C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool
Figures
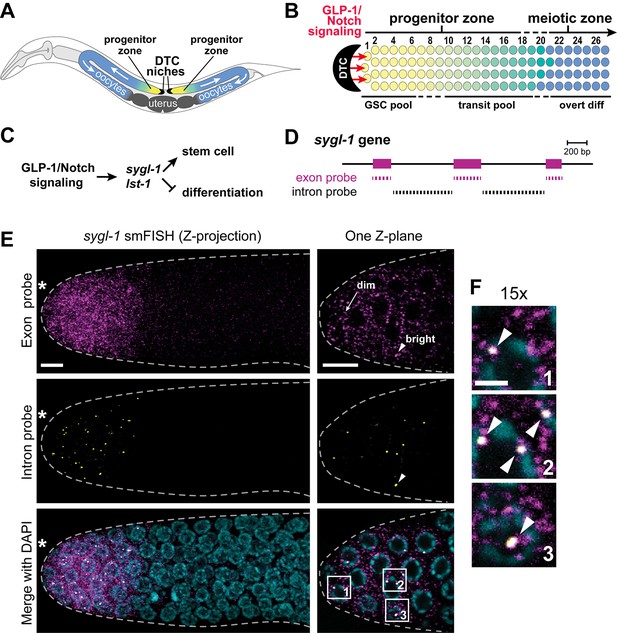
Visualization of sygl-1 transcripts using smFISH.
(A) Schematic of adult C. elegans hermaphrodite with two U-shaped gonadal arms, each with a single-celled niche (DTC, black crescent) and a progenitor zone of mitotically dividing germ cells at the distal end. Germ cell movement is from distal to proximal (white arrows). Somatic gonadal structures are located centrally (dark grey). (B) Organization of germ cells in distal gonad. The only somatic cell in the distal gonad is the DTC; diagrammed here is its cell body (see Introduction for more about DTC architecture). The progenitor zone includes a distal pool of naïve undifferentiated germ cells (yellow), which have been proposed to constitute the GSC pool, and more proximal germ cells (yellow to green transition), which have been triggered to differentiate and are maturing as they transit towards overt differentiation (Cinquin et al., 2010). Transit germ cells divide only once or twice before entering the meiotic cell cycle (Fox and Schedl, 2015). The boundary between progenitor and meiotic zones is not sharp (dashed line), and similarly, the boundaries of GSC and transit pools are not sharp (dashed lines). Positions of germ cells are conventionally designated as the number of 'germ cell diameters' along the distal-proximal axis from the distal end, with position 1 being immediately adjacent to the DTC cell body; the transition from GSC to transit pools is proposed to occur at position 6–8 (Cinquin et al., 2010), and from progenitor to meiotic zone at position 19–22 (Crittenden et al., 1994). (C) The sygl-1 and lst-1 genes are direct targets of GLP-1/Notch signaling and key regulators of germline stem cell maintenance (Kershner et al., 2014). (D) Schematic of sygl-1 exon/intron structure. Exon-specific (magenta) and intron-specific (black) probes for single-molecule RNA FISH (smFISH) were labeled with different fluors (see Materials and methods). (E-F) sygl-1 smFISH in distal gonad. Exon probes (magenta); intron probes (yellow). DAPI marks nuclei (blue). Nuclei have DAPI-free centers because of their large nucleoli. Merge (bottom) is an overlay of exon probe, intron probe and DAPI channels. Figure 1—figure supplement 1A shows sygl-1 smFISH in a whole gonad. (E) Distal gonad dissected from wild-type adult (24 hr post mid-L4 stage), showing dim spots in the cytoplasm (arrow) and bright spots in the nucleus (arrowhead). Grey dashed line marks gonadal outline; asterisk marks distal end of gonad; scale bar is 5 µm. (F) 15X magnification of nuclei within boxes in Figure 1E, bottom panel. The merged images show overlap of exon and intron signals as white spots (arrowheads), which not only overlap with DAPI, as shown here, but are also within nuclei as assayed using a nuclear lamin (Figure 1—figure supplement 1B). Scale bar: 2 μm. The specificity of smFISH probes is confirmed in Figure 1—figure supplement 1C,D and Notch dependence of smFISH signals in Figure 1—figure supplement 2.
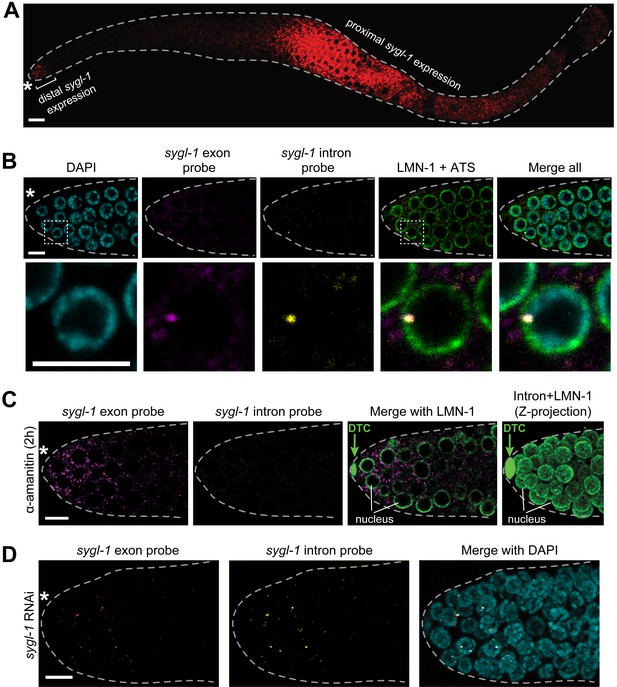
sygl-1 smFISH visualization of nascent nuclear transcripts and mature cytoplasmic transcripts.
