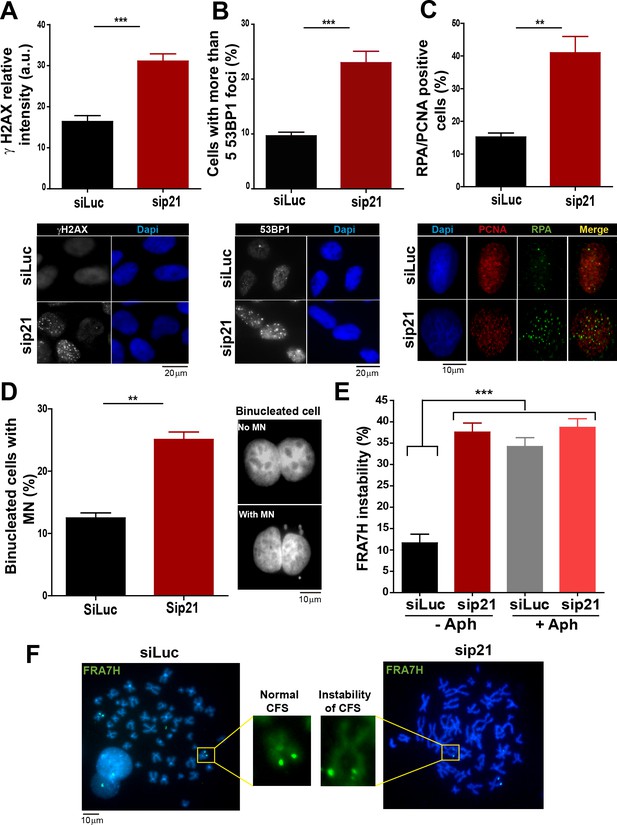
In the absence of DNA damage, replication stress markers and genomic instability increase when p21 is depleted.
(A) Quantification of γ H2AX intensity in the nucleus of U2OS transfected with the indicated siRNA. 200 nuclei/sample were examined in three independent experiments. Representative images are shown in the lower part of the panel. (B) U2OS cells with more than five 53BP1 foci were quantified. 200 cells/sample were analysed in three independent experiments. Representative images are shown in the lower part of the panel. (C) Quantification of cells with RPA foci. 150 nuclei positive for PCNA-resistance to CSK extraction/sample were examined in three independent experiments. (D) Quantification of cells with perinuclear DNA (MN) accumulation. 300 binucleated U2OS cells/samples were inspected in three independent experiments. Representative images are shown on the right. (E) Quantification of CFS expression for the indicated conditions. APH treatment corresponds to 0.2 µm for 24 hr. 50 metaphases from HCT116 cells/sample were examined in three independent experiments.
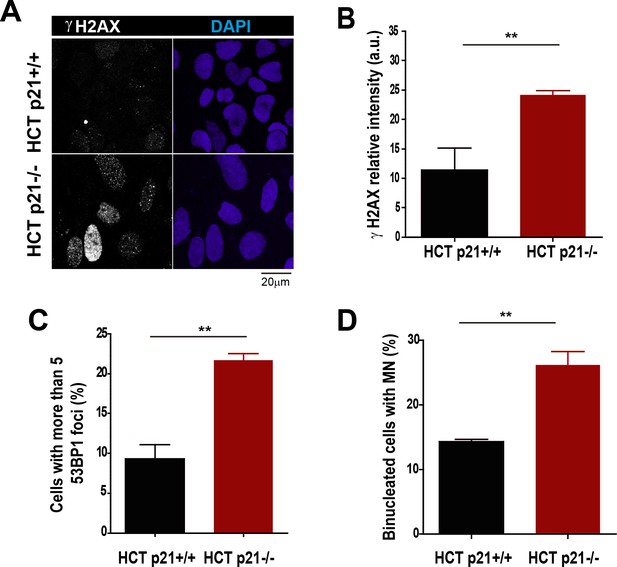
Stable p21 depletion cause alterations in the genomic stability of HCT116 cells.
