BRAF activates PAX3 to control muscle precursor cell migration during forelimb muscle development
Figures
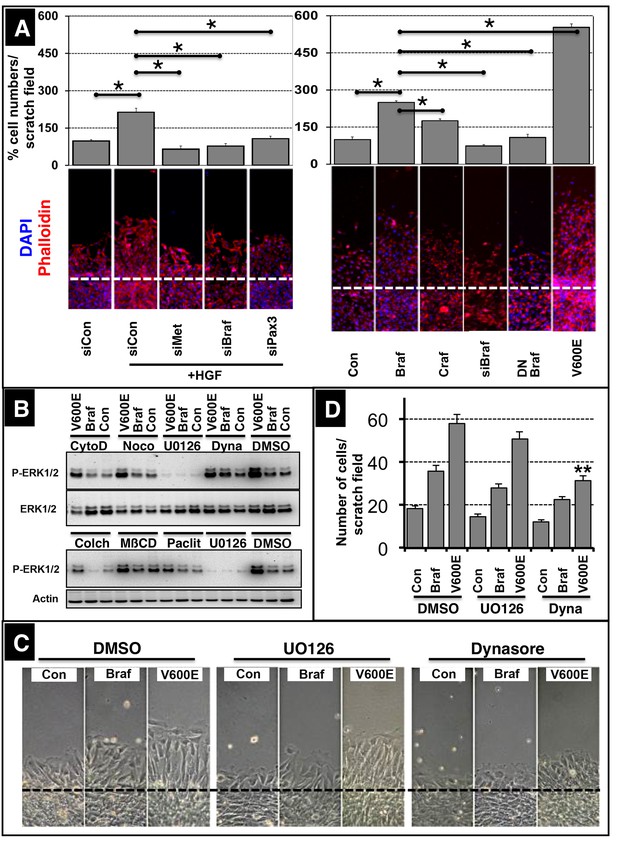
BRAF mediates muscle precursor cell migration independent of MEK/ERK signaling.
(A) Immunofluorescence staining of migrating muscle cells (C2C12) after knock down of the hepatocyte growth factor (HGF) receptor (siMet), Braf (siBraf) and Pax3 (siPax3) or after transfection of cultures with Craf, Braf, a dominant negative form of Braf (DN Braf) and CA Braf (V600E) in the presence or absence of HGF. Con indicates control vector and siCon represents a scrambled siRNA control. Cultures were analyzed 4 hr after scratching excluding effects of cell proliferation. Cell numbers were determined by counting the number of DAPI-stained nuclei. A statistical assessment is shown in the upper part of the panel (n = 15; Mann-Whitney-U test, p*<0.05). siRNA knock-down efficiencies for Met (80%), Braf (70%) and Pax3 (85%) were determined by Western blot analysis. (B) Phosphorylation of ERK1/2 after transfection of C2C12 cells with control vector (Con), WT Braf and CA Braf (V600E) in the presence or absence of UO126 (5 µM) or cytochalasin D (5 µM; CytoD), noco (5 µM; nocodazole), Dynasore (80 µM; dyna), colchicine (0.01%; colch), methyl-β-cyclodextrin (3 mM; MβCD), and paclitaxel (5 µg/ml; paclit). Addition of DMSO served as an additional control. (n = 2). Cytochalasin D disrupts actin filaments. Nocodazole, colchicine, and paclitaxel interfere with microtubuli assembly or disassembly. U0126 inhibits the MEK/ERK pathway. Dynasore blocks dynamin-dependent endocytosis. Methyl-β-cyclodextrin removes cholesterol from cultured cells and disrupts lipid rafts. (C) Statistical assessment of the experiments shown in (D) (n = 15; Mann-Whitney-U test, p**<0.01). (D) Microscopic imaging of migrating C2C12 cells after transfection with a control vector (Con), WT Braf and CA Braf (V600E) in the presence or absence of DMSO, UO126 and Dynasore. Cultures were analyzed 4 hr after scratching excluding effects of cell proliferation.
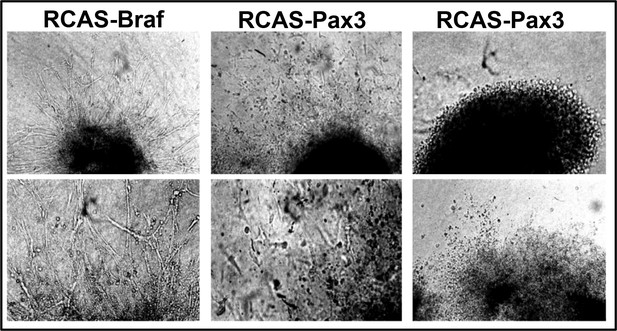
BRAF and PAX3 stimulate limb muscle precursor cell migration.
