Genetic specification of left–right asymmetry in the diaphragm muscles and their motor innervation
Figures
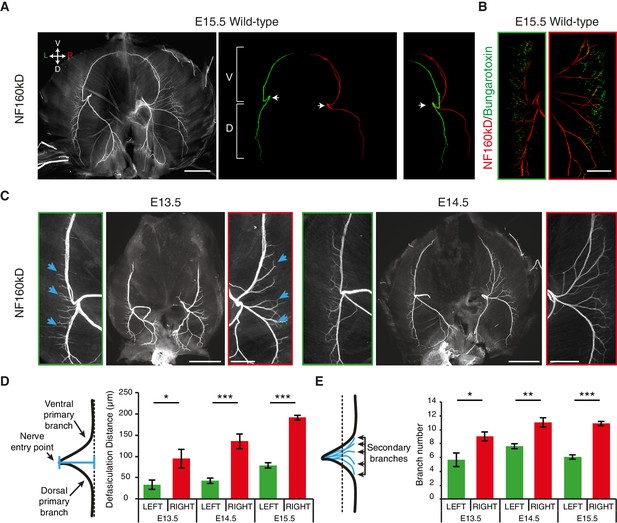
L/R asymmetries of the phrenic nerve patterns are established from the onset of diaphragm innervation.
(A) Neurofilament (NF) staining showing the branching patterns of the left and right phrenic nerves in whole-mount E15.5 mouse diaphragm. Left and right primary branches are pseudocolored (middle panel) in green and red, respectively. (See Figure 1—figure supplement 1A, for complete branch traces). L/R asymmetry is especially apparent after superimposing the left and right primary branches (right panel). Arrows point to the nerve entry points. Images are top views of the whole diaphragm, oriented as indicated in the top left hand corner of the left panel (V, Ventral; D, Dorsal; L, Left; R, Right). (B) NF and Bungarotoxin staining showing the asymmetry of acetylcholine receptor clusters and nerve domains on the left (left panel, green frame) and right (right panel, red frame) diaphragm muscles of an E15.5 embryo (see Figure 1—figure supplement 2 for quantification). (C) NF staining showing the patterns of left and right phrenic nerves at E13.5 and E14.5. Green- and red-framed panels show enlarged images of the left and right phrenic nerves, respectively. (D) Schematics showing the method used to quantify the defasciculation distance (shown in blue), from the nerve entry point to the dotted line and histogram of the defasciculation distance at E13.5, E14.5 and E15.5 (E13.5 — left 32.76 ± 11.01, right 94.82 ± 21.94, N = 9, p=0.0106; E14.5 — left 42.56 ± 4.16, right 135.71 ± 10.20, N = 8, p=0.00015; E15.5 — left 77.16 ± 7.32, right 188.51 ± 7.01, N = 18, p=4 E-10, Mann-Whitney). (E) Schematics showing the method used to quantify the secondary branch number by counting the number of NF-positive fascicles that crossed the dotted line positioned at 80% of the defasciculation distance and histogram of the secondary branch number at E13.5, E14.5 and E15.5 (E13.5 — left 5.55 ± 0.96, right 8.88 ± 0.65, N = 9, p=0.0288; E14.5 — left 7.5 ± 0.38, right 10.88 ± 0.69, N = 8, p=0.00117; E15.5 — left 5.94 ± 0.31, right 10.7 ± 0.3, N = 18, p=2.35 E-7, Mann-Whitney). Histograms show the mean ± SEM for each stage. Scale bars: 200 μm (A,C); 100 μm (B). Numerical values used to generate the graphs are accessible in Figure 1—source data 1.
-
Figure 1—source data 1
Left and right measures of the defasciculation distance and branch number in E13.5, E14.5 and E15.5 mouse embryos.
This file provides the mean, SEM, statistical report and individual measures used to create the histograms shown in Figure 1D, E. Defasciculation distances measured on left and right hemi-diaphragms are shown in the first sheet and numbers of secondary branches between the two primary branches on the second sheet.
- https://doi.org/10.7554/eLife.18481.004
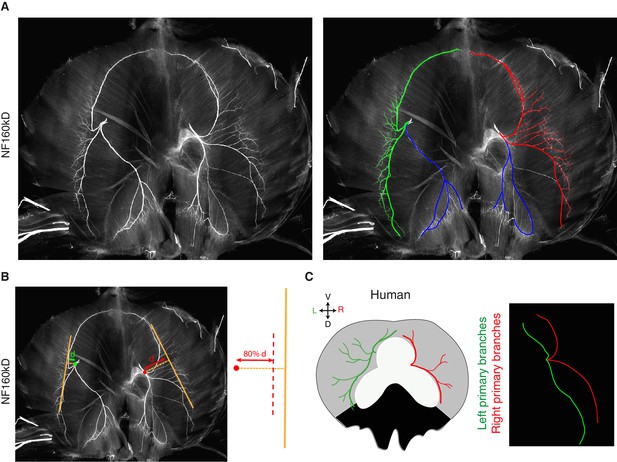
Phrenic nerve patterns and quantification in mice and L/R nerve asymmetry in a human diaphragm.