(A) sygl-1 smFISH using exon probe set shows RNAs (red) in both the distal and proximal regions of a wild-type dissected gonad, as seen previously with conventional in situ hybridization (Kershner et al., 2014). The proximal sygl-1 expression is not GLP-1/Notch dependent (Kershner et al., 2014) and was not pursued further in this work. Asterisk, distal end; grey dashed line, gonadal outline; scale bar, 20 µm. One Z-plane is shown. (B-D) sygl-1 smFISH in distal gonad dissected from an adult (24 hr post mid-L4 stage), stained as in Figure 1E. Exon probes (magenta); intron probes (yellow); DAPI (blue). Grey dashed line, gonadal outline; asterisk, distal end of gonad; scale bar, 5 µm. (B) Gonad stained with sygl-1 exon and intron probes plus an α-LMN-1 antibody against nuclear lamin to highlight nuclear boundaries (green). Bottom row, 20X magnification of boxed region in top row. One Z-plane is shown. (C) Gonad after a 2 hr treatment with 100 µg/ml α-amanitin and stained with sygl-1 exon and intron probes plus α-LMN-1 antibody staining (green). Left and middle, a single Z-plane. Right, Z-projection of intron probes with LMN-1. DTC, nucleus of DTC. (D) Gonad from an animal whose mother was treated with sygl-1 RNAi at L4 stage. Z-projection is shown.
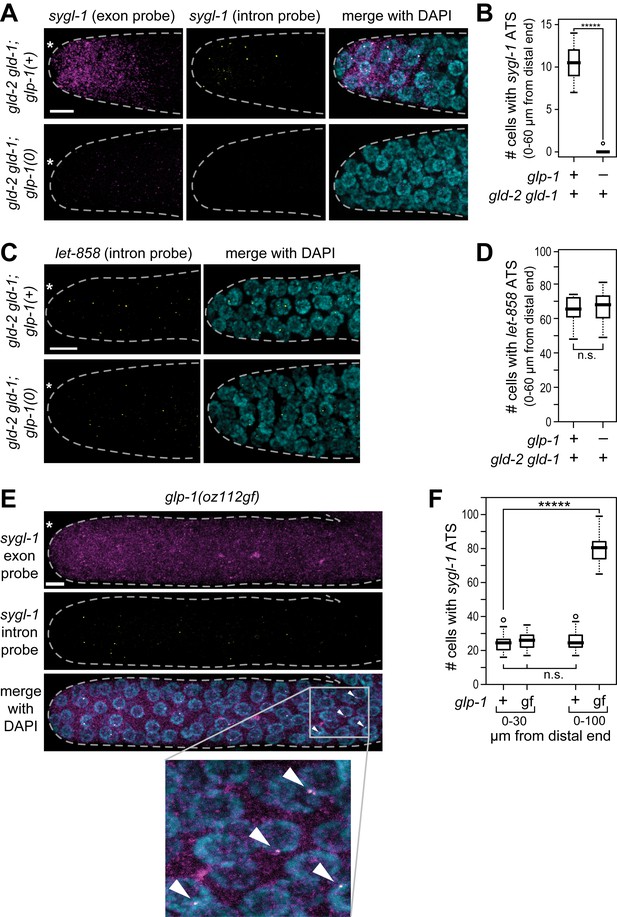
GLP-1/Notch-dependence of sygl-1 ATS.
(A,C,E) Z-projections of distal gonads stained using smFISH to sygl-1 (A,E) or let-858 (C). Exon probes (magenta); intron probes (ATS; yellow); and DAPI marks nuclei (blue). Conventions and scale as in Figure 1E. (B,D,F) number of germ cells with sygl-1 ATS (B,F) or let-858 ATS (D). *****p<0.00001; n.s.: not significant by t-test. For gld-2 gld-1 and gld-2 gld-1; glp-1(0), n = 16 (B,D); for glp-1(oz112gf), n = 10 (F); for wild-type, n = 21 (F). For box-and-whisker plots, the bold line in the box shows the median; top and bottom of box are the third and first quartiles, respectively; whiskers, maximum and minimum of data points; circles, outliers (value greater than 1.5X first or third quartile from the median). (E) Arrowheads mark ATS in proximal gonad.