Microscopic images of chicken somitic explants after infection with retroviral vectors expressing Braf, Pax3 or human alkaline phosphatase (AP). BRAF and PAX3 both stimulated migration of cells out of explants while AP, which served as a negative control, caused no effects. n = 3.
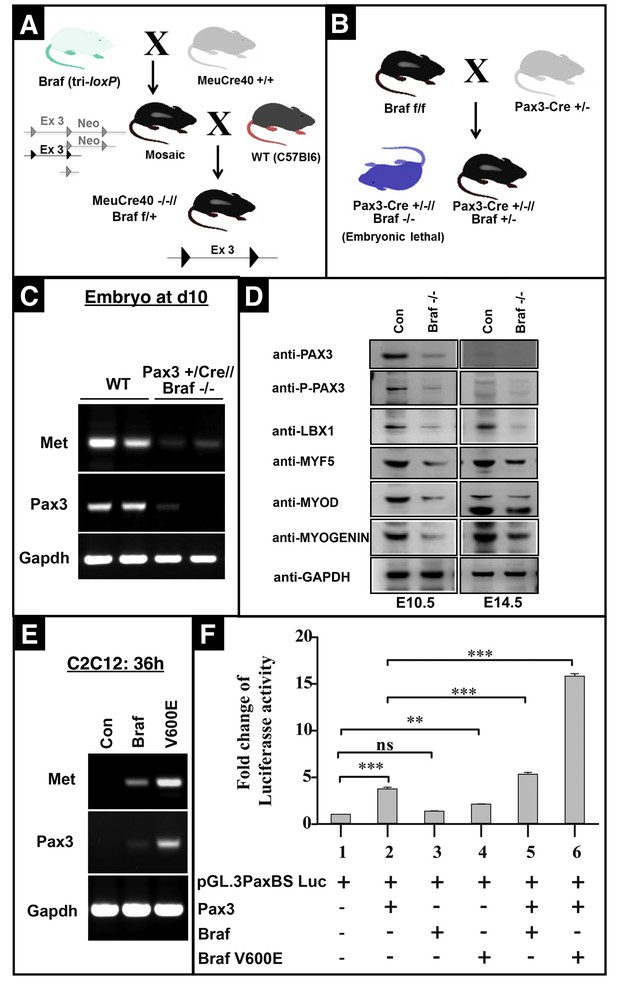
Inactivation of Braf in limb muscle precursor cells.
(A) Strategy for generation of Braf floxed mice (Brafnfl). Breeding with MeuCre mice yielded Braffl mice, in which the neomycin cassette was removed but exon three is flanked by loxP-sites. (B) Braffl mice were bred with Pax3-Cre mice to generate animals lacking Braf in Pax3 expressing cells. Pax3-Cre//Brafdel/del embryos die around E15.5. Mutant embryos were analyzed between E10.5 and E14.5. (C) RT-PCR analysis of Met and Pax3 expression in limb buds of Pax3-Cre//Brafdel/del embryos at E10.5 n = 2. (D) Western blot analysis of expression of different markers in limb buds of Pax3-Cre//Brafdel/del embryos at E10.5 and E14.5 n = 2. (E) Expression of WT and CA Braf (VE600E) in C2C12 cells increases expression of Met and Pax3. n = 3. (F) BRAF enhances PAX3-dependent transcriptional responses. The pGl.3 Pax3BS luc reporter construct containing two PAX3 binding sites in front of a minimal promoter was co-transfected with different combinations of Pax3, Braf, and CA Braf (V600E) expression vectors into HEK293T cells. The activity of Firefly Luciferase was normalized by a co-transfected renilla luciferase in all experiments. Data represent the mean ± SEM and analyzed using ANOVA with a Tukey-Kramer post-hoc comparison test. ***p<0.001.
