Maintenance of age in human neurons generated by microRNA-based neuronal conversion of fibroblasts
Figures
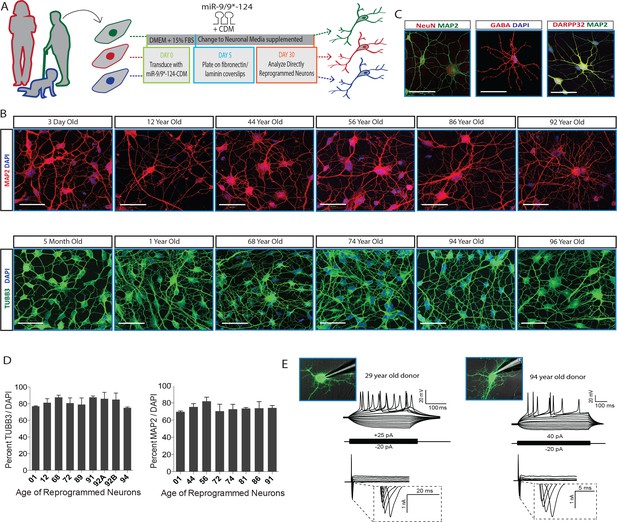
MicroRNA-mediated direct neuronal conversion applied to fibroblasts across the age spectrum.
(A) Schematic diagram of neuronal conversion of human fibroblast samples from individuals ranging from three days to 96 years of age. Primary fibroblasts were transduced with microRNA-9/9*-124 and the transcription factor cocktail CTIP2, DLX1/2, MYT1L (CDM) and analyzed for neuronal properties after 30 days. (B) Expression of pan-neuronal markers MAP2 (top) and TUBB3 (bottom) after neuronal conversion of human fibroblasts ranging in age. Scale bar = 50 µm. (C) Expression of pan-neuronal marker NeuN and medium spiny neuron-specific markers GABA and DARPP32 in reprogrammed neurons from fibroblasts aged 91-, 72-, 92-years respectively. (D) Immunostaining analysis of percentage of reprogrammed neurons positive for neuronal markers TUBB3 and MAP2 over DAPI signals (n = 200–300 per cell line). (E) Representative whole-cell current clamp recording of converted neurons from young (29 years old, left) and old (94 year old donor, right) donors. Converted human neurons in monoculture displayed multiple action potentials in response to step current injections at four weeks post-transduction. All reprogrammed neurons from old fibroblasts recorded (n = 11) fired APs in response to current injections (top). Representative traces of fast-inactivating inward currents recorded in voltage-clamp mode. Voltage steps ranged from +10 to +70 mV (bottom).
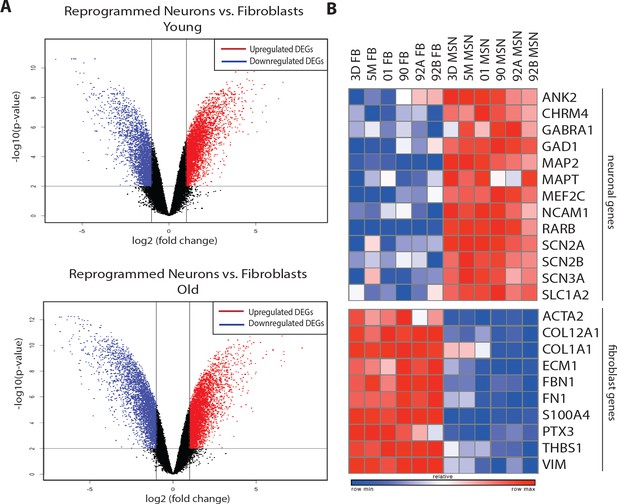
Transcriptome analyses between converted neurons and fibroblasts of young and old age groups.
(A) Volcano plots show global transcriptomic differences between reprogrammed neurons compared to starting human fibroblasts from young (three day, five month and one year-old, top) and old (90, 92 and 92 year-old, bottom) donors. Upregulated DEGs (in red) and downregulated DEGs (in blue) after direct neuronal reprogramming are shown. (B) Heatmap of representative DEGs: upregulation of neuronal genes (top) and downregulation of fibroblast associated genes (bottom) in reprogrammed neurons from both young and old fibroblasts (right) when compared to corresponding fibroblasts (left).
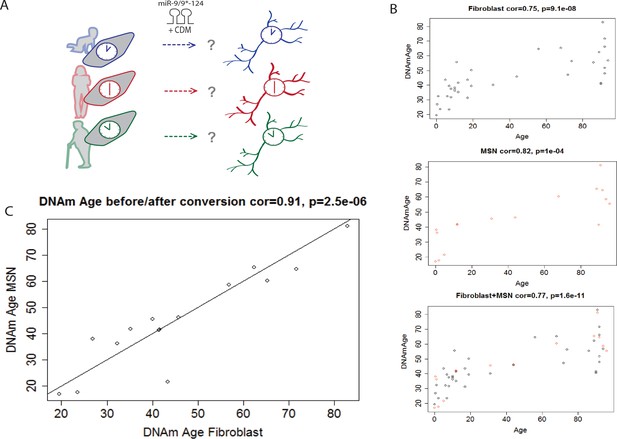
Conservation of the epigenetic clock of reprogrammed neurons from human fibroblasts.