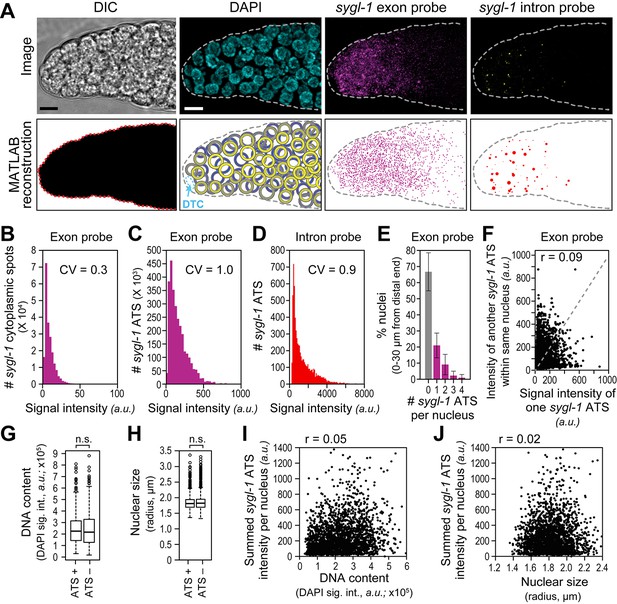
The sygl-1 transcriptional response to Notch signaling.
(A) MATLAB reconstruction of distal gonad. Top row, DIC image is one Z-plane; all others are Z-projections. Far left, DIC was used to determine gonadal outline; middle-left, DAPI reveals nuclei (blue); middle-right, signal from sygl-1 exon probes shows sygl-1 cytoplasmic mRNAs and nuclear active transcription sites (ATS) (magenta); far right, signal from sygl-1 intron probes show only ATS (yellow). Bottom row, outputs from custom MATLAB code (Source code 1). Far left, gonadal outline (red dashed line), middle-left, germ cell nuclei false-colored in a depth gradient (yellow-blue) according to Z-position and with DTC (cyan) excluded; middle-right, cytoplasmic mRNAs (magenta) with nuclear ATS excluded computationally; far right, ATS (red) with smFISH signals scaled according to intensity. Scale bar: 5 μm. (B) Signal intensities of cytoplasmic dots from sygl-1 exon probes. A total of 222,260 spots were analyzed from 78 gonads. Raw values from Z-planes were normalized to background levels in the same plane and the mean intensity value set to 10 arbitrary units (a.u.) for each gonad. CV: coefficient of variation (CV <1, significantly narrow distribution). (C,D) Signal intensities of sygl-1 ATS. A total of 2627 spots were analyzed from 78 gonads. (C) Intensities of sygl-1 ATS using exon probes. We first normalized raw values to background levels in the same Z-plane and then normalized to mean intensity of sygl-1 cytoplasmic spots seen with the same probe in the same gonad. Mean intensity of sygl-1 ATS was 172.0 a.u., or roughly 17-fold more than the mean intensity of sygl-1 individual mRNAs. (D) Intensities of sygl-1 ATS using intron probes. Raw values were normalized to background levels in the same Z-plane. (E) Number of sygl-1 ATS per nucleus in 7018 nuclei (78 gonads). Error bars: standard deviation. (F) Pair-wise comparisons of sygl-1 ATS intensities within one nucleus (78 gonads), using normalized values from exon probe. Each black dot represents one pairing. Grey dashed line indicates a perfect correlation (Pearson’s correlation coefficient r = 1); r indicates the correlation coefficient from data in the graph. (G-J) sygl-1 transcriptional activity is independent of cell cycle stage. The cell cycle stage was monitored for DNA content (summed DAPI signal) or nuclear size, which are correlated (Figure 2—figure supplement 1A), in all nuclei located 0–30 µm (1–7 gcd) from the distal end (n = 6979 nuclei total); n.s.: not significant (p>0.05) by t-test. The cell cycle is also independent from nuclear location in the gonad (Figure 2—figure supplement 1B,C). (G,H) DNA content (G) or nuclear size (H) was compared between ATS-positive and ATS-negative cells. For all box-and-whisker plots in this study, the bold line in the box shows the median; top and bottom of box are the third and first quartiles, respectively; whiskers, maximum and minimum of data points; circles, outliers (value greater than 1.5X first or third quartile from the median). (I, J) Summed sygl-1 ATS intensity was estimated by pooling all ATS signal intensities (a.u.) within the same nucleus. Each black dot represents a single nucleus.
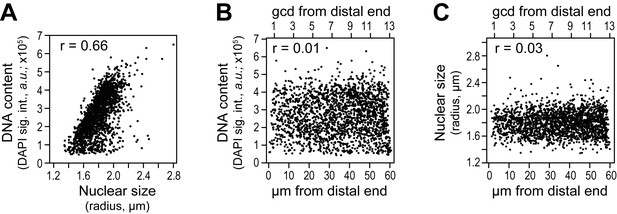
Cell cycle analysis in reconstructed gonads.
