GUY1 confers complete female lethality and is a strong candidate for a male-determining factor in Anopheles stephensi
Figures
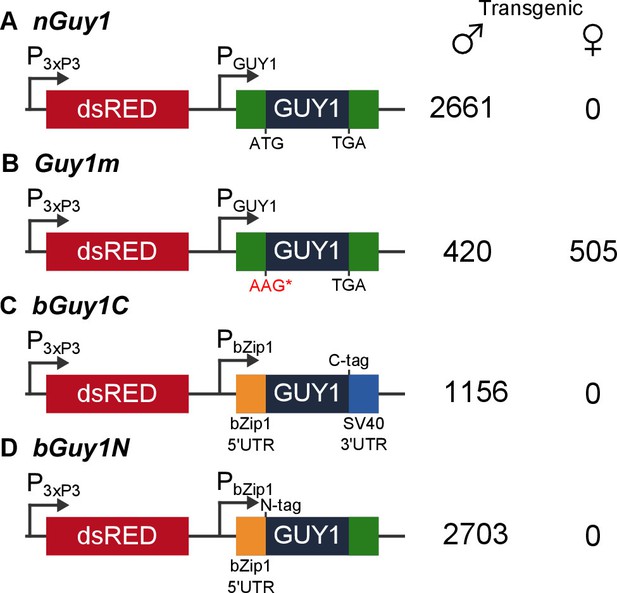
Four donor plasmids were used to generate transgenic Anopheles stephensi.
nGuy1 and Guy1m were also used in the transient assays described in Table 1. All constructs shown in the figure were flanked by the piggyBac arms to facilitate piggyBac-mediated integration into the An. stephensi genome (Horn et al., 2000). The DsRed fluorescent marker gene under the control of the 3xP3/Hsp70 promoter (3xP3) was the transformation marker. PGUY1 refers to the native Guy1 promoter (Criscione et al., 2013). Note that the only difference between nGuy1 and Guy1m is the point mutation in the first ATG. PbZip1 refers to a promoter derived from an An. stephensi gene (Genbank JQ266223) and this promoter is used to drive early zygotic expression of the transgene (Figure 3). The C-tag and N-tag refer to the eight residue Strep II tag (Lichty et al., 2005) placed at either the C- or N-terminus of the GUY1 protein. A stretch of eight glycine residues were placed between the Strep II tag and the GUY1 protein (Supplementary file 3). The number of transgenic males and females were total counts from screens performed on all lines of each construct (Supplementary files 1 and 2).
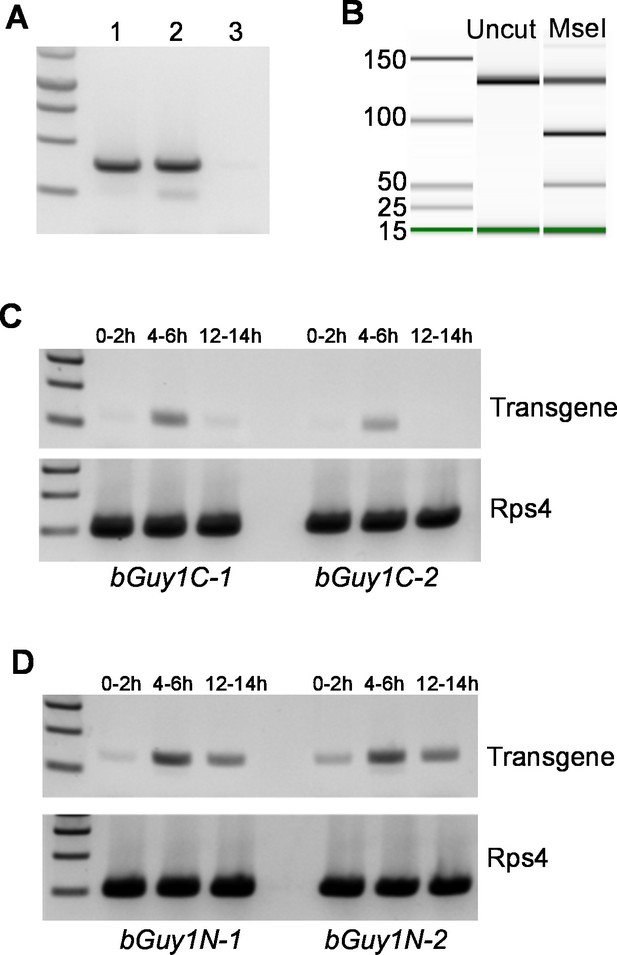
Early embryonic expression of the Guy1 transgene in nGuy1 (A), Guy1m (B), bGuy1C (C), and bGuy1N (D) lines.