(A) Representative images of γH2AX intensity in HCT116 p21+/+ and p21−/− cells. (B) Quantification of γH2AX intensity in HCT116 p21+/+ and p21−/− cells. 200 nuclei were counted in three independent experiments. (C) Quantification of cells with more than five 53BP1 foci in HCT116 cells. 200 nuclei were counted in three independent experiments. (D) Quantification of MN accumulation. 300 binucleated cells were analyzed in three independent experiments.
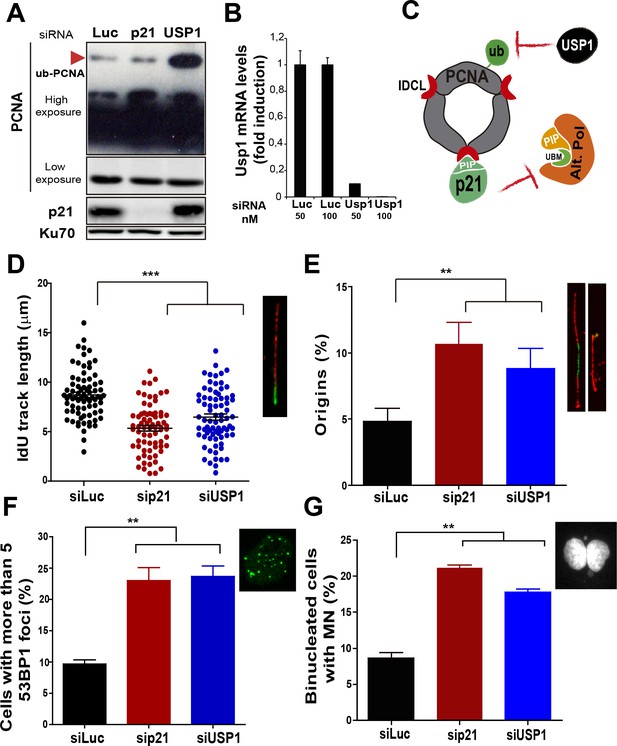
Mechanistically distinct regulators of alt DNA pols are required to facilitate unperturbed DNA replication.
(A) W.B. analysis revealing PCNA, ubi-PCNA and p21 levels in U2OS cells transfected with the indicated siRNAs. (B) USP1 mRNA levels were examined by quantitative RT-PCR. (C) Model depicting the different mechanisms of PCNA regulation by p21 and USP1. (D) IdU track length was measured in 85 fibers/sample in three independent experiments. (E) Origin firing frequency. 150 fibers/sample were analysed in three independent experiments. (F) Quantification of cells with more than five 53BP1 foci. 200 U2OS cells/sample were analysed in three independent experiments. (G) MN accumulation. 300 binucleated cells/sample were analyzed in three independent experiments. Data on the effect of USP1 downregulation on the modulation of nascent DNA elongation (Figure 4D) and accumulation of cells with 53BP1 foci (Figure 4F) and micronuclei (Figure 4G) was reproduced from Jones et al, EMBO.
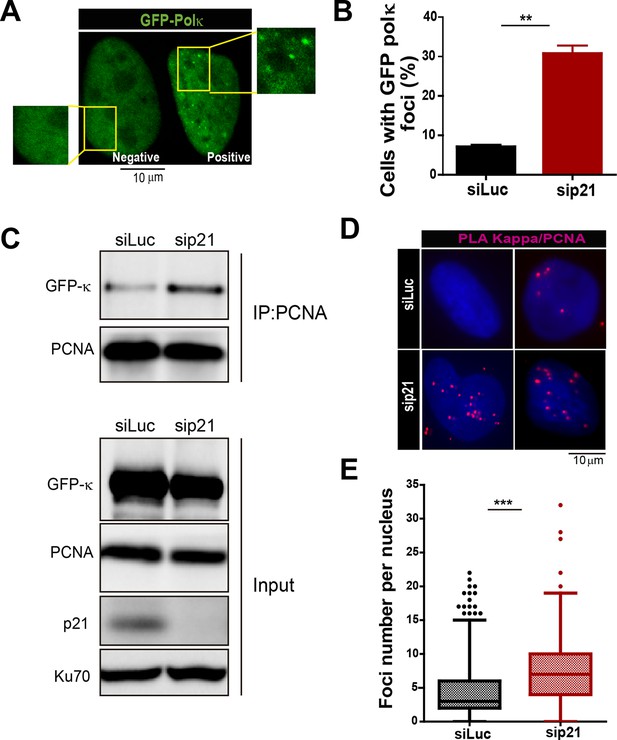
The recruitment of pol κ to replication-associated structures increases in the absence of p21.