DNA content was estimated as total DAPI signal intensity in a 3-D reconstructed spherical nucleus, and nuclear size as the radius of a 3-D reconstructed spherical nucleus. Each dot in the scatter plots represents a single nucleus. Pearson’s correlation coefficient (r) is shown in each plot (r values approaching 0 indicate no correlation while r values approaching 1 indicate a strong correlation). (A-C) DNA content and nuclear size were assessed in all germ cell nuclei located 0–60 µm (1–13 gcd) from the distal end in 78 wild-type adult gonads (15,468 nuclei total). (A) DNA content is plotted against nuclear size of corresponding nucleus. (B,C) DNA content (B) or nuclear size (C) as a function of position.
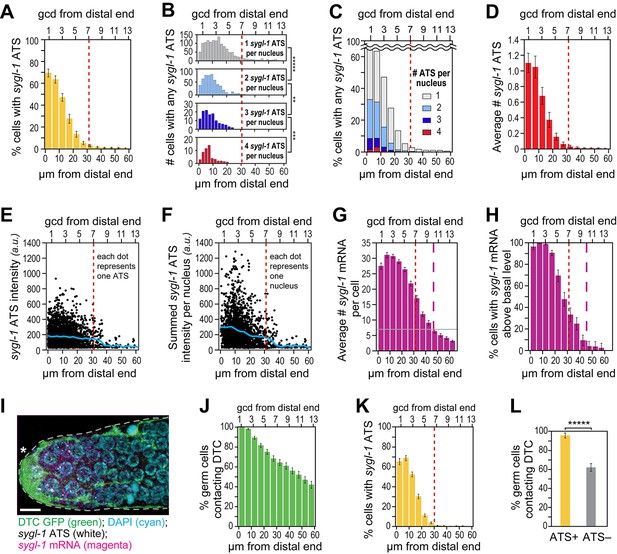
The sygl-1 transcriptional response is spatially graded.
(A–H and K,L) sygl-1 smFISH signals as a function of position along the distal-proximal axis of adult gonads. Positions were measured at 5 μm (A,C–K) or 2 µm (B) intervals from the distal end (x-axis below each graph), and translated to the conventional measure of germ cell diameters (gcd) from the distal end (x-axis above each graph). Error bars: standard error of the mean (SEM). n = 78 gonads. Red dashed line marks the site where the mean percentage of ATS-positive germ cells falls lower than 5%; purple dashed line marks the site where the mean percentage of mRNA-positive germ cells falls lower than 5%. (A) Gradient in percentage of germ cell nuclei with any number of sygl-1 ATS as a function of distance from the distal end in the wild-type adult gonad. By contrast, that percentage is essentially uniform in glp-1(gf) gonads (Figure 3—figure supplement 1A). (B) Numbers of cells with one, two, three or four sygl-1 ATS per nucleus as a function of distance from distal end. Total n = 2058 nuclei. **p<0.01, ***p<0.001, *****p<0.00001 by t-test. (C) Percentages of total ATS-positive nuclei that have one, two, three or four sygl-1 ATS per nucleus as a function of distance from distal end. Total n = 2058 nuclei. (D) Average number of sygl-1 ATS as a function of distance from the distal end. (E) Signal intensities of individual sygl-1 ATS do not change substantially in region of graded ATS (1–7 gcd, border marked with red line). Each dot represents a single sygl-1 ATS. The blue curve indicates mean ATS intensity. n = 2978 ATS. Signal intensities of individual sygl-1 ATS are comparable in glp-1(gf) gonads (Figure 3—figure supplement 1B). (F) Summed sygl-1 ATS intensities per nucleus were used as a measure for total sygl-1 nascent transcripts per nucleus and then plotted as a function of distance from the distal end, with each dot representing a single nucleus. The blue curve shows the mean for summed ATS intensities at each position relative to the distal end. n = 2058 nuclei. (G) The number of sygl-1 cytoplasmic mRNA per cell as a function of distance from the distal end. Boundaries of cells were estimated using 3-D Voronoi diagram (Figure 3—figure supplement 2A; see Materials and methods for details). The grey line marks the average basal sygl-1 mRNA level, which was calculated by averaging sygl-1 mRNA density over a region where mRNAs are both least abundant and no longer graded (the 40–60 μm interval in each gonadal image). A cell with abundant sygl-1 mRNA can reside next to a cell with few mRNA (Figure 3—figure supplement 2B–E). (H) Percentage of germ cells with sygl-1 cytoplasmic mRNAs above basal level as a function of distance from the distal end. (I) sygl-1 smFISH in adult distal gonad with DTC and its processes visualized with myristoylated GFP (green) and nuclei seen with DAPI (blue). sygl-1 exon probes mark cytoplasmic mRNAs (magenta) and intron probes (yellow) highlight ATS. Overlap is in white. Conventions and scale as in Figure 1E. Z-projection is shown. (J-L) Quantitative analyses of sygl-1 response in animals where DTC was marked with myristoylated GFP, n = 60 gonads at 24 hr post mid-L4 stage. Error bar: SEM. (J) Percentage of germ cells in contact with DTC or its processes as a function of distance from the distal end. (K) Percentage of germ cells with one or more sygl-1 ATS as a function of distance from the distal end. Red dashed line marks region defined as in Figure 3A. (L) Percentage of germ cells in contact with DTC or its processes within 30 µm from the distal end. Left bar, ATS-positive cells; right bar, ATS-negative cells or without ATS. *****p<0.00001 by chi-square test for independence.