(A) Schematic diagram representing the hypothesis that the epigenetic clock of fibroblasts from different age groups is conserved in reprogrammed neurons after miR-mediated neuronal conversion. (B) Top: Predicted ages based on the methylation status (DNAm age) of fibroblasts plotted against the actual ages of fibroblasts (correlation = 0.75, p=9.1e-08). Middle: Predicted DNAm ages of reprogrammed neurons against actual ages of starting fibroblast, correlation = 0.82, p=1e-04. Bottom: Combined plot of DNAm ages of fibroblasts and DNAm ages of reprogrammed neurons against actual ages (correlation = 0.77, p=1.6e-11). (C) DNAm age of reprogrammed neurons plotted against the DNAm ages of the corresponding, starting fibroblast ages, correlation = 0.91 p=2.5e-06.
-
Figure 2—source data 1
Output for sample information and DNAm ages for fibroblasts and reprogrammed neurons compared to original age.
- https://doi.org/10.7554/eLife.18648.006
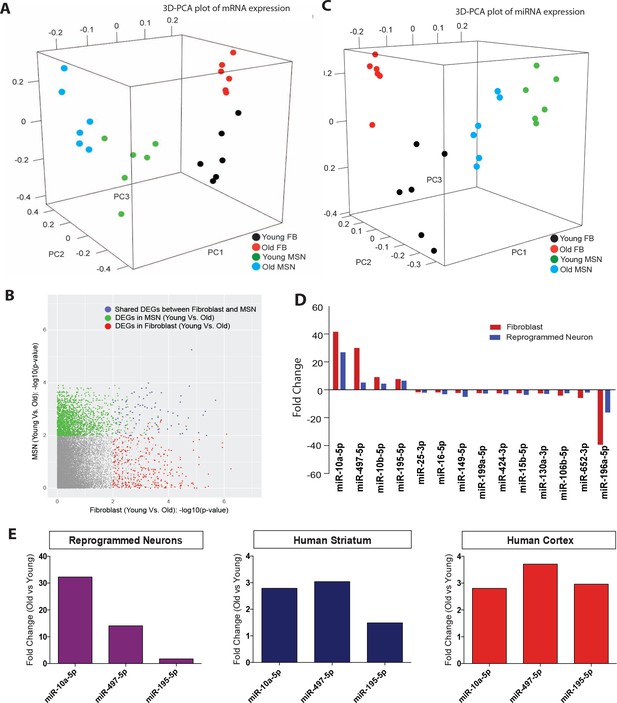
Age-associated changes in transcriptome and microRNA profiles in reprogrammed neurons.
(A) Principle component analysis (PCA) of transcriptome profiling of reprogrammed neurons from young fibroblasts aged three days, five months one year (green) and from old fibroblasts aged 90, 92, and 92 years (blue) alongside corresponding young fibroblasts (black) and old fibroblasts (red) (FDR < 0.05). (B) Differentially expressed genes (DEGs) with age in fibroblasts (x axis) and in reprogrammed neurons (y axis). Age-associated DEGs in reprogrammed neurons shown in green, DEGs in fibroblasts shown in red, and commonly shared DEGs with age in both fibroblasts and reprogrammed neurons shown in blue. (C) PCA of miRNA profile in reprogrammed neurons from young fibroblasts aged three days, five months one year (green) and from old fibroblasts aged 90, 92, and 92 years (bue) alongside the corresponding young fibroblasts (black) and old fibroblasts (red) (FDR < 0.05). (D) MicroRNAs that are differentially regulated with age in both fibroblasts (red) and reprogrammed neurons (blue). Four microRNAs, miR-10a, miR-497, miR-10b, and miR-195, are upregulated with age, while 10 microRNAs are shown to be downregulated with age, p<0.05. (E) Validation of expression of miRNA expression upregulated with aging (miR-10a, miR-497, miR-195) in reprogrammed neurons from old fibroblasts over reprogrammed neurons from young fibroblasts (left). Validation of expression changes of miR-10a, miR-497, miR-195 in human striatum slices (middle) and human cortex slices (right) from old individuals compared to young individuals.
-
Figure 3—source data 1
Raw data for qPCR for microRNA expression analysis.
- https://doi.org/10.7554/eLife.18648.008
-
Figure 3—source data 2
Full GO terms for age-regulated genes in reprogrammed neurons and for predicted targets of miR-10a-5p and miR-497-5p.