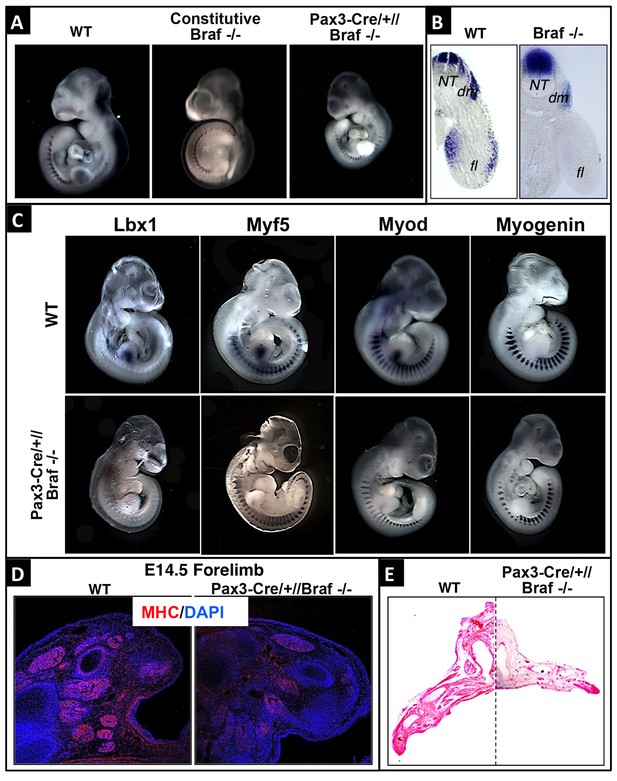
BRAF is required for limb muscle precursor cell migration during mouse embryogenesis.
(A) Pax3 whole mount in situ hybridization of WT, germ line Brafdel/del and Pax3-Cre//Brafdel/del mutant embryos at E10.5. (B) Transverse sections of WT and Pax3-Cre//Brafdel/del mutant embryos at E10.5 after Pax3 whole mount in situ hybridization. Inactivation of Braf results in loss of PAX3+ cells in forelimbs. NT: neural tube; dm: dermomyotome; fl: forelimb. (C) Whole mount in situ hybridization of E10.5 WT and Pax3-Cre//Brafdel/del mutant embryos using Lbx1, Myf5, Myod, and Myogenin probes. (D) Immunofluorescence staining of forelimbs from WT and Pax3-Cre//Brafdel/del mutants for myosin heavy chain (MHC) at E14.5. (E) Hematoxylin and eosin staining of forelimbs from WT and Pax3-Cre//Brafdel/del mutants at E14.5.
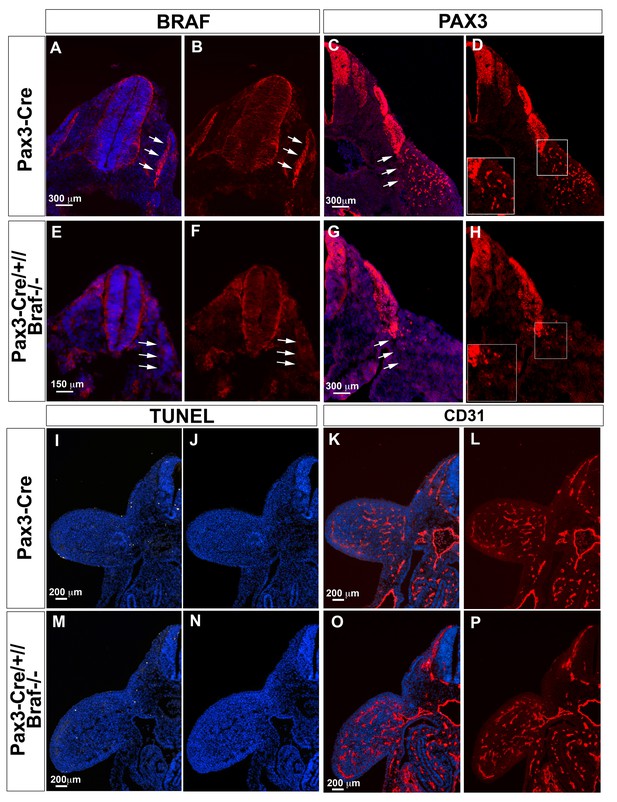
Inactivation of Braf in Pax3 expressing cells impairs limb muscle precursor cell but not endothelial cell migration during mouse embryogenesis.