(A) U2OS cells were transfected with GFP-polκ. Representative images of cells with and without with GFP-pol κ foci. (B) Percentages of cells with GFP-pol κ focal organization. 150 nuclei/sample were analyzed in two independent experiments. (C) siLuc and sip21 depleted samples were subjected to chromatin immunoprecipitation using a monoclonal PCNA antibody. GFP-pol κ recruitment to chromatin was revealed by using GFP antibodies. The result was reproduced in three independent experiments. (D) Proximity Ligation Assay (PLA) between PCNA and endogenous pol κ was performed in U2OS cells. Two representative images of PLA in siLuc and sip21 cells are shown. (E) Quantification of PLA foci per nuclei. More than 1000 nuclei were analysed in three independent experiments.
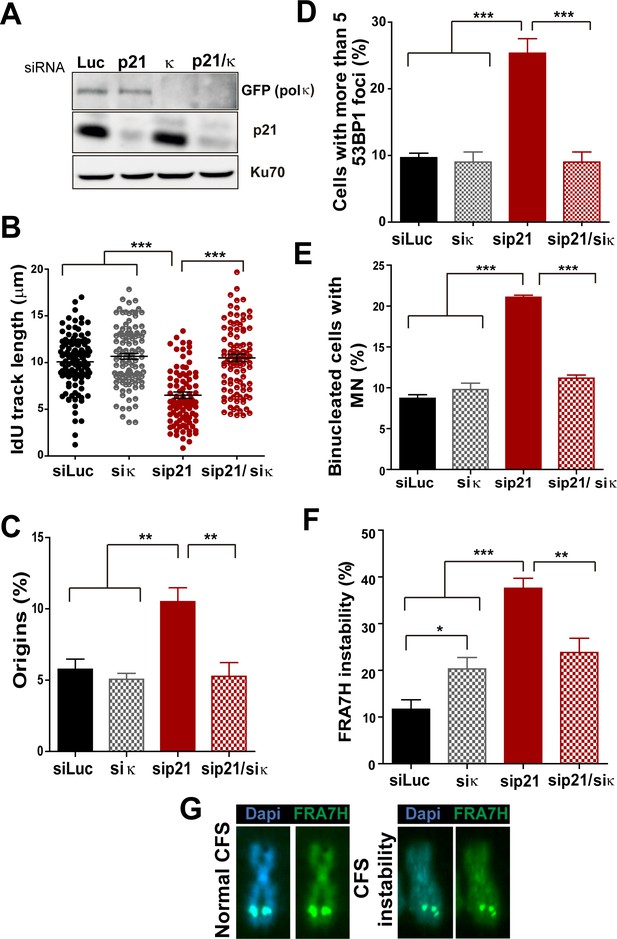
Pol κ depletion prevents the DNA replication defects and the genomic instability caused by p21 downmodulation.
(A) Western blots showing GFP-pol κ and p21 levels in U2OS cells transfected with the indicated siRNAs. (B) Total IdU track length was evaluated in 100 fibers/sample in three independent experiments. (C) Frequency of origin firing. 200 fibers were analysed in three experiments. (D) Quantification of cells with 53BP1 foci. 200 cells were analysed in three independent experiments. (E) Quantification of MN accumulation. 300 binucleated cells/sample were analysed in three independent experiments. (F) Quantification of CFS instability in HCT116 cells transfected with the indicated siRNAs. 50 metaphases/sample were analysed in three independent experiments. (G) Representative images of the CFS analysed in F.
