Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction
Figures
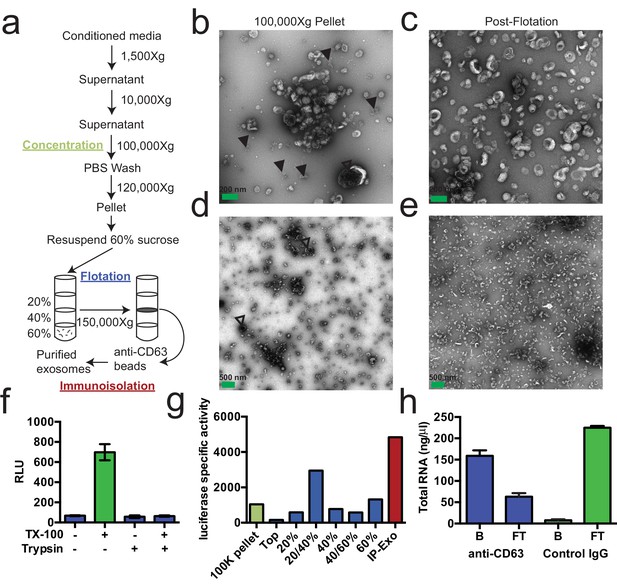
Purified CD63-positive exosomes contain RNA.
(a) Exosome purification schematic. (b–e) Representative electron micrographs of negative stained samples from the 100,000 ×g pellet fraction (b,d) and post-flotation fractions (c,e) at either 9300X (a,b) or 1900X (c,e) magnification. Open arrows indicate large (>200 nm) vesicle contaminants and closed arrows indicate protein aggregates. (f) CD63-luciferase activity in purified exosomes after treatment with 1% Triton X-100 (TX-100) and/or 100 µg/ml trypsin for 30 min at 4°C. Error bars represent standard deviations from 3 independent samples. (g) Specific activity of CD63-luciferase (RLU/µg of total protein) at each stage of purification (green: 100,000 ×g pellet, purple: post-flotation, red: post-immunoisolation a-CD63 beads). (h) Total RNA recovered from conditioned medium after immuno-isolation with a-CD63 or an IgG control. B – bound to beads, FT – flow-through not bound to beads. Error bars represent standard deviations from 3 separate purifications (biological replicates).
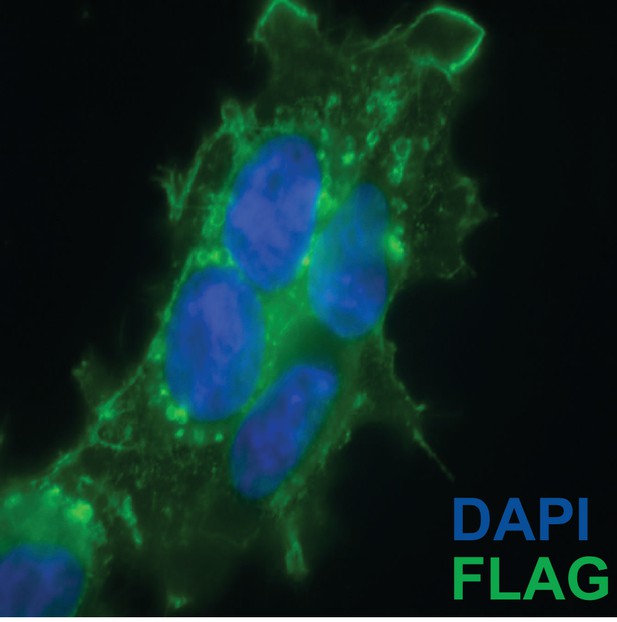
Sub-cellular localization of C-terminal CD63-luciferase-FLAG fusion.
CD63-luciferase-FLAG cells induced for 48 hr were fixed with 4% paraformaldehyde blocked with 5% BSA in PBS and stained with M2-Flag antibody (1:500) and then Alexa-488 conjugated anti-mouse secondary. Cells were mounted with prolong gold (containing DAPI stain) and imaged at 400 X total magnification.