(A, B, E, F) Immunofluorescence staining of E10.5 Pax3-Cre (A, B) and Pax3-Cre//Brafdel/del embryos (E, F) for BRAF (red). White arrows in (A, B) indicate the presence of BRAF in the dermomyotome. (C, D, G, H) Immunofluorescence staining of E10.5 Pax3-Cre (C, D) and Pax3-Cre//Brafdel/del embryos (G, H) for PAX3. White arrows in (C) indicate the delamination of PAX3+ cells from the dermomyotome in Pax3-Cre control embryos whereas delamination of PAX3+ cells is compromised in Pax3-Cre//Brafdel/del embryos (White arrows in (G)). Inserts in (D) and (H) represent magnification of boxed areas. (I, J, M, N) TUNEL staining of E10.5 Pax3-Cre (I) and Pax3-Cre//Brafdel/del embryos (M) to detect apoptotic cells. DAPI stained sections of Pax3-Cre (J) and Pax3-Cre//Brafdel/del embryos (N) without the TUNEL fluorescence channel are shown for comparison. Inactivation of Braf in Pax3-expressing cells does not lead to an increase of apoptotic cells. (K, L, O, P) Immunofluorescence staining of E10.5 Pax3-Cre (K, L) and Pax3-Cre//Brafdel/del embryos (O, P) for CD31 to visualize endothelial cells. Inactivation of Braf in Pax3-expressing cells does not compromise formation of the vascular network in limb buds.
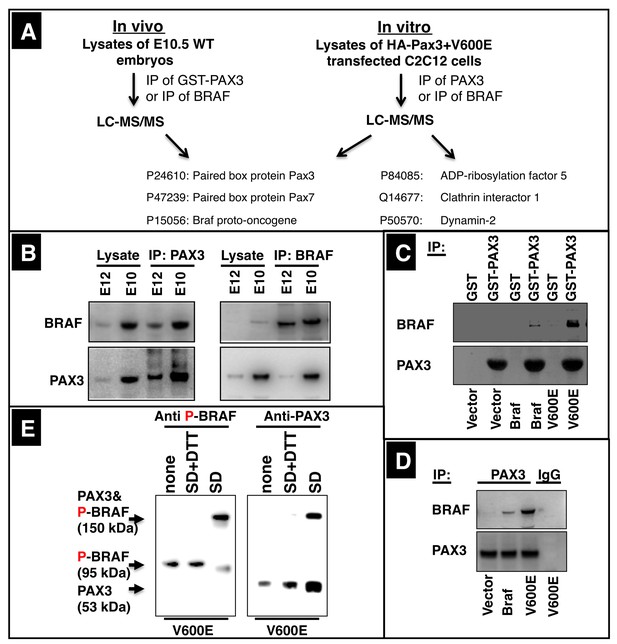
BRAF directly interacts with PAX3 in migrating muscle precursor cells.
(A) PAX3, GST-PAX3 and PAX3 were immunoprecipitated from protein extracts of E10.5 wild type embryos or from C2C12 muscle cells after transfection with HA-Pax3 or CA Braf (V600E). Immunoprecipitations were analyzed by mass spectrometry after SDS-PAGE and in gel digestions. A selected list of proteins identified by Mascot search analysis is presented. (B) Analysis of the interaction of PAX3 and BRAF in E10.5 and E12.5 WT embryos by coupled immunoprecipitation/Western blot analysis. n = 3. (C) Western blot analysis of GST and GST-PAX3 immunoprecipitations after transfection of C2C12 cells with empty vector, Braf and CA Braf (V600E). n = 3. (D) Western blot analysis of PAX3 or BRAF immunoprecipitations from C2C12 transfected with CA Braf (V600E) after chemical cross-linking (SD) and cleavage of the cross-linker with DTT (SD+DTT). None = no cross-linker added. n = 3. (E) Western blot analysis of PAX3 immunoprecipitations from C2C12 transfected with WT Braf, CA Braf (V600E) and vector control (vector). n = 3.
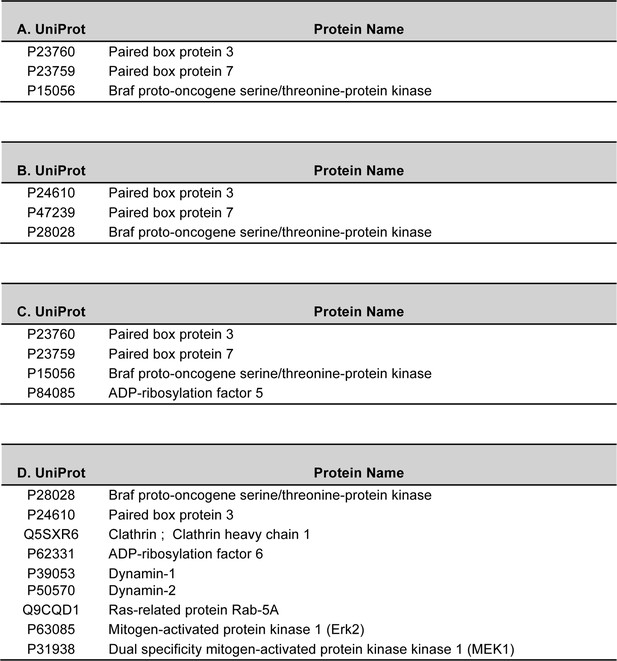
Selected lists of BRAF interaction partners identified by mass spectrometry analysis.