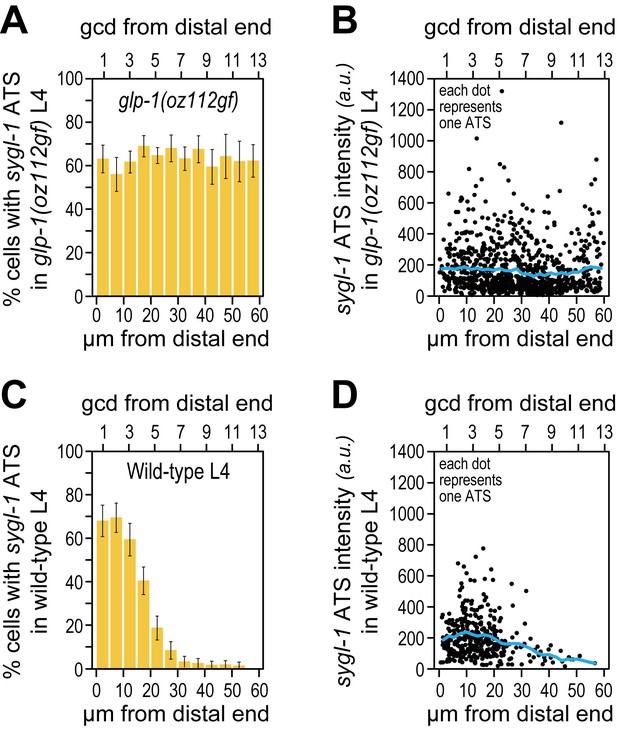
The sygl-1 transcriptional response is not spatially graded in glp-1(oz112gf) mutant germ cells.
(A,B) 3528 nuclei from 10 glp-1(oz112 gf) late larval (mid-L4) gonads were used for this analysis. Adult glp-1(GF) were not assayed here, because their distal germlines are mitotically quiescent and therefore not similar to wild type. (C,D) 1902 nuclei from 24 wild-type (mid-L4) gonads were used. (A,C) Percentage of germ cell nuclei with any sygl-1 ATS as a function of distance from distal end in glp-1(oz112 gf) (A) or wild type (C). (B,D) Intensities of individual sygl-1 ATS as a function of position in glp-1(oz112 gf) (B) or wild type (D), as in Figure 3E.
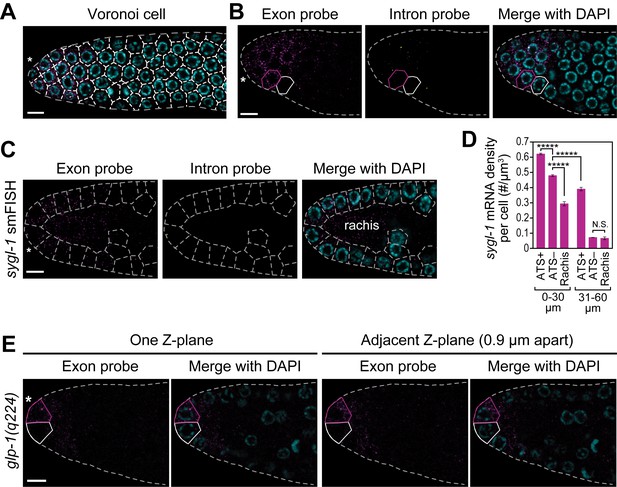
Cells with abundant sygl-1 mRNA can reside next to cells with little mRNA.
(A,C) Germ cells are interconnected via cytoplasmic bridges to a central cytoplasmic core or ‘rachis’ that extends the length of the gonad (Hirsh et al., 1976). Germ cell boundaries (dashed lines) were determined using a 3-D Voronoi diagram (Ledoux, 2007; Yan et al., 2010) in MATLAB. A representative section of germ cell boundaries (Voronoi cells) in one Z-plane is shown. DAPI marks nuclei (blue). Conventions and scales as in Figure 1E. (B,E) Germ cells with abundant sygl-1 mRNA (magenta outline) can reside next to a neighbor with very little sygl-1 mRNA (white outline) in wild-type (B) or glp-1(q224) (E) adult gonad. Conventions and scale as in Figure 1E. One Z-plane is shown. (D) sygl-1 mRNA density (total number of mRNAs in cell or region divided by total cytoplasmic volume in same cell or region). Cytoplasmic volume excludes nuclear volume; volume in rachis was calculated from non-cellular germline tissue in region spanning 0–20 µm (4–5 gcd). *****p<0.00001; n.s.: not significant by t-test.