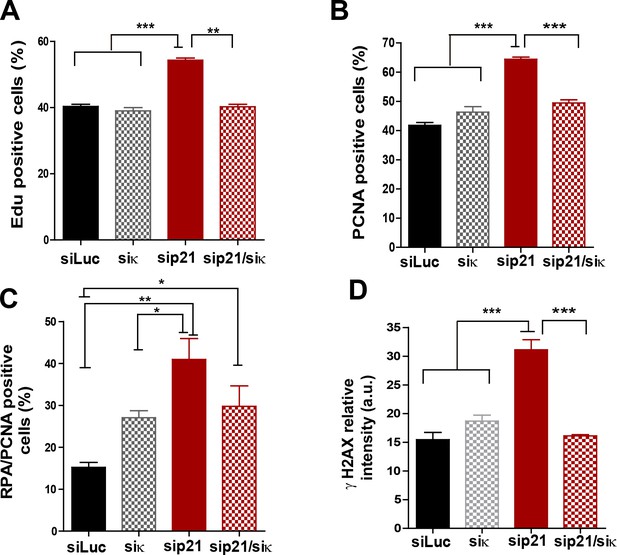
Pol κ depletion prevents the accumulation of DNA replication stress markers caused by p21 downmodulation.
(A) Quantification of EdU positive cells transfected with the indicated siRNAs. 200 nuclei per sample were analyzed in three independent experiments. (B) Quantification of CSK-resistant, PCNA positive U2OS cells transfected with the indicated siRNAs. 250 nuclei per sample were analyzed in three independent experiments. (C) Quantification of nuclei with more than 10 RPA foci in PCNA positive cells after transfection with the indicated siRNAs. 150 nuclei per sample were analyzed in three independent experiments. (D) Quantification of nuclear γH2AX intensity in cells transfected with the indicated siRNAs. 200 nuclei per sample were analyzed in two independent experiments.
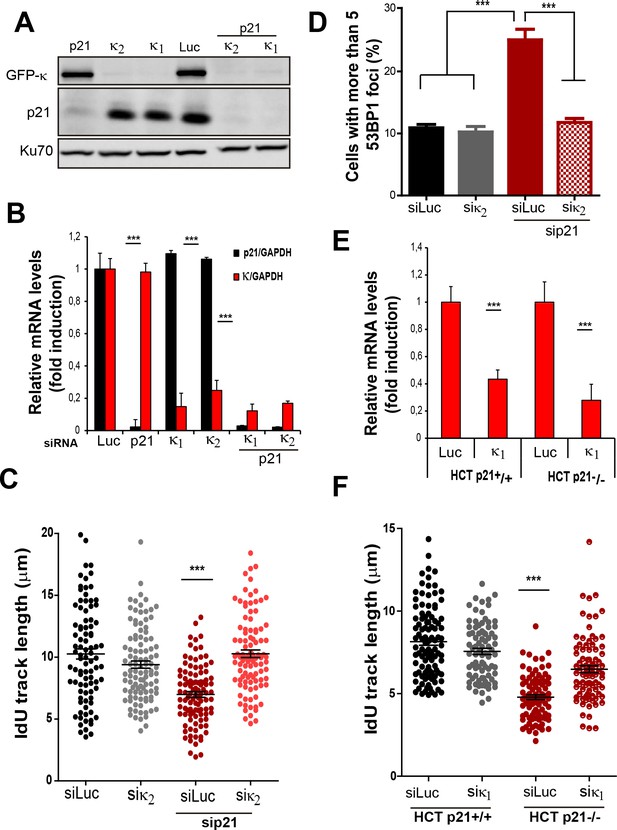
Pol κ prevents the accumulation of DNA replication stress in p21 depleted cells independently of the siRNA used and in cells stably lacking p21.