Mass spectrometry was performed after in-gel digestion of Coomassie-stained bands from polyacrylamide gels. (A) Proteins identified after GST-PAX3 pull down of protein extracts from WT mouse embryonic forelimbs (E10.5). (B) Proteins identified after immunoprecipitation of BRAF (antibody from BD Biosciences (612375)) from extracts of WT mouse embryonic forelimbs (E10.5). (C) Proteins identified after immunoprecipitation of PAX3 form C2C12 myoblasts transfected with HA-tagged Pax3. (D) Proteins identified after immunoprecipitation of BRAF (antibody from BD Biosciences (612375)) from C2C12 myoblast cells after transfection with HA-tagged Pax3. Selected lists of proteins identified in the different co-immunoprecipitation experiments are shown.
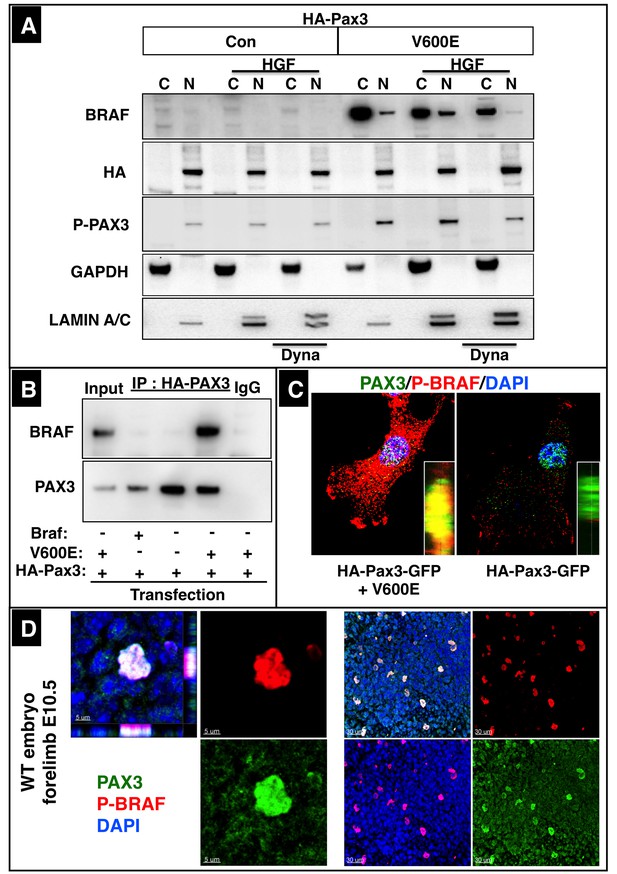
A fraction of BRAF co-localizes with PAX3 in nuclei of muscle cells.
(A) Western blot analysis of cytoplasmic (C) and nuclear fractions (N) of C2C12 cells transfected with CA Braf (V600E), WT Braf, HA-Pax3 or HA-Pax3-GFP. n = 3. Successful fractionation was monitored by cytoplasmic GAPDH and the nuclear protein LAMIN A/C. Some cultures were treated with Dynasore (Dyna) for 30 min before fractionation as indicated. (B) Western blot analysis of immunoprecipitations of nuclear fractions isolated from migrating C2C12 cells after transfection with WT Braf, CA Braf (V600E), or HA-Pax3. n = 3. (C) High resolution confocal images of C2C12 cells transfected with WT, CA Braf (V600E) and HA-Pax3-GFP. (D) High resolution confocal image of a PAX3 and BRAF positive forelimb muscle precursor cell at E10.5 (left panel). A lower magnification is shown in the right panel.