(A) NF staining showing the branching patterns of the left and right phrenic nerves in a whole-mount E15.5 mouse diaphragm. In the right panel, the primary, secondary and tertiary branches of the left and right phrenic nerves are traced in green and red, respectively. The left and right crural phrenic nerves are traced in blue. (B) Example of quantification on an NF-labelled wholemount diaphragm. (C) L versus R differences of nerve pattern in human diaphragms, the left (green) and right (red) branches are innervating the lateral muscle (grey regions). Reproduced from the original figure of Hidayet et al. (1974). The L/R asymmetry is especially apparent after superimposing the left and right nerve pattern (right panel).
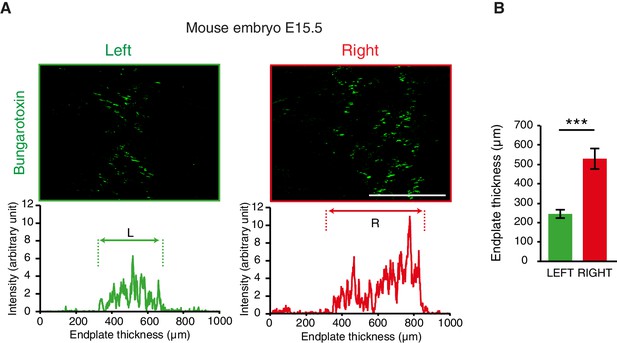
L/R differences of acetylcholine clusters during synaptogenesis.
(A) Bungarotoxin staining on the left and right sides of an E15.5 mouse diaphragm and plot profile showing the asymmetry of the clusters of acetylcholine receptor indicative of the endplate thickness (left and right in green and red, respectively). (B) Histogram showing the quantification of the endplate thickness (left: 254.9 ± 22.2, right: 529.3 ± 53.0, N = 11, p=0.00097, Wilcoxon signed rank). Scale bars: 200 μm. Numerical values used to generate the graphs are accessible in Figure 1—figure supplement 2—source data 1.
-
Figure 1—figure supplement 2—source data 1
Left and right endplate thicknesses measured from Bungarotoxin labeling in E15.5 mouse embryos.
This file provides the mean, SEM, statistical report and individual measures used to create the histograms shown in Figure 1—figure supplement 2B.
- https://doi.org/10.7554/eLife.18481.007
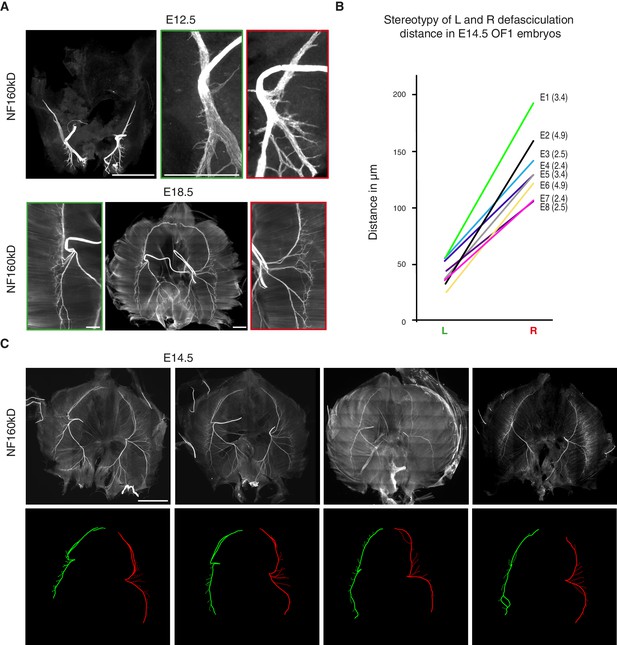
Stereotypy and variability of L/R asymmetry of the phrenic nerve patterns.