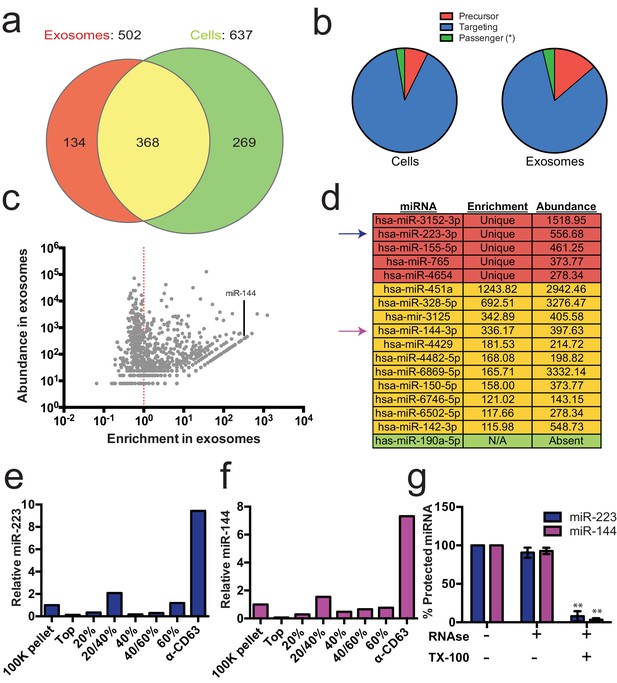
Enrichment of select miRNAs in exosomes.
(a) Venn diagram showing the number of total (above diagram), unique (inside red or green circles) and shared miRNAs (inside yellow) from each library. (b) Pie charts showing the relative proportion of reads mapping to each miRNA species (Precursor - red, passenger strand – green, targeting strand - blue) in cellular and exosome small RNA libraries (c) Scatterplot showing the enrichment (reads per million miRNA mapped reads (RPM) in exosomes/cells) and relative abundance in exosomes (RPM) of all miRNAs found in both libraries. (d) Table showing the enrichment and abundance (RPM) of relevant miRNAs in exosomes. (Red - unique to exosomes, Yellow - highly enriched in exosomes, Green - unique to cells) (e,f) Relative miR-223 (e) and miR-144 (f) per ng of RNA as quantified by qRT-PCR during each stage of the purification. The 100K pellet was set to 1. (g) RNase protection of exosomal miRNAs quantifed by qRT-PCR. Purified exosomes treated with or without RNase I and/or Triton X-100. Errors bars represent the standard deviation from 3 biological replicates. Statistical significance was performed using Student's t-test (**p<0.01).
-
Figure 2—source data 1
Mapping statistics for small RNA-seq libraries to the human genome (hg19).
Reads were processed (see Materials and methods) and mapped to the human genome (hg19) using Bowtie 2. Total counts for reads mapped to the genome, to rRNA and to miRNA (using miRdeep2 - see Materials and methods) are shown. Percent of total reads are shown in parenthesis.
- https://doi.org/10.7554/eLife.19276.006
-
Figure 2—source data 2
miRNA counts from cell and exosomes libraries using miRdeep2.
Number of reads mapped to each miRNA annotated in miRBase version 21 using the quantifier program of the miRdeep2 package. Reads per million miRNA mapped reads (RPM) were calculated and the quotient was taken to determine enrichment in exosomes.
- https://doi.org/10.7554/eLife.19276.007
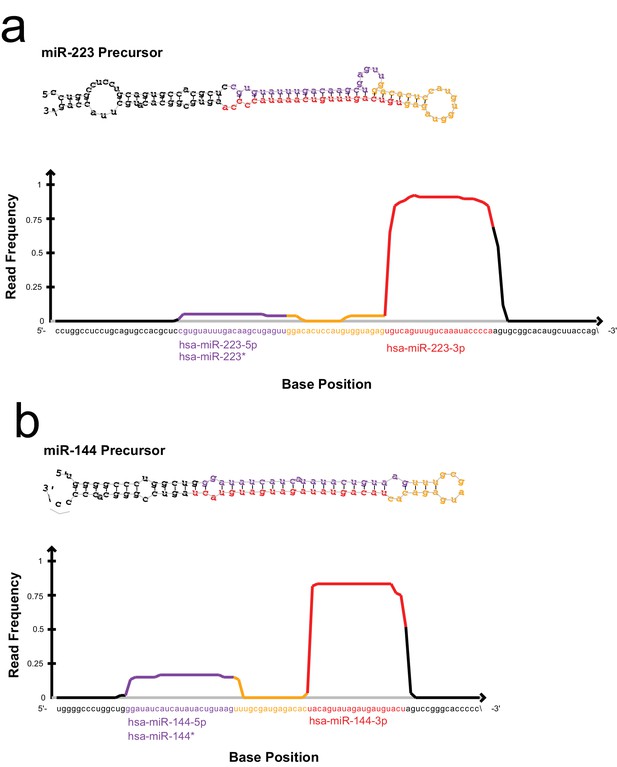
Read frequency distribution along miR-223 and miR-144 precursors.