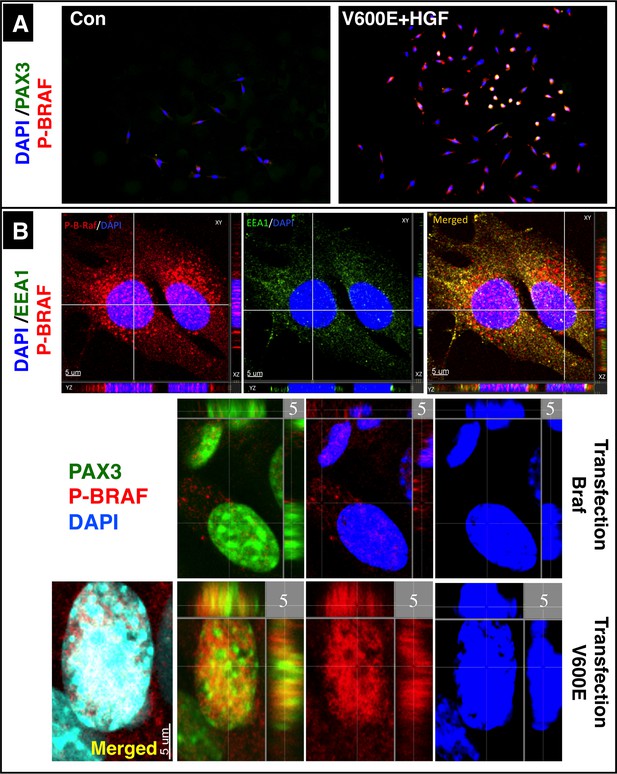
Intracellular localization of BRAF in C2C12 cells.
(A) Immunofluorescence images of C2C12 cells in a Boyden micro chemotaxis chamber. Cultures were either not stimulated (Con) or treated with HGF after transfection with CA Braf (V600E). Note: Overlay of red BRAF and green PAX3 results in a yellow color of the nuclei. (B) High resolution confocal images of nuclei of migrating C2C12 cells transfected with WT and CA Braf (V600E). Cells were co-stained for P-BRAF and EEA1 as well as for P-BRAF and PAX3. Nuclei were visualized by DAPI. Note the absence of the endosomal marker EEA1 in nuclei, while P-BRAF is clearly visible within nuclei.
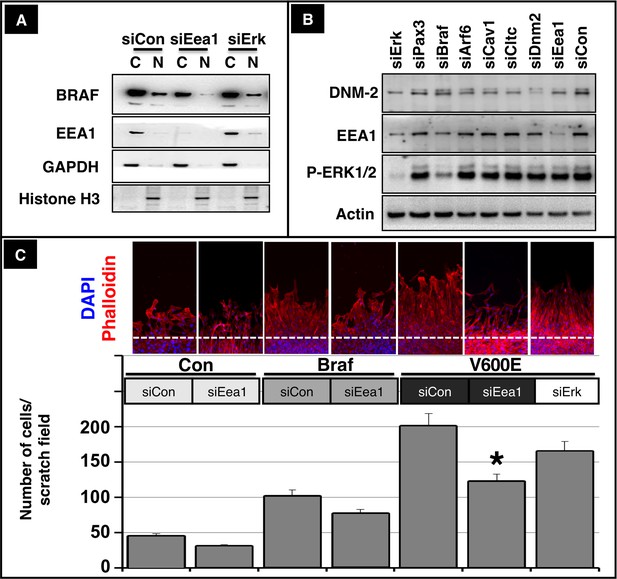
Nuclear translocation of BRAF and migration of muscle cells depend on intact endosomal trafficking.
(A) Western blot analysis of isolated subcellular fractions of C2C12 cells after transfection of CA Braf V600E and knockdown of Eea1 or Erk1/2. n = 3. Knockdown of Eea1 prevented accumulation of BRAF in the nucleus (N). C: cytoplasm. siRNA knock-down efficiencies for Eea1 (60%) and Erk (95%) were determined by Western blot analysis. (B) Knock down of Braf (siBRAF) but not of Eea1 (siEea1), Dnm-2 (siDnm2), Cltc (siCltc), Cav1 (siCav1) and Arf6 (siArf6) did prevent phosphorylation of ERK1/2. Western blot analyses of siRNA transfected C2C12 cells are shown. n = 3. Actin served as loading control. siRNA knock-down efficiencies for Pax3 (85%), Braf (70%), Arf6 (80%), Cav1 (75%), Cltc (70%), Dnm2 (75%) and Eea1 (60%) were determined by Western blot analysis. (C) Immunofluorescence staining of migrating C2C12 cells after knockdown of Eea1 or Erk-1/2. Cultures were transfected with control vector, Braf or CA Braf (V600E) as indicated. Con indicates control cultures and siCon means control siRNA. siRNA knock-down efficiencies for Eea1 (60%) and Erk (95%) were determined by Western blot analysis. Cell numbers were determined by counting the number of DAPI-stained nuclei. A statistical analysis of siEea1 versus siCon in CA Braf (VE600E) transfected cultures is shown. n = 12; Mann-Whitney-U test, (p*<0.05).