(A) NF staining showing the patterns of left and right phrenic nerves at E12.5 and E18.5. Green- and red-framed panels show enlarged images of the left and right phrenic nerves, respectively. Note that at E12.5, the dorsal and ventral branches have already split with different angles on the left and right sides (left: 166° ± 4°; right: 132° ± 4°; N = 8). (B) Ladder graph showing the stereotypy of the left and right defasciculation distances for eight E14.5 embryos (E1 to E8) (ratio shown in brackets). (C) NF staining of whole-mount diaphragms from E14.5 mouse embryos showing the phrenic nerve pattern variability at that stage. Left (green) and right (red) primary branch traces are shown in the lower panels. Scale bars: 200 μm, 100 μm for enlargement panels. The numerical values used to generate the graphs are accessible in Figure 1—figure supplement 3—source data 2.
-
Figure 1—figure supplement 3—source data 2
Paired analysis of left and right defasciculation distances in E14.5 mouse embryos.
This file provides the individual measurements used to create the ladder graph shown in Figure 1—figure supplement 3B.
- https://doi.org/10.7554/eLife.18481.009
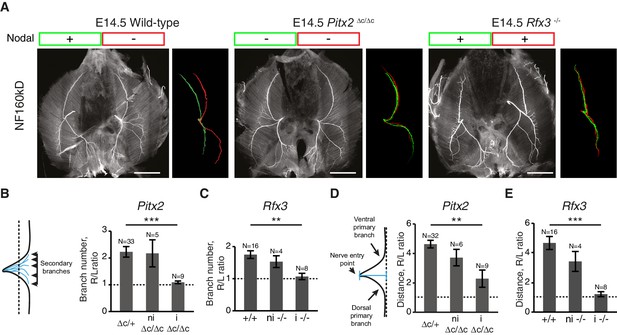
L/R asymmetries of the phrenic nerve patterns require Nodal signaling.
(A) NF staining of E14.5 diaphragms from wild-type, Pitx2∆C/ΔC and Rfx3–/– embryos with the respective superimposed L/R nerve pattern and the Nodal expression. (B–C) Schematic of the secondary branches quantification and histograms of the R/L ratios of secondary branches: Pitx2∆C/+and Pitx2+/+ 2.23 ± 0.20, versus Pitx2∆C/∆C with lung isomerism 1.09 ± 0.05, p=4.493E-5 (B); Rfx3+/+ and Rfx3–/+ 1.75 ± 0.12, versus Rfx3–/– with lung isomerism 1.07 ± 0.10, p=0.002884, Mann-Whitney (C). (D–E) Schematic of the defasciculation distance measurements and histograms of the R/L ratios of defasciculation distance for: Pitx2∆C/+and Pitx2+/+ 4.63 ± 0.26, versus Pitx2∆C/∆C with visceral isomerism: 2.28 ± 0.59, p=0.001268, Mann-Whitney (D); Rfx3+/+ and Rfx3–/+ 4.62 ± 0.43, versus Rfx3-/- with visceral isomerism 1.35 ± 0.19, p=2.719E-6, Mann-Whitney (E). Note that there is no lung isomerism in wild-type embryos. Histograms show the mean ± SEM. Numbers above bars indicate the number of embryos analysed. ni, non-isomeric (embryos that did not exhibit visceral isomerism); i, isomeric. Scale bars: 200 μm. Numerical values used to generate the graphs are accessible in Figure 2—source data 1.
-
Figure 2—source data 1
Ratios of the defasciculation distance and branch number in E14.5 mouse embryos of Pitx2C and Rfx3 lines.
The file provides the mean, SEM, statistical report and individual values used to create the histograms shown in Figure 2B, C, D and E. Branch numbers ratios found in Pitx2C∆C/+ and Pitx2C∆C/∆C embryos as well as those found in Rfx3+/+ and Rfx3–/– embryos are shown on the first and second sheet, respectively. Defasciculation distance ratios measured in Pitx2C∆C/+ and Pitx2C∆C/∆C embryos or in Rfx3 +/+ and Rfx3–/– embryos are shown on the third and fourth sheet, respectively.
- https://doi.org/10.7554/eLife.18481.011
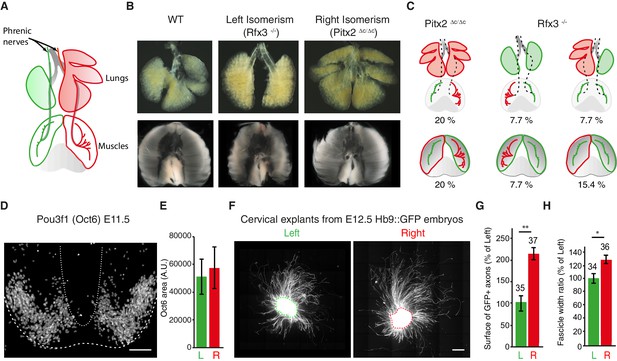
The asymmetry of phrenic circuits results from an intrinsic neuronal program.