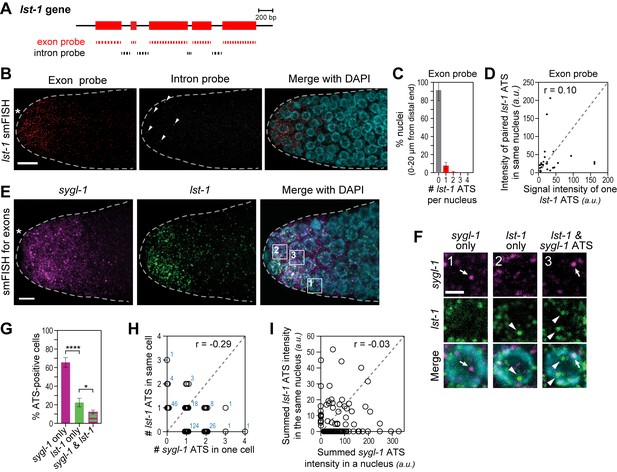
The lst-1 transcriptional response to Notch signaling.
(A) Schematic of lst-1 exon/intron structure. Exon-specific (red) and intron-specific (black) smFISH probes were labeled with different fluors (see Materials and methods). (B) lst-1 smFISH in distal gonad. Exon probes (red); intron probes (white). DAPI marks nuclei (blue). Arrowheads indicate ATS. Conventions and scale as in Figure 1E. (C) Percentage of nuclei with 0–4 lst-1 ATS per nucleus, calculated from 2107 nuclei in 36 gonads: 162 nuclei had one ATS, 15 had two ATS, 3 had three ATS and 2 had four ATS. Error bars: standard deviation. (D) Pair-wise comparisons of lst-1 ATS intensities within one nucleus (total of 30 ATS from 36 gonads). Each black dot represents one pairing. Grey dashed line indicates a perfect correlation (Pearson’s correlation coefficient r = 1); r indicates correlation coefficient from data. (E) Double-labeled smFISH against sygl-1 (magenta) and lst-1 (green) exons using distinct fluors (see Materials and methods). Conventions and scale as in Figure 1E. Full Z-projection is shown. (F) 10X magnification of nuclei within boxes in Figure 4E, right panel. Each panel shows a restricted Z-projection that only includes the corresponding nucleus. Arrow: sygl-1 ATS; arrowhead: lst-1 ATS. Scale bar: 2 μm. (G) Percentage of cells with sygl-1 ATS only, lst-1 ATS only or both sygl-1 and lst-1 ATS, out of all ATS-positive cells identified (n = 233 cells from 15 gonads). ****p-value<0.0001 and *p<0.05 by t-test. (H) Plot of nuclei possessing both sygl-1 and lst-1 ATS, with each open circle representing one nucleus (n = 233; 15 gonads). Overlapping data points (open circles) are spread using ‘jitter’ function in MATLAB. Blue numbers show how many nuclei of each type were found. For example, three nuclei had two lst-1 ATS and one sygl-1 ATS. Pearson’s correlation coefficient (r) is shown on top. Grey dashed line indicates a perfect correlation (r = 1). (I) Comparison of summed ATS intensities of sygl-1 and lst-1 within the same nucleus (data from 128 nuclei in 9 gonads). Each open circle represents a nucleus. r indicates Pearson’s correlation coefficient from data. Grey dashed line indicates a perfect correlation (r = 1).
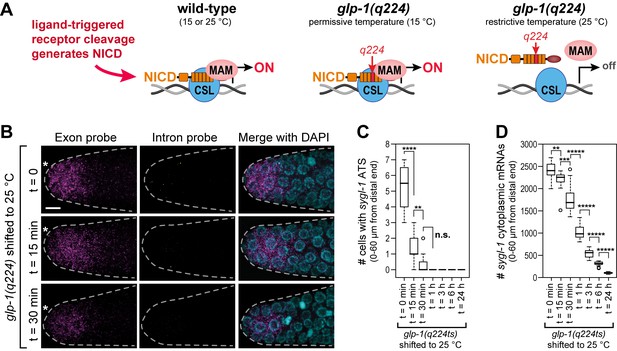
The sygl-1 transcriptional response is abolished in glp-1(q224) mutant germ cells at the restrictive temperature.
(A) Schematics of GLP-1/Notch-dependent nuclear complex in wild-type and glp-1(q224) temperature-sensitive mutant. Left, wild-type GLP-1/Notch intracellular domain (NICD) is cleaved from receptor and assembles into a complex in the nucleus to activate transcription; middle, glp-1(q224) NICD has a missense mutation in its fourth ankyrin repeat (red) but can assemble into the complex at the permissive temperature; right, glp-1(q224) NICD fails to assemble into the complex at the restrictive temperature. Effect of glp-1(q224) on complex formation from Petcherski and Kimble (2000). CSL, Notch-specific DNA-binding protein; MAM, mastermind-like transcriptional coactivator. (B-D) glp-1(q224) homozygous adults were raised at the permissive temperature (15°C) and then shifted to the restrictive temperature (25°C) for defined time intervals. Dissected gonads were probed for sygl-1 transcripts with smFISH. n ≥ 20 gonads for each time point. The wild type sygl-1 transcriptional response is essentially the same at 15, 20 and 25°C (Figure 5—figure supplement 1). (B) Z-projections after shift to the restrictive temperature for 0 min (top), 15 min (middle) and 30 min (bottom). sygl-1 exon probes (magenta); sygl-1 intron probes (yellow); and DAPI marks nuclei (blue). Conventions and scale as in Figure 1E. (C,D) smFISH signals for sygl-1 ATS (C) or mRNAs (D) were analyzed in all cells in the region of 0–60 µm (1–13 gcd) from distal end. Asterisks indicate p-value range by t-test. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; *****p<0.00001; n.s.: not significant (p>0.05).