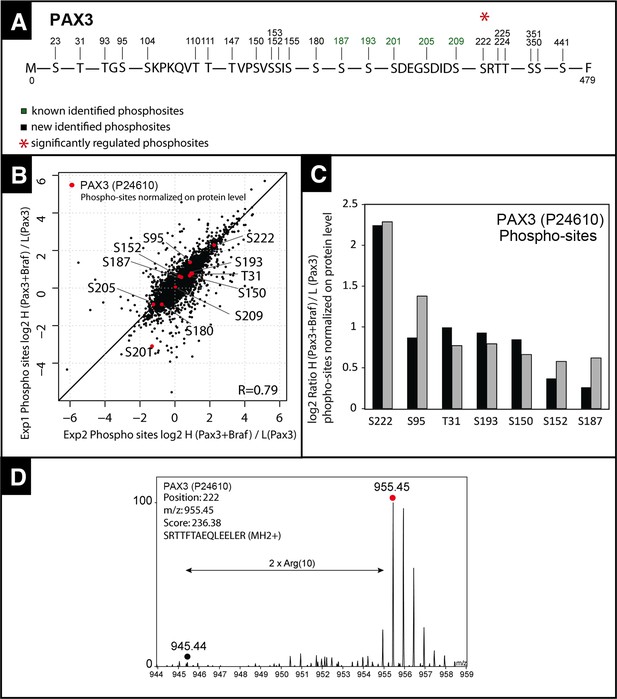
Quantitative mass spectrometry analysis of PAX3 phosphorylation sites.
(A) Map of PAX3 phosphorylation sites determined by mass spectrometry. Sites shown in green have been published before (phosphosite.org) and sites displayed in black were newly identified. The red asterisk marks a phosphorylation, which was strongly induced by co-transfection with CA Braf (V600E). (B) Scatter plot showing the distribution of SILAC log2 phosphosite ratios normalized to PAX3 protein expression levels after transfection with Pax3 and Braf (heavy) and Pax3 (light). (C) Histogram of log2 PAX3 phosphosite ratios in two independent experiments (exp1: light grey; exp2: black) normalized to PAX3 protein expression ratios. (D) Representative MS SILAC spectra for identification of S222. Andromeda Score for corresponding MS/MS spectra is 236.38. The mass deviation is given in p.p.m.
-
Figure 7—source data 1
Source data for mass spectrometry analysis.
- https://doi.org/10.7554/eLife.18351.013
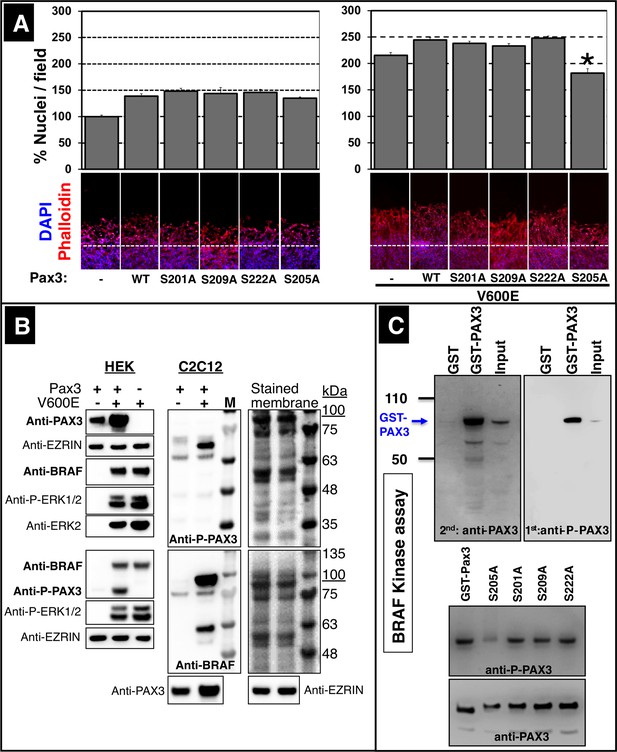
Phosphorylation of PAX3 at Ser205 is critical for stimulation of muscle cell migration.