(A) U2OS cells were transfected with sip21 and 2 different siRNA for Pol κ. Western Blot analysis was performed to detect endogenous p21 and GFP- Pol κ. (B) RT-PCR was performed to detect mRNA levels of p21 and Pol κ using the indicated siRNAs. (C) IdU track length was measured using 2 different siRNAs for Pol κ in siLuc and sip21 depleted cells. 100fibers/sample were counted in 2 independent experiments. (D) Cells with more than five 53BP1 foci were analyzed in cells depleted of p21 and using 2 different siRNAs for Pol κ. (E) RT-PCR was performed in HCT116 p21+/+ and p21−/− cells to determine mRNA levels of Pol κ. (F) IdU track length in HCT116 p21+/+ and p21−/− depleted of Pol κ.100fibers/sample were counted in 2 independent experiments.
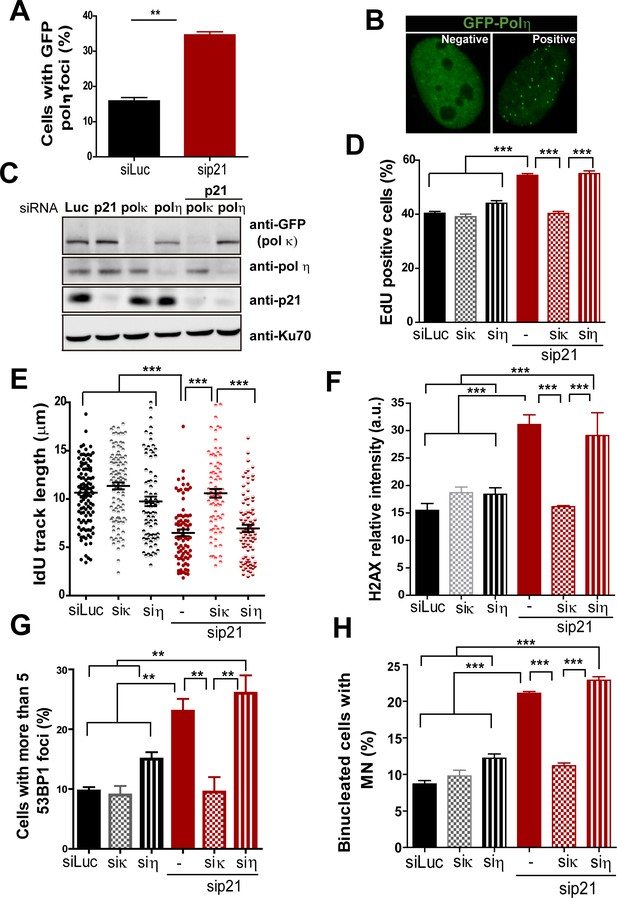
Pol κ but not pol η depletion prevents the DNA replication defects and the genomic instability caused by p21 downmodulation.
(A) U2OS cells were transfected with the indicated siRNA and GFP-pol η. The percentage of cells with GFP-pol η foci was calculated after analyzing 250 nuclei in three independent experiments. (B) Representative images of cells with GFP-pol η foci. (C) U2OS cells were transfected with GFP-pol κ and the indicated siRNAs. Western blots were performed to detect GFP-tagged Pol κ, endogenous pol η and p21. (D) Percentage of EdU positive cells were determined with the indicated siRNA. (E) DNA fiber analysis was performed in the indicated samples as described in Figure 2. The total length of the IdU track was evaluated in 100 fibers in three independent experiments. (F) Samples in E were used to analyse the frequency of origins as described in Figure 2. 200 fibers were analysed in three independent experiments. (G) Quantification of cells with more than five 53BP1 foci. 200 cells were scored in three independent experiments. (H) Quantification of MN accumulation. 300 binucleated cells were scored in three independent experiments.