(A) Schematic representation of the organisation of the phrenic nerves as they pass through the lungs and reach the diaphragm. (B) Photomicrographs of the expected L/R asymmetry of lungs and diaphragm muscles at E14.5 in wild-type embryos and the altered L/R asymmetry observed in the Rfx3–/– and Pitx2∆C/∆C mutant embryos. Quantification of diaphragm muscle asymmetry: Pitx2+/+ and Pitx2∆C/+ 6.25 ± 0.68, N = 20, versus Pitx2∆C/∆Ciso 0.26 ± 0.6, N = 6; Rfx3 +/+ and Rfx3-/+ 7.02 ± 0.74, N = 17 versus Rfx3–/-–iso 0.72 ± 1.6, N = 7 (see methods). (C) Schematic representation of L/R asymmetries in the lungs, diaphragm muscles and phrenic nerves. A colour code is used to show the uncoupling occurring between phrenic nerve and lung asymmetries or phrenic nerve and diaphragm muscle asymmetries. Any structure represented in green is indicative of its left characteristics, whether it is observed on the left or the right side of the embryo, whereas red structures represent right characteristics. (D) Pou3f1 (Oct6) staining showing the pool of phrenic motoneurons, projection formed by serial sections of the entire cervical region of an E11.5 spinal cord embryo. (E) Histogram showing the area positive for the Pou3f1 (Oct6) labeling in the left and right cervical motoneuron domains (N = 3, p=0.5, Wilcoxon signed rank). (F) GFP staining of ventral cervical spinal cord explants from E12.5 HB9::GFP embryos; the dashed line is indicative of the explant border. (G) Quantification of the area occupied by GFP-positive axons for left and right explants (left — 100% ± 17.4; right — 214% ± 30.2, p=0.0045, Mann-Whitney). (H) Quantification of the width ratio (see Figure 3—figure supplement 1 for quantification details) (left —100% ± 7.3; right — 127% ± 8.0, p=0.0127, Mann-Whitney). Numbers above bars indicate the numbers of explants analysed. Histograms show the mean ± SEM. Scale bars: 100 μm (D), 200 μm (F). Numerical values used to generate the graphs are accessible in Figure 3—source data 1.
-
Figure 3—source data 1
Pool size and in vitro axon growth from left and right motoneurons.
This file provides the mean, SEM, statistical report and individual values used to create the histograms shown in Figure 3D, G and H. Left and right Oct6-labeled surfaces are shown on the first sheet. The surface and the defasciculation index of motoneurons axons (GFP+) growing from left and right explants are shown on the second and third sheet, respectively.
- https://doi.org/10.7554/eLife.18481.013
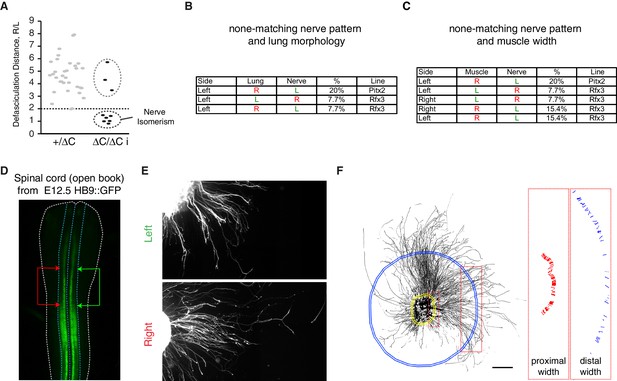
Uncoupling between lung or muscle and nerve asymmetry and intrinsic L/R differences of axon growth from cultured cervical motoneuron explants.
(A) Distribution of the defasciculation distance ratio amongst E14.5 Pitx2+/ΔC and Pitx2∆C/ΔC embryos. Values for Pitx2+/+ and Pitx2+/ΔC embryos are pooled. Two clearly separated groups are visible amongst the Pitx2∆C/ΔC embryos, one below the dashed line composed of embryo with nerve isomerism and one above the line composed of normally asymmetric nerves. (B–C) Table showing the uncoupling observed between the nerve pattern and lung morphology (B) or the nerve pattern and muscle morphology (C) in the Pitx2C and Rfx3 mutant embryos and its frequency. (D) Photomicrograph of the GFP signal observed from HB9::GFP spinal cord. The blue dashed line outlines the ventral spinal cord and the white dashed line outlines the spinal cord. The arrows delimit the area of interest. (E) Photomicrographs illustrating the defasciculation behaviours of GFP-labelled axons extending from left (top panel) or right (bottom panel) ventral cervical spinal cord explants from E12.5 HB9::GFP embryos. (F) Quantification method used to calculate the area occupied by GFP=positive axons and defasciculation index. The binary image (left panel) shows the GFP-positive area extracted with the ImageJ plugin NeuriteJ that was used to calculate the area. The proximal (yellow in left panel) and the distal (blue in left panel) selections are created using the same plugin. The width of each fascicule crossing the proximal and distal selections was measured and the defasciculation index calculated. Scale bar: 300 μm (F). Numerical values used to generate the graphs are accessible in Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
Distribution of defasciculation ratios in the Pitx2C mouse line.