(A) Exchange of serine by alanine in PAX3 at Ser205 reduces C2C12 myoblast migration after transfection with CA Braf (V600E). Cultures were either transfected with a control vector (-), with wildtype Pax3 (WT) or with mutated Pax3 constructs. Mutated serine residues are indicated (S201A, S205A, S209A, S222A). Cell numbers were determined by counting the number of DAPI-stained nuclei. n = 10; Mann-Whitney-U test, (p*<0.05). (B) Phosphorylated PAX3 proteins were isolated from Pax3 and mutant Braf (V600E) transfected HEK293T or C2C12 cells by phospho-column affinity purification and detected by immunoblotting. Phosphorylated ERK1/2 (P-ERK1/2), ERK2 and EZRIN served as controls. Two sets of independent experiments are shown for HEK293T cells. Molecular sizes (in kDa) are indicated by the overlay of the colored marker with the PAX3 and BRAF bands visualized by chemiluminescence. Note that PAX3 shows a shift in the molecular weight due to the HA-tag. Stained membranes with marker are shown. (C) In vitro kinase assay of GST, GST-PAX3, PAX3 and different PAX3 mutants demonstrating phosphorylation of PAX3 by BRAF. Bacterially produced GST-PAX3 (WT and mutants) was purified by GST-affinity chromatography. Column eluates and inputs were incubated in vitro together with BRAF. n = 2. Phosphorylated PAX3 was detected using an antibody specific for p-Ser205 PAX3. Specificity of the p-Ser205 PAX3 antibody was assessed by the failure to detect the S205A PAX3 mutant (bottom panel).
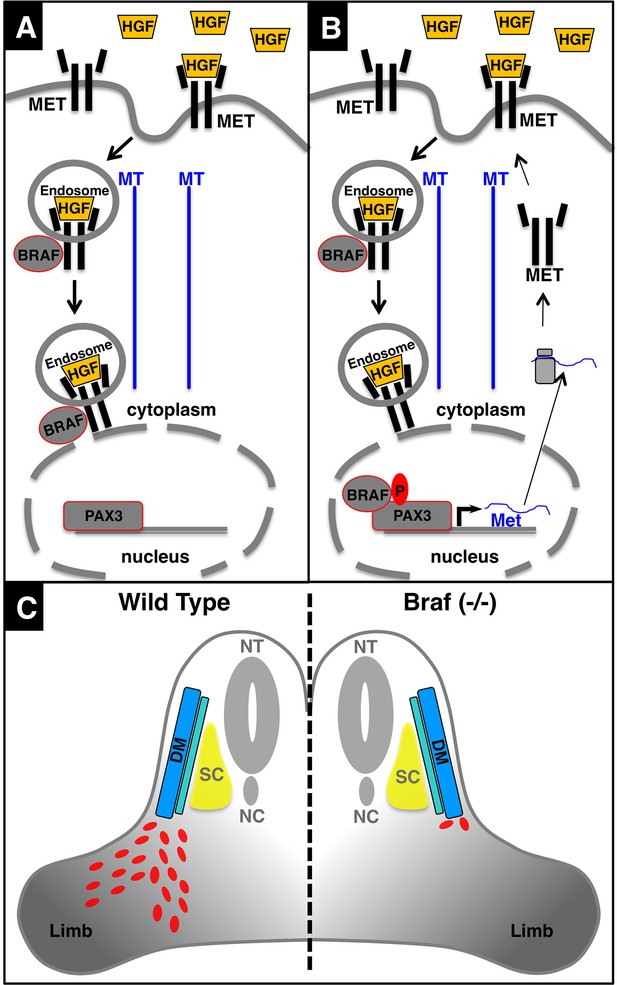
Schematic model of the MET-BRAF-PAX3 feedback loop enabling migration of limb muscle precursor cells.
(A) Activation and subsequent internalization of MET leads to activation of BRAF and transport via endosomal trafficking to a perinuclear position. (B) After translocation of BRAF into the nucleus BRAF phosphorylates and activates PAX3, which promotes sustained expression of Met and other PAX3 target genes enabling limb muscle precursor cell migration. (C) Inactivation of Braf in PAX3-expressing cells disrupts the regulatory feedback loop leading to arrest of limb muscle precursor cell migration and defective limb muscle development. NT: neural tube; DM: dermomyotome; SC: sclerotome; NC: notochord.