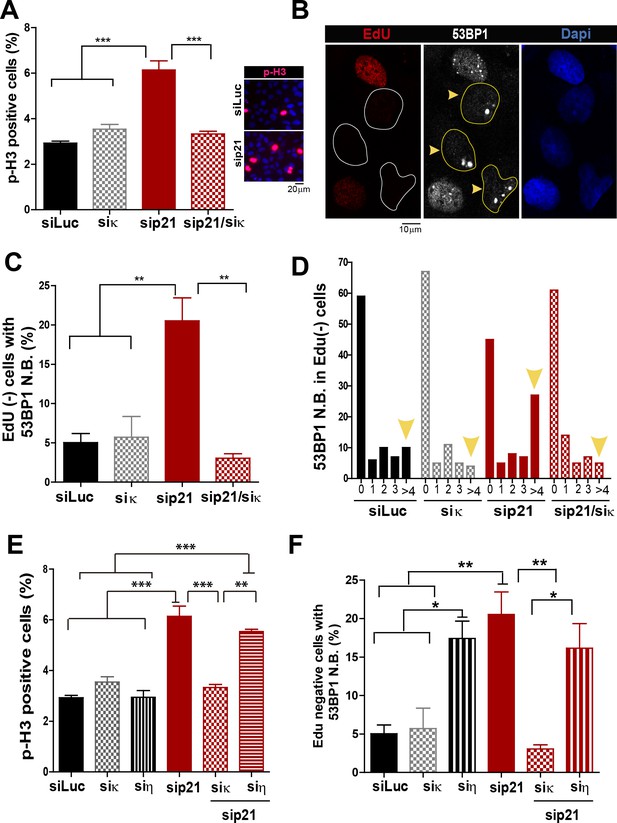
Pol κ-mediated replication defects of p21-depleted cells are transmitted to the M and G1 phases of the cell cycle.
(A) Quantification of phospho-H3 positive U2OS. 200 cells/sample were analysed in three independent experiments. (B) Representative images of 53BP1 bodies outside S phase. Yellow arrows indicate EdU negative cells with 53BP1 foci (C) Percentages of EdU negative cells which are positive for 53BP1 bodies. 200 nuclei/sample were analysed in three independent experiments. (D) Distribution of EdU negative cells with increasing number of 53BP1 bodies per cell in the experiments showed in C. 100 nuclei/sample were analysed in three independent experiments. (E) Phospho-H3 accumulation was quantified in 200 nuclei/sample of three independent experiments. Samples depleted from pol κ and pol η were compared. (F) Percentages of cells with more than five 53BP1 foci were determined after analysing 200 nuclei/sample in three independent experiments. Samples depleted from pol κ and pol η were compared.
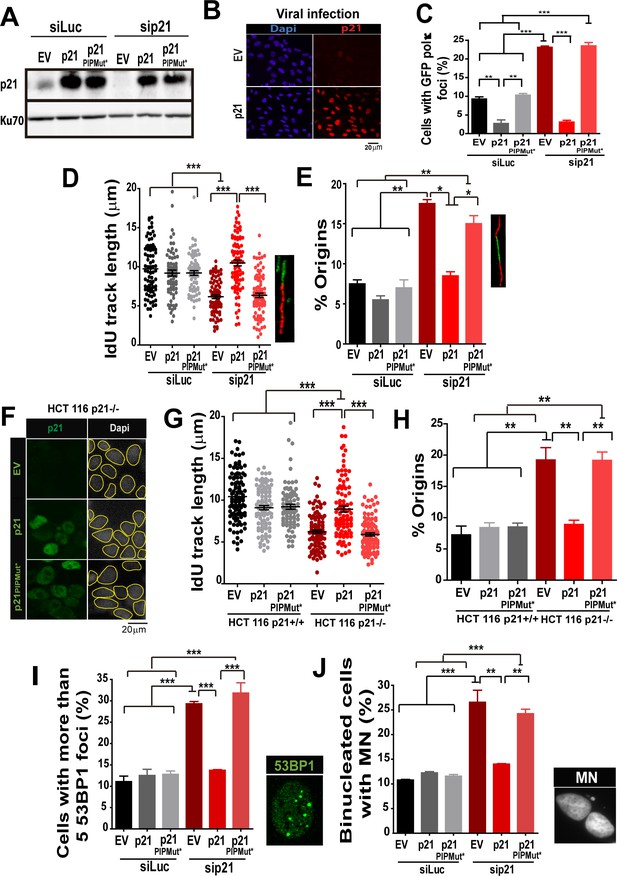
The PIR domain of p21 and not the CDK-binding domain is required to prevent DNA replication defects in p21 depleted cells during the unperturbed S phase.