All reads detected for miR-223 (a) and miR-144 (b) in the exosome small RNA-seq libraries were aligned to the hairpin precursor sequences and the read frequency distribution across the precursor was determined using the quantifier program of the miRdeep2 package.
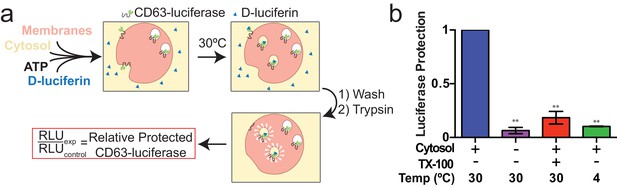
Cell-free exosome biogenesis reaction.
(a) Schematic illustrating the in vitro biogenesis reaction. (b) Exosome biogenesis measured by relative protected CD63-luciferase. Reactions with or without cytosol, 1%Tx-100 and incubation temperature are indicated. All quantifications represent means from three independent experiments and error bars represent standard deviations. Statistical significance was performed using Student's t-test (**p<0.01).
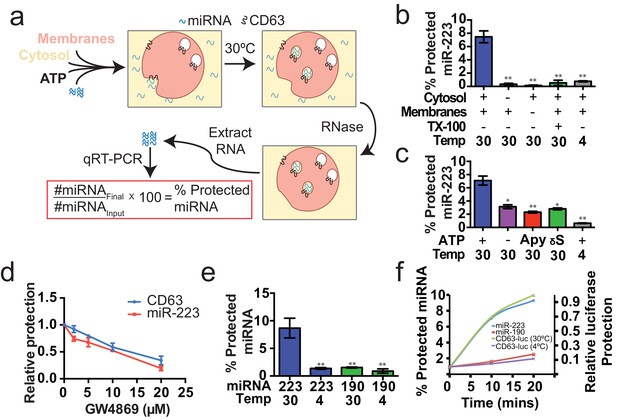
Cell-free selective sorting of miRNA into exosomes.
(a) Schematic illustrating the in vitro packaging reaction. (b) Cell-free packaging of miR-223 measured as percent protected by qRT-PCR. Reactions with or without membranes (15,000 ×g pellet), cytosol (100,000 ×g supernatant) and 1% Triton X-100 (TX-100), and incubated at 4 or 30°C are indicated. (c) ATP requirements for miR-223 packaging (Apy - Apyrase, (1 U/ml) γS – ATPγS (10 mM)). (d) Dose dependent effect of neutral sphingomyelinase 2 inhibitor (GW4869). Measured as relative protection of miRNA and CD63-luciferase normalized to vehicle only control (DMSO). (e) Protection of miR-223 or miR-190 measured as a percent protected by qRT-PCR. (f) Relative CD63-luciferase (right axis) and percent miRNA protection (left axis) measured over a 20-min time course using the indicated miRNA cargo and incubation temperature. All quantifications represent means from three independent experiments and error bars represent standard deviations. Statistical significance was performed using Student's t-test (*p<0.05, **p<0.01).
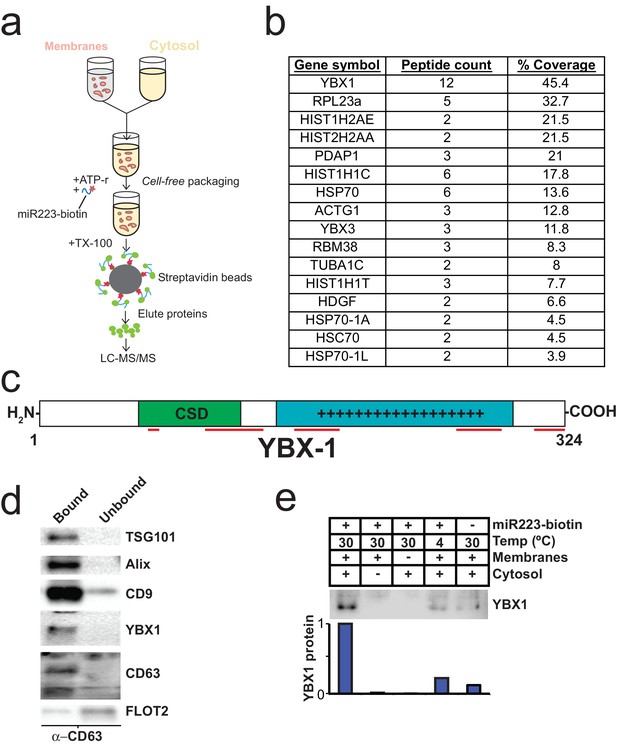
Identification of YBX1 as a candidate exosomal miRNA sorting protein.