This file provides the individual values used to generate the graph plot shown in Figure 3—figure supplement 1A.
- https://doi.org/10.7554/eLife.18481.015
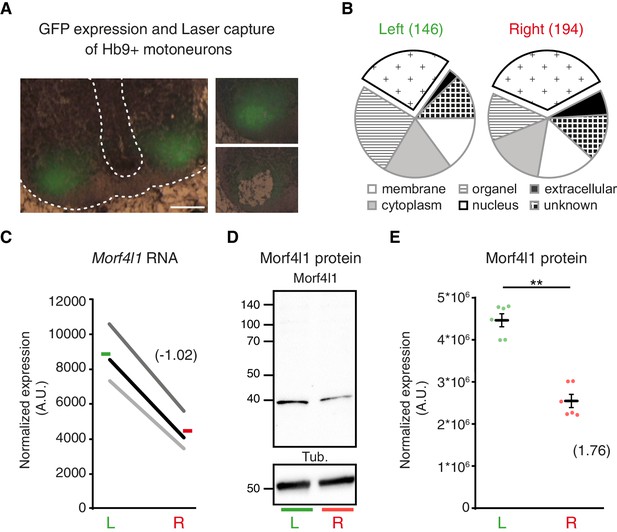
L/R molecular signature of cervical motoneurons.
(A) Transverse sections of E11.5 Hb9::GFP embryo cervical spinal cord, illustrating the areas used for laser-capture microdissection. (B) Pie charts showing the proportion of left-enriched and right-enriched genes according to their Gene Ontology ‘cellular component’ terms. The ‘nucleus’ component is detached from the pie. (C) Ladder graph showing the left and right expression of Morf4l1 in three embryos. Average L/R fold-change shown in brackets. (D) Immunodetection of Morf4l1 and loading control tubulin (Tub.) in left and right ventral cervical spinal cord tissues. (E) Graph showing normalized protein levels of Morf4l1 in left and right ventral cervical spinal cords from E11.5 mouse embryos. Individual values observed for the six western-blots (dots) and mean ± SEM are represented (L/R ratio: 1.81 ± 0.163, L versus R; p=0.0022, Wilcoxon signed rank). Average L/R fold-change shown in brackets. Scale bar: 100 μm. Numerical values used to generate the graphs are accessible in Figure 4—source data 3.
-
Figure 4—source data 1
List of enriched genes in the left cervical motor neurons of HB9::GFP embryos at E11.5.
Genes are included on this list if the average change in expression was > 1.5 (or −0.58< in log2) between the left and right sides. The listed genes had the same enrichment trend in all embryos with a fold-change > 1.5 in at least in two embryos.
- https://doi.org/10.7554/eLife.18481.017
-
Figure 4—source data 2
List of enriched genes in the right cervical motor neurons of HB9::GFP embryos at E11.5.
Genes are included in the list if the average change in expression was > 1.5 (or > 0.58 in log2) between the right and left sides. The listed genes had the same enrichment trend in all embryos with a fold-change superior to 1.5 in at least in two embryos.
- https://doi.org/10.7554/eLife.18481.018
-
Figure 4—source data 3
Lateralization expression of Morf4l1 in cervical motoneurons.
This file provides the statistical reports and individual values used to create the ladder graphs shown in Figure 4C and E. RNA expression is shown on the first sheet. Normalized protein levels are shown on the second sheet.