(A) W.B. analysis was performed to evaluate p21 expression in U2OS cells transfected with control or p21 siRNA and infected with p21 or p21PIPMut* lentiviruses 6 hr later. (B) Representative panel showing the efficiency of lentiviral infection (>90%). (C) GFP-pol κ focal organization in samples depleted from p21 and infected with p21 or p21PIPMut*. (D) IdU track length was measured in 100 fibers/sample in three independent experiments. (E) Frequency of origin firing. 100 fibers/sample were analysed in three experiments. (F) Infection were performed in HCT116 p21+/+ and p21−/− cells. Representative panel showing the efficiency of lentiviral infection in HCT116 p21−/− s. (G) IdU track length was measured in 100 fibers/sample in three independent experiments. (H) Frequency of origin firing was measured in the experiments shown in G. 200 fibers/sample were analysed. (I) Quantification of the percentage of cells with more than 5 53BP1 foci/cell. 200 cells/sample were evaluated in three independent experiments. (J) Quantification of MN accumulation. 200 binucleated cells were analysed in three independent experiments.
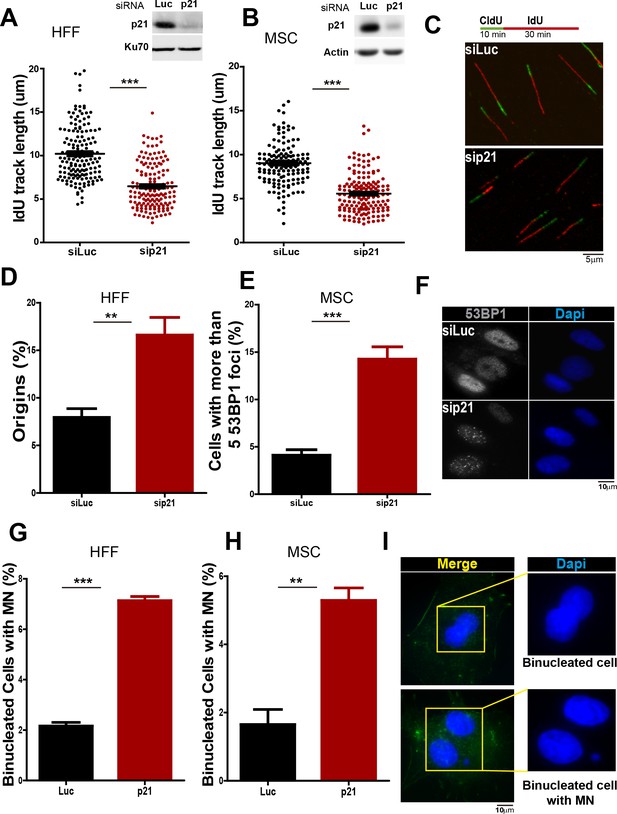
p21 depletion promotes replication stress and genomic instability in primary human cells.
(A and B) IdU track length in Human Foreskin fibroblasts (HFF) and Umbilical Cord Mesenchymal Stem cells (MSC). 100 fiber/sample were analysed in three independent experiments. Western blot showing p21 depletion in the indicated cell line. (C) Representative fibers in siLuc and sip21 HFF cells. (D) Origin frequency in HFF cells. 200 fiber/sample were analyzed in three independent experiments. (E) Cells with more than 5 53BP1 foci were analyzed in MSC cells. 200 nuclei/sample were analyzed in three independent experiments. (F) Representative images of 53BP1 in siLuc and sip21 MSC cells. (G and H) Quantification of MN accumulation. 200 binucleated cells were analyzed in HFF and MSC cells respectively in three independent experiments. (I) Representative images of Binulceated cells with and without MN.