(a) Scheme to identify candidate miRNA sorting proteins (b) Proteins identified by tandem mass spectroscopy from the experiment illustrated in (a). (c) Schematic of YBX1 protein. The cold-shock domain (green) and positively charged low-complexity region (blue) are highlighted. Red lines indicate detected unique peptides from mass spectroscopy. (d) Immunoblots for the indicated protein markers in the CD63 immuno-isolated (bound) or unbound fractions. Exosomes were purified as in Figure 1a. (e) Immunoblot for YBX1 following cell-free packaging reactions performed according to the conditions indicated and immobilized with streptavidin beads as shown in (Figure 5a). Bar graph represents densitometry values for the blot shown.
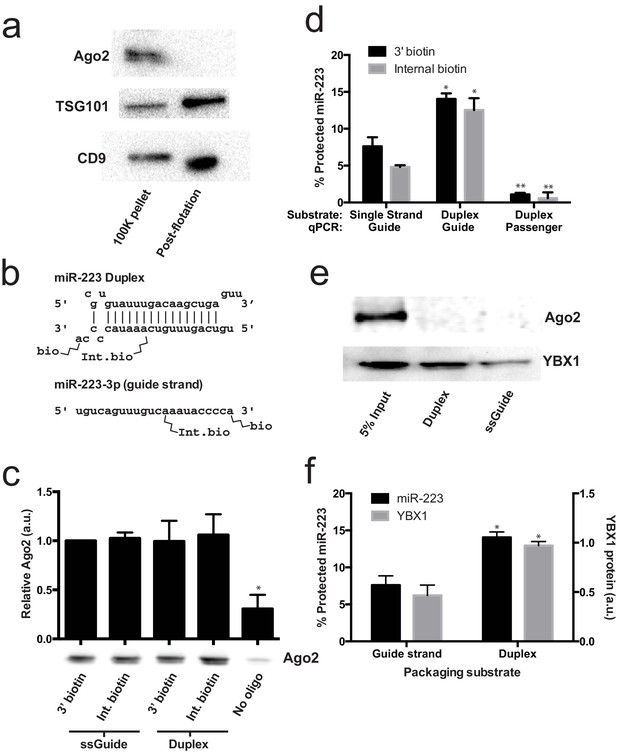
Lack of evidence for a specific role for Ago2 in sorting miR-223 into exosomes.
(a) Immunoblots for Ago2 and exosome markers TSG101 and CD9 in 100,000 ×g (100K) pellet and the 20/40% sucrose interface fractions. (b) Schematics showing 3' and internally biotinylated miR-223 duplex and mature guide strand substrates. (c) Immunoblots for Ago2 from substrates mixed with cytosol alone for 30 min at 30°C and then absorbed on streptavidin-conjugated beads. (d) Percent protected miR-223 (either guide strand or passenger strand) from 3' biotinylated or internally biotinylated single strand or duplex substrates. (e) Immunoblots for Ago2 and YBX1 from substrates packaged in the complete in vitro reaction and then absorbed on streptavidin-conjugated beads. (f) Percent RNAse protected miR-223 and relative level of streptavidin-absorbed YBX1 protein (normalized to duplex). MiR-223 and YBX1 quantification comes from data in d) and e), respectively. All quantifications represent means from 3 independent samples and errors bars represent standard deviations. All quantifications represent means from three independent experiments and error bars represent standard deviations. Statistical significance was performed using Student's t-test (*p<0.05, **p<0.01).