- https://doi.org/10.7554/eLife.18648.009
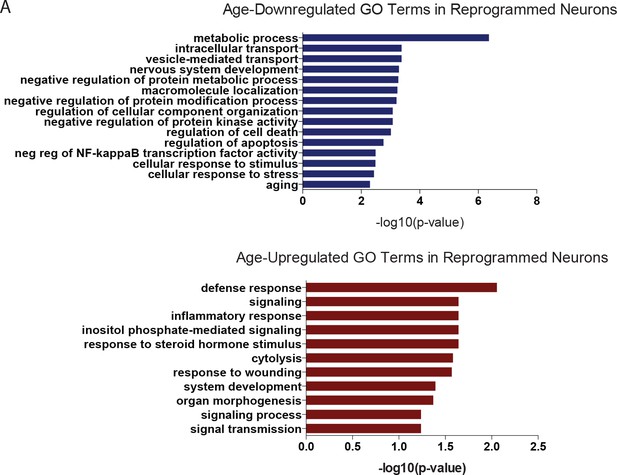
Gene ontology of DEGs in reprogrammed neurons.
(A) GO analysis of age-downregulated transcripts (top) and upregulated transcipts (bottom) in reprogrammed neurons, log fc > 1, FDR < 0.01 using BiNGO from Cytoscape version 3.3.0. (FDR 0.05). GO terms related to aging-associated processes are represented here.
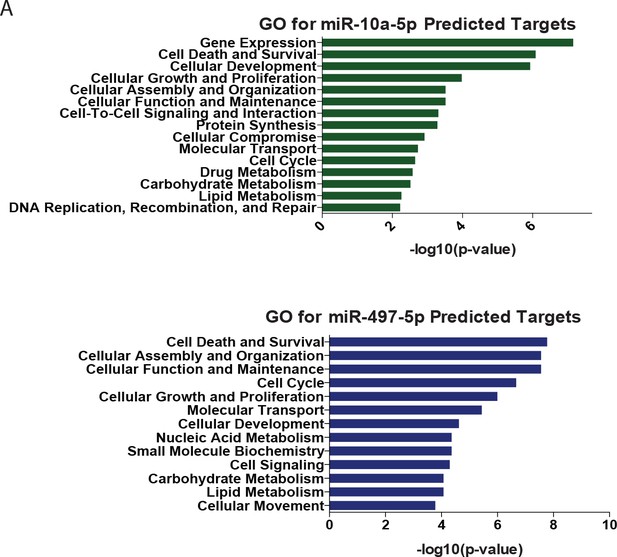
Gene ontology of predicted targets of age-resulted microRNAs.
(A) GO analysis of list of age-downregulated transcripts predicted to be targeted by upregulated microRNAs, miR-10a-5p (top) and miR-497-5p (bottom). A list of original downregulated transcripts were filtered by fc<-1.5, FDR < 0.05. Target prediction analysis with Ingenuity Pathway Analysis, MicroRNA-mRNA Interactions.
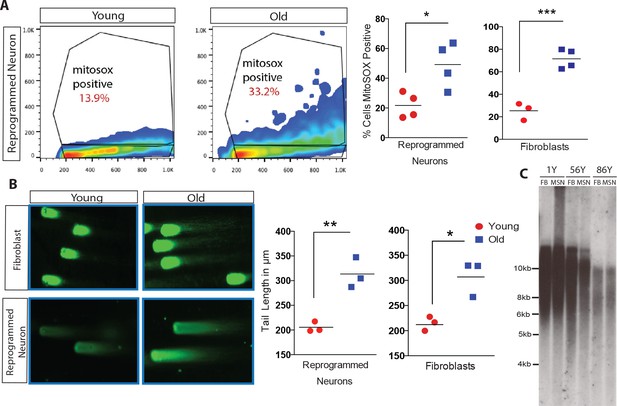
Analysis of cellular biomarkers of age reveals conservation of neuron-specific aging in reprogrammed neurons.
(A) ROS levels visualized by MitoSOX and analyzed by FACs. Representative dot plot of reprogrammed neuron from 1-year-old fibroblast (left) and from 91-year-old fibroblast (right), y axis = FL2 channel (MitoSOX), x axis = FSC. Quantification of percent of cells positive for MitoSOX reveals a significant difference in fibroblasts with age in addition to reprogrammed neurons with age. ***p-value=0.0008; *p-value=0.019. (B) Representative images of comets indicating DNA damage from five month old fibroblasts (top left) versus old fibroblasts (top right) alongside reprogrammed neurons from 5-month-old fibroblasts (bottom left) and 92-year-old fibroblasts (bottom right). Quantification of tail lengths in fibroblasts and reprogrammed neurons with age *p-value=0.013, **p-value=0.004. (C) Telomere length analyses of reprogrammed neurons are maintained from corresponding starting fibroblasts from one year old, 56 year old, and 86 year old donors.
-
Figure 4—source data 1
Raw data for mitoSOX and comet assay.
- https://doi.org/10.7554/eLife.18648.013