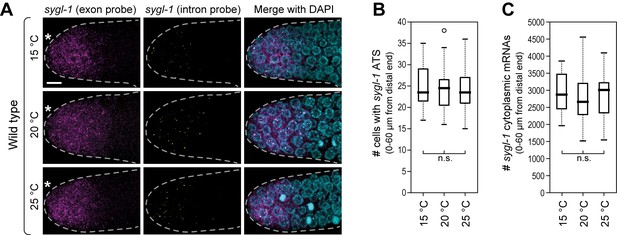
Wild-type sygl-1 transcriptional response is essentially the same at 15, 20 or 25°C.
(A) Gonads were dissected from wild-type adults raised at 15, 20 or 25°C (hours to adulthood vary at these temperatures; see Materials and methods for staging). Exon probes (magenta); intron probes (ATS; yellow); DAPI marks nuclei (blue). Conventions and scale as in Figure 1E. Z-projection is shown. (B,C) Analysis of smFISH signals in germ cells residing 0–60 μm (1–13 gcd) from distal end. n = 16 gonads for each temperature. n.s., not significant (p>0.05) by t-test.
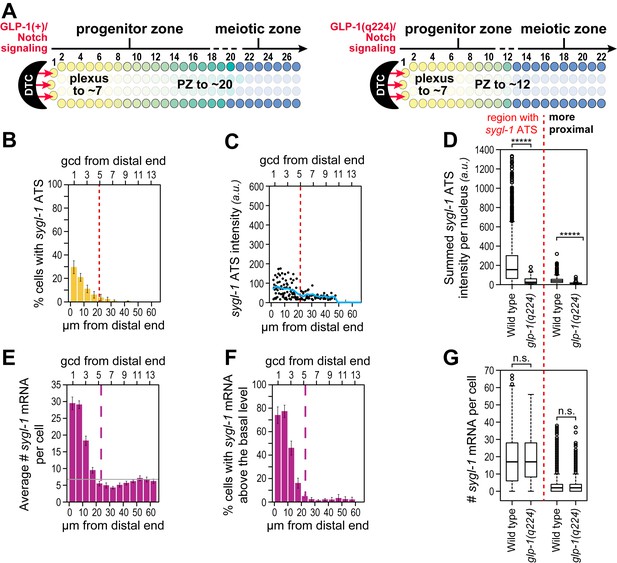
Weakened sygl-1 transcriptional response in glp-1(q224) at permissive temperature.
(A) The glp-1(q224) progenitor zone is smaller than wild-type (Cinquin et al., 2010; Fox and Schedl, 2015) though its plexus of DTC processes is essentially the same (Byrd et al., 2014). (B,C,E,F) sygl-1 transcriptional response in glp-1(q224) at 15°C (n = 20 gonads). Format, conventions and scales as in Figure 3A,E,G,H. (B) Percent cells with sygl-1 ATS as a function of position. (C) Individual sygl-1 ATS intensities as a function of position. (D) Summed sygl-1 ATS intensities per nucleus compared between wild type and glp-1(q224). Left of red line, comparison from region with graded sygl-1 ATS, which for wild-type was 0–30 µm and for glp-1(q224) was 0–20 µm; right of red line, comparison from region proximal to graded sygl-1 ATS, which for wild-type was 30–60 µm and for glp-1(q224) was 20–60 µm. *****p<0.00001 by t-test. (E) Average number sygl-1 mRNAs per cell as a function of position. The grey line marks the average basal sygl-1 mRNA level (see Materials and methods). (F) Percent cells with sygl-1 mRNAs as a function of position. (G) Number of sygl-1 mRNAs per cell compared between wild type and glp-1(q224). Regions compared were same as described for Figure 6D. n.s.: not significant by t-test.
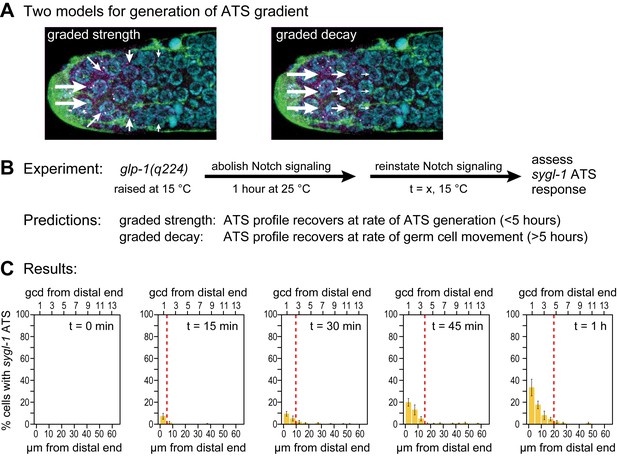
Graded sygl-1 transcriptional response likely reflects graded signaling strength.