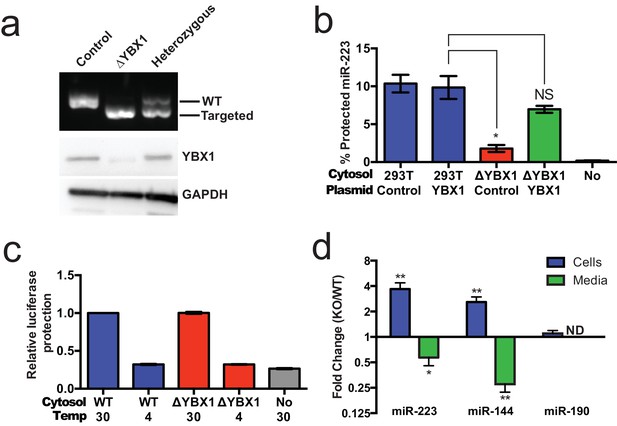
YBX1 is necessary for exosomal miRNA packaging and secretion.
(a) Analysis of wild-type and CRISPR/Cas9 genome edited HEK293T clones by PCR flanking the genomic target site (top) and immunoblot for YBX1 (middle) and GAPDH (bottom). (b) In vitro miR-223 packaging into exosomes from ΔYBX1 or WT cytosol transfected with control (pCAG) or YBX1 plasmid. (c) Cell-free exosome biogenesis with cytosol from ΔYBX1 or WT cells and membranes from CD63-luciferase cells. (d) Fold change of miR-223 and miR-144 in cells and media from by ΔYBX1 (KO) and WT cells (KO/WT) ND = Not detected. All quantifications represent means from three independent experiments and error bars represent standard deviations. Statistical significance was performed using Student's t-test (*p<0.05, **p<0.01 and NS = not significant).
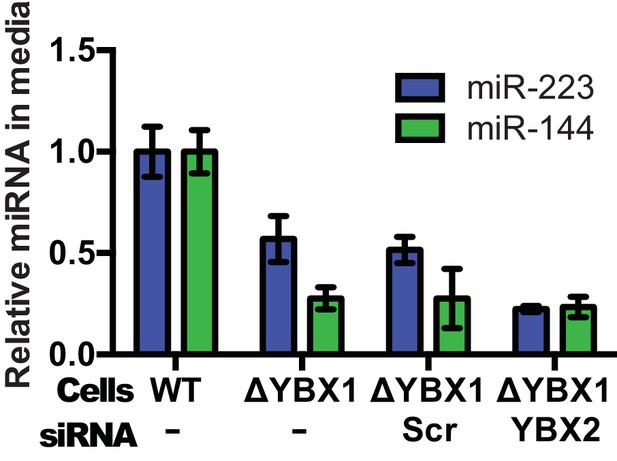
Partial redundancy for YBX2 for the secretion of miR-223 in cells.
Relative quantity of miR-223 secreted into the medium by WT and ΔYBX1 cells after 24 hr with or without transfection with control or YBX2 siRNA.
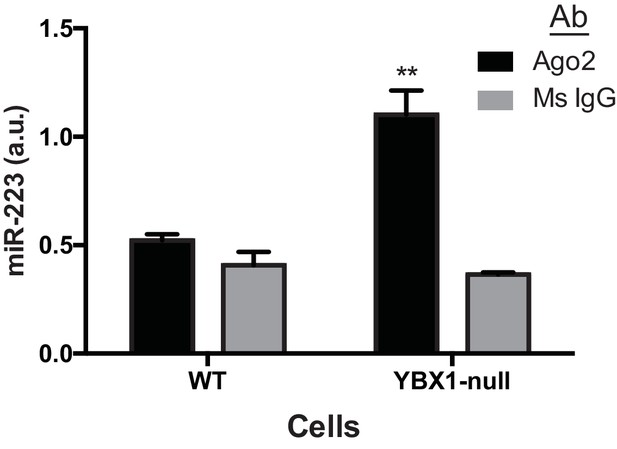
miR-223 association with Ago2 in WT and ΔYBX1 cells.
RNA immunoprecipitation was performed using mouse anti-Ago2 antibody or a mouse IgG isotype control antibody in lysates of WT or ΔYBX1 cells. Ago2-associated miR-223 was detected by qPCR. Statistical analysis was performed using Student's t-test (**p<0.01, NS = not significant).