- https://doi.org/10.7554/eLife.18481.019
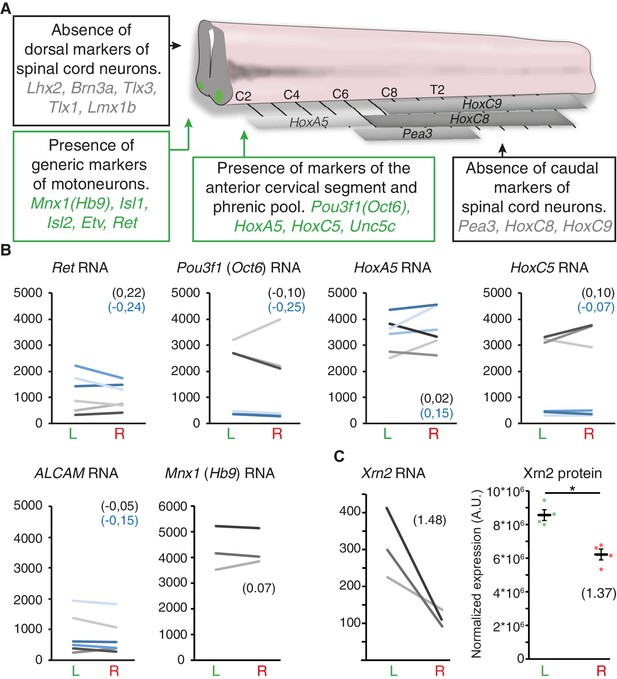
Symmetric expression of phrenic motoneuron markers, and lateralized Xrn2 expression.
(A) Schematic representation of the spinal cord that depicts the expression domain of Hox genes and brachial-specific Pea3 (Etv4) transcription factor. Boxes represent present/absent call tests indicating that markers of dorsal spinal cord and brachial motoneurons are absent. By contrast, generic markers of motoneurons as well as markers that are enriched in cervical and phrenic motoneurons are present in all three embryos. (B) Ladder graphs showing the normalized expression signals in the left and right laser-captured samples of the three embryos for probes that detect Ret, Pou3f1 (Oct6), HoxA5, HoxC5, ALCAM and Mnx1 (Hb9) RNA. The average log2 (R/L ratios) are indicated in brackets in black for probe one and blue for probe 2. None of these probes showed significant L/R difference according to the threshold used (see Materialsand methods). (C) Ladder graph of Xrn2 RNA expression in left and right samples from the three embryos (left panel), average log2 (R/L ratio) indicated in brackets. Graph showing the left and right normalized protein levels of Xrn2 in ventral cervical spinal cord from E11.5 embryos. Values of the four western-blots (dots) and mean ± SEM are represented (L/R fold-change 1.37 ± 0.13, L versus R, p=0.028; Mann-Whitney) (right panel). Numerical values used to generate the graphs are accessible in Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
RNA level of motoneuron markers and asymmetric expression of Xrn2.
This file provides the individual values used to create the ladder graphs shown in Figure 4—figure supplement 1B and C. RNA expression of motoneuron markers is shown on the first sheet. RNA and normalized protein levels of Xrn2 are shown on the second and third sheets.
- https://doi.org/10.7554/eLife.18481.021
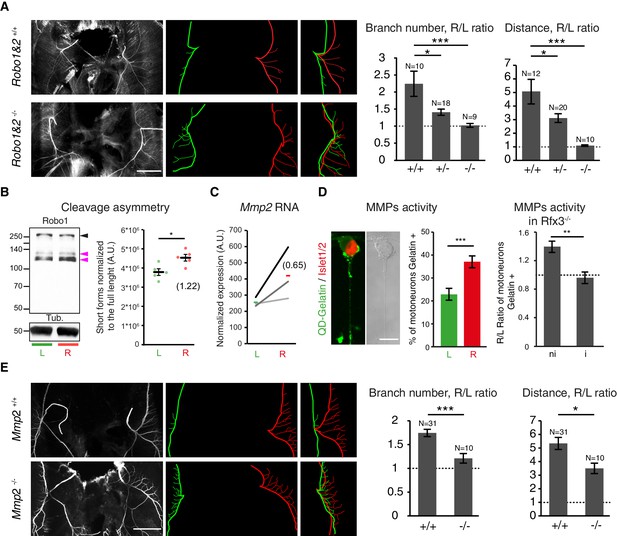
Slit/Robo signalling and MMP2 control asymmetry of L/R phrenic nerves.