(A) Two models to generate graded sygl-1 transcriptional response. Left, DTC signaling is graded within the niche; right, signaling is primarily at distal end but decays as cells move proximally within the niche. (B) Above, experiment to monitor rate of ATS pattern establishment after being abolished. Below, prediction made by each model. (C) Graded response is re-established within 1 hr. Percentage germ cells with sygl-1 ATS as a function of distance at 15 min intervals during reformation. Red line marks the region with sygl-1 ATS, as in Figure 3A. Conventions as in Figure 3A; n = 21 gonads for each time point.
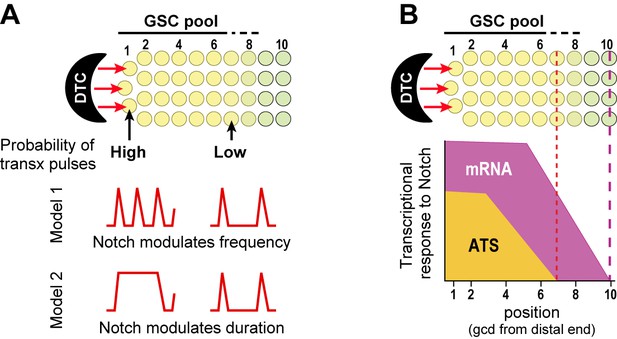
Gradient of Notch-dependent active transcription sites: models for underlying mechanism (A) and relation to mRNA distribution (B).
(A,B) Top, Notch signaling from the DTC niche maintains a pool of germline stem cells (GSCs) (Cinquin et al., 2010). (A) Notch signaling generates a gradient in the probability of transcriptional activation across the stem cell pool. ATS probability differences may result from modulation of ATS frequency (Model 1) or duration (Model 2). Red lines illustrate theoretical effects on transcriptional bursting at high (left) or low (right) probability. (B) Notch signaling from niche generates differently-shaped gradients of ATS and mRNA: mRNA abundance is essentially ungraded in the region where ATS probability is graded; the mRNA gradient extends further proximally than the ATS gradient; and more cells have mRNAs than ATS at any given position.
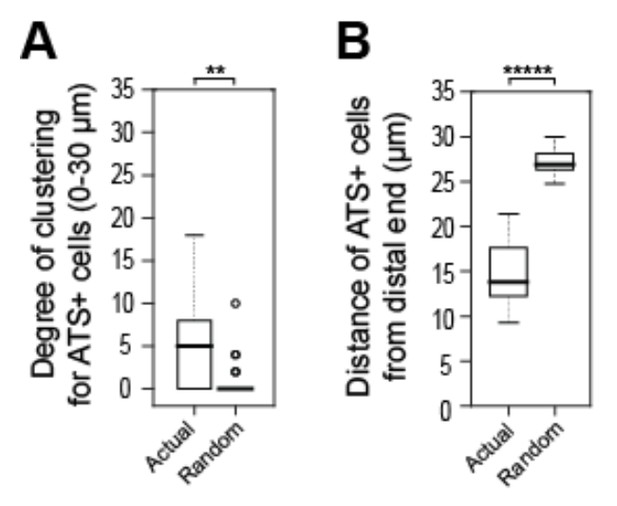
Tests to ask if positions of sygl-1 ATS is stochastic among cells in the GSC pool.
A, B) We recorded positions of sygl-1 ATS-positive and -negative cells within the region harboring ATS (0-30 µm from distal end) for each gonad examined (n=60 gonads) and then used a Poisson distribution to position ATS-positive cells randomly. We conducted this random simulation 100 times for each gonad to generate a dataset of “Complete Spatial Randomness” (CSR) for ATS-positive cells, which establishes a baseline for true stochasticity. (A) We compared the random CSR simulations with actual ATS data using 3-D Ripley’s H function, a widely used method for analyzing spatial patterns that assesses degrees of clustering. This analysis shows that the CSR clustering was 0, which differs significantly from the clustering seen with the actual ATS data. **p-value < 0.01 by t-test. n = 78 gonads. (B) We compared distance of ATS-positive cells from the distal end for the random CSR simulations and the actual data, and find that the CSR data differ significantly from the actual data. The actual ATS-positive cells are generally closer to the distal end than CSR data, which supports the non-random ATS clustering seen in A. *****p < 0.00001 by t-test. n = 2759 nuclei from 78 gonads.
Additional files
-
Source code 1
MATLAB codes for smFISH analysis.
- https://doi.org/10.7554/eLife.18370.016