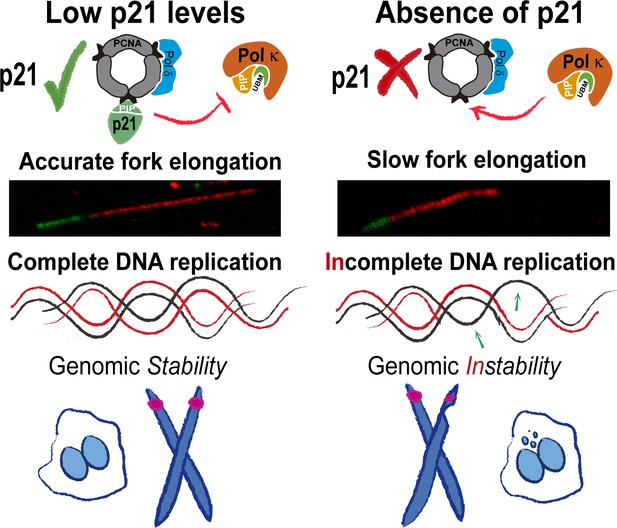
Model depicting the implication of our findings.
p21 levels in S phase are low but sufficient to prevent pol κ loading to replication forks. When p21 is absent, the abnormal use of pol κ during unperturbed replication impairs nascent DNA elongation. While origin firing increases to compensate the slow fork progression, late replicating and origin-poor DNA regions, that is. CFS, are inefficiently duplicated. The replication defects accumulated in p21-depleted samples lead to genomic instability and transmission of replication defects to G1.
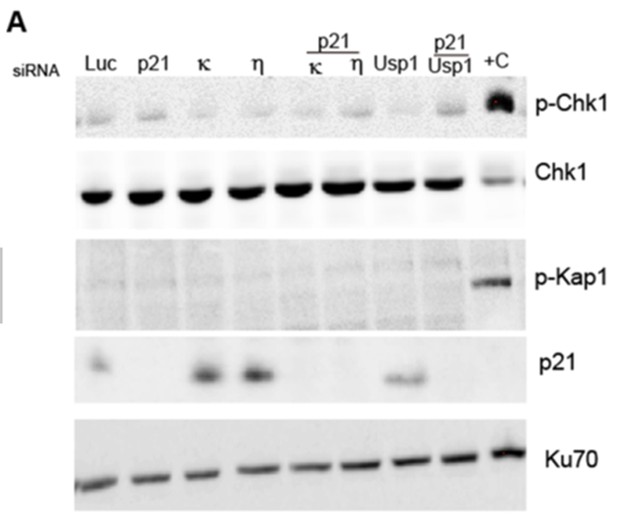
U2OS cells were transfected with the indicated siRNAs and 48hs.
Later, a western blot was performed to analyse DNA damage markers such as p-Chk1, and p-Kap1. Cells irradiated with 20J/m2 were used as a positive control of the DNA damage response.
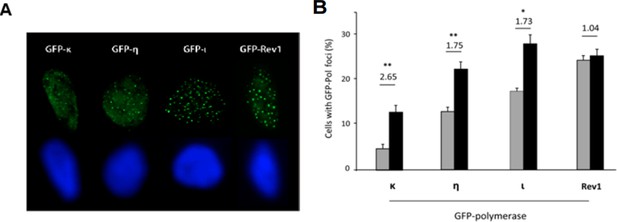
(A) U2OS cells were transfected with the indicated GFP-pols in siLuc and sip21 depleted cells. Samples were fixed with methanol/acetone and analyzed for focal organization of GFP-pols at replication factories. Representative images for each polymerase are shown. (B) Transfected cells were fixed with Methanol/Acetone and the number of cells with detectable foci was quantified.