(A) NF staining of E14.5 diaphragm from Robo1+/+ and Robo2+/+ and Robo1–/– and Robo2–/–- embryos, left and right primary branches are pseudocolored in green and red,respectively, and superimposed to show the lack of asymmetry in the Robo1 and 2–/– embryos. Histogram showing the branch number and the defasciculation distance in Robo1+/+ and Robo2+/+, Robo1+/– and Robo2+/– and Robo1–/– and Robo2–/– embryos (R/L branch ratio: Robo1+/+ and Robo2+/+ 2.30 ± 0.37, versus Robo1–/– and Robo2–/– 1.06 ± 0.06; p=0.00048; R/L distance ratio: Robo1+/+ and Robo2+/+ 4.99 ± 0.89, versus Robo1–/– and Robo2–/– 1.05 ± 0.07; p=3E-6, Mann-Whitney). (B) Immunodetection of Robo1 and loading control (Tub) in left and right HB9::GFP ventral cervical spinal cord and distribution of the relative amount of the two shorter forms (pink arrowheads) to the full-length form (black arrowhead). The graph shows the normalized left and right values obtained for the five western-blots (dots, 6–8 embryos per sample) and the mean ± SEM (R versus L: p=0.01587, Wilcoxon singed rank); average fold-change is shown in brackets (1.22 ± 0.10). Normalization between lines was done on the Robo1 long form. (C) Ladder graph showing the left and right expression of Mmp2 detected by microarray in three embryos. Average Log2(R/L ratio) shown in brackets. (D) Photomicrograph of cultured ventral cervical spinal cord motoneuron. The combination of in situ zymmography with DQ-Gelatin and Islet1/2 staining enables the identification of motoneuron with MMP gelatinase activity. Histogram showing the amount of motoneuron with gelatinase activity in left and right samples (left 23.37% ± 2.7, N = 792 versus right 37.94% ± 2.1, N = 797; p=0.00109, Mann-Whitney). Histogram showing the gelatinase activity measured in cultures from Rfx3–/– embryos with symmetric lungs (Iso) and in cultures from Rfx3+/+, Rfx3+/– embryos (Rfx3 wt: — 1.4 ± 0.08; Rfx3 iso — 0.96 ± 0.08, p=0.0013, Mann-Whitney). (E) NF staining of E14.5 diaphragms from wild-type and Mmp2–/– embryos. Left (green) and right (red) primary and secondary branch traces shown in the middle panel are superimposed in the right panel to compare the left and right patterns. Histograms showing the R/L ratios of branch number and defasciculation distances. Ratio of secondary branches: Mmp2+/+ and Mmp2–/+ 1.74 ± 0.07, versus Mmp2–/– 1.21 ± 0.10; p=0.00029; defasciculation distance: Mmp2+/+ and Mmp2–/+ 5.33 ± 0.44, versus Mmp2–/– 3.49 ± 0.38; p=0.022, Mann-Whitney. Scale bar: 200 μm (A,E), 10 μm (D). Numerical values used to generate the graphs are accessible in Figure 5—source data 1.
-
Figure 5—source data 1
Slit/Robo signalling controls asymmetry of L/R phrenic nerves and Robo1 exhibits different processing levels in left and right cervical motoneurons.
This file provides the statistical report and individual values used to create the histograms and ladder graphs shown in Figure 5A, B, C, D and E. The ratios of branch numbers and defasciculation distances in Robo1+/+ and Robo2+/+, Robo1+/– and Robo2+/– and Robo1–/– and Robo2–/– embryos are shown on the first and second sheets. The third sheet contains left and right normalized values of short Robo1 forms presented in the graph of Figure 5B. RNA levels of Mmp2 are shown on fourth sheet. The percentage of left and right motoneurons (Islet+) exhibiting gelatinase activity from wild-type embryos are shown on fifth sheet and the ratio found in Rfx3–/– embryos with lung isomerism and Rfx3+/+ and Rfx3+/– on the sixth sheet. Branch number and defasciculation distance ratio measured in Mmp2+/+ and Mmp2–/– embryos are shown on the seventh and eighth sheets.
- https://doi.org/10.7554/eLife.18481.023
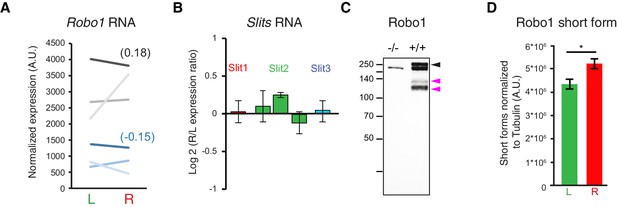
Post-translational regulation of Robo1.
(A) Ladder graph of Robo1 RNA expression detected by the two probes present on the microarray for each of the three embryos (probe1 — black; probe2 — blue; average Log2(R/L ratio) shown in brackets). (B) Histogram showing the average R/L ratio of expression (in log2) assessed by the microarray probes targeting Slit1, Slit2 and Slit3. Error bars represent SEM. (Note that 1.5 fold-change gives 0.5849 in log2.) (C) Immunodetection of Robo1 in spinal cord lysates of Robo1–/– and Robo2–/– and wild-type tissues. The antibody detects three specific bands. Black arrowhead points to the expected full-length Robo1 and the two pink arrowheads point to the two shorter forms. (D) Graph shows the normalized left and right values obtained for the four western-blots (6–8 embryos per sample) and the mean ± SEM (R versus L: p=0.028, Mann-Whitney; average fold-change is 1.22 ± 0.11). Normalization between lines was done on the tubulin band. Numerical values used to generate the graphs are accessible in Figure 5—figure supplement 1—source data 1.
-
Figure 5—figure supplement 1—source data 1
Post-translational regulation of Robo1 and biased expression of Mmp2.
This file provides the statistical report and individual values used to create the histograms and ladder graphs shown in Figure 5—figure supplement 1A and B. Left and right levels or ratios for the Robo1 and the Slits transcripts are shown on the first and second sheet, respectively. The third sheet contains left and right tubulin normalized values of short Robo1 forms presented in the graph of Figure 5—figure supplement 1D.
- https://doi.org/10.7554/eLife.18481.025
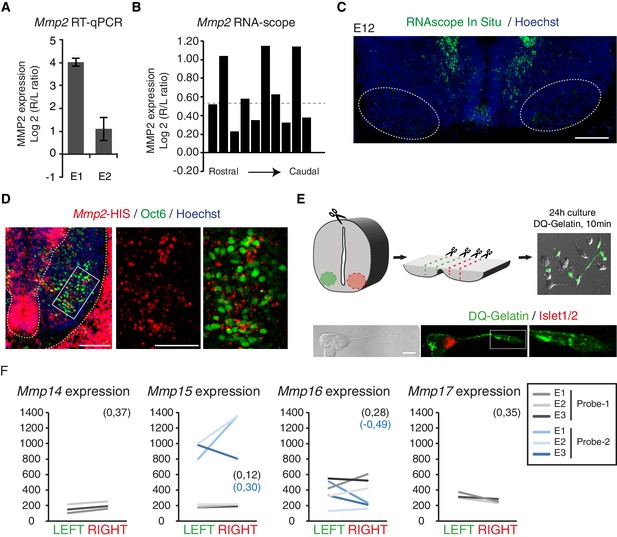
Asymmetric expression of MMP2.
(A) Histogram showing the average ratio of Mmp2 RNA expression assessed by qPCR in cervical motoneurons from two E11 embryos. Expression normalized to GAPDH. (B) Histogram showing the R/L ratio (in log2) of the surface labelled by the RNAscope MMP2 probe in the motoneuron region. Each bar shows the R/L ratio for one section of a series of serial sections that cover the entire cervical spinal cord region. The dashed line highlights the log2 value that corresponds to a 1.5-fold change. (C) Mmp2 RNAscope in situ hybridization on an E12 transversal spinal cord section at cervical levels. (D) Mmp2 in situ hybridization combined with Pou3f1 immunolabeling on E11 transversal spinal cord sections. Enlarged panels of the motoneuron domain (right) show that Mmp2 transcripts are detected within the Pou3f1-positive domain. (E) Schematics of the dissection and the in situ zymography (ISZ) procedure for E12.5 ventral cervical spinal cord. Cleavage-induced fluorescence of DQ-Gelatin (green) is overlaid over the phase contrast image. Islet1-positive motoneurons exhibit gelatinase activity in different cellular regions, including the axon and the growth cone (lower right panel). (F) Ladder graph showing the expression signals in the left and right laser-captured samples of the three embryos detected with the microarray Mmp14, Mmp15, Mmp16 and Mmp17 probes, average Log2(R/L ratio) shown in brackets. Scale bars: 100 μm (C) and (D left panel); 200 μm (D right panel) and 10 μm (E). Numerical values used to generate the graphs are accessible in Figure 5—figure supplement 2—source data 2.
-
Figure 5—figure supplement 2—source data 2
Asymmetric expression of Mmp2 in cervical motoneurons and expression of other MMPs.
Mmp2 expression ratios quantified by qRT-PCR and quantitative in situ hybridization (RNAscope) are presented on the first and second sheets, respectively. The third sheet shows the microarray data used to generate the ladder graphs shown in Figure 5—figure supplement 2F.
- https://doi.org/10.7554/eLife.18481.027
Additional files
-
Supplementary file 1
MMP2 addition on cervical ventral spinal cord increases Robo1 short form.
This file provides the statistical report and individual values used to calculate the fold change presented in the discussion.
- https://doi.org/10.7554/eLife.18481.028
-
Supplementary file 2
Robo1 short form ratio in Mmp2–/– cervical spinal cord.
This file provides the statistical report and individual values for the fold changes presented in the discussion.
- https://doi.org/10.7554/eLife.18481.029