(A) RT-PCR from 5–6 hr old genotyped single embryos. Lane 1, Wild type male; Lane 2, Transgenic female; Lane 3, Wild type female. Genotyping was performed according to Criscione et al. (2013). nGuy1 transcription is detected in transgenic females where there is no Y chromosome or endogenous Guy1. Detecting nGuy1 expression in transgenic female embryos is the only way we can show that the nGuy1 transgene is expressed because there is no sequence difference between the nGuy1 transgene transcript and the endogenous Guy1 transcript. (B) A pseudo-gel image from the bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) showing the presence of RT-PCR products from the mutant Guy1m transgene. We took advantage of the Guy1m mutation (ATG to AAG) that introduced an MseI site so we do not have to perform RT-PCR on genotyped single embryos. RT-PCR was done using pooled 5–6 hr embryos. The three fragments in the MseI lane are 130 bp (uncut wildtype Guy1), 90 bp (cut Guy1m), and 40 bp (cut Guy1m). The presence of the MseI digested 90 and 40 bp fragments indicates that there were transcripts from theGuy1m transgene. Panels C and D show RT-PCR products using a Guy1 primer and a bZip1 UTR primer, which only amplify cDNA from the bGuy1C and bGuy1N transgenes (Figure 1). Eggs of 0–2 hr, 4–6 hr and 12–14 hr post oviposition were collected from A. stephensi bGuy1C-1 and bGuy1C-2 (panel C) and bGuy1D-1 and bGuy1D-2 (panel D) lines. RPS4, ribosomal protein subunit 4. All primers can be found in Table 3.
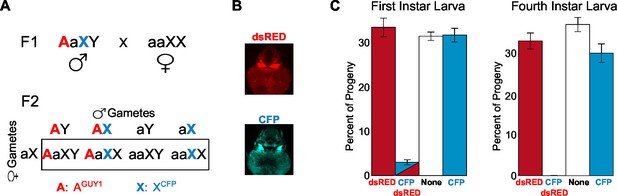
Analysis of the transgenic nGuy1-1 line indicates 100% female lethality prior to or soon after egg hatching.
(A) A schematic and Punnett square showing the cross performed to genotype F2 progeny based on expression of fluorescent transformation markers. The red uppercase A indicates an autosome that carries the Guy1 transgene and the dsRED transformation marker gene, or AGUY1. The cyan uppercase X indicates an X chromosome that carries a Cyan fl protein (CFP) transformation marker gene, or XCFP. AGUY1aXY males and aaXCFPXCFP females were initially crossed to obtain F1 AGUY1aXCFPY progeny. AGUY1aXCFPY were then crossed with wild type females (aaXX) to obtain progeny of four possible genotypes: AGUY1aXY, transgenic Guy1 males; AGUY1aXCFPX, transgenic Guy1 females; aaXCFPX, wild type females; aaXY, wild type males. (B) Images of transgenic L3 instar showing DsRed positive and CFP positive, respectively. (C) Distribution of the four genotypes in the F2 progeny at L1 and L4 instar stages, respectively. Analysis of the L1 and L4 instar is from two independent experiments. Percentages of each genotype were shown as the average of four replicates with standard error. The actual count of each genotype is provided in Figure 3—source data 1. Note that all CFP-DsRed double positive L1 instars died within 8 hr after hatching.
-
Figure 3—source data 1
Number of the four types of progeny from wild type males mated with nGuy1-1 (DsRed) and CFP positive males.
The mating strategy and progeny genotypes are described in Figure 3A.
- https://doi.org/10.7554/eLife.19281.008
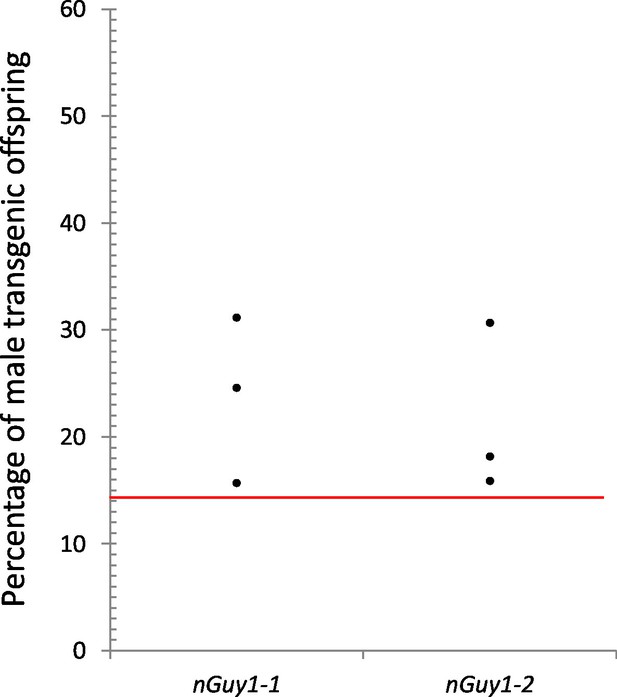
Reproductive competitiveness of transgenic males compared to their non-transgenic male siblings in two independent lines, nGuy1-1 and nGuy1-2.
Sibling cohorts of 20 transgenic and 20 non-transgenic males were mated with 10 wild type females. The resulting progeny were screened for transgenes at L3 instar stage as indicated by the DsRed marker and sexed by the presence or absence of testes. Shown in the figure percentages of male transgenics in biological triplicates for both lines. The red line indicates the expected percentage (1/7 or 14.29%) of transgenic male progeny, assuming that the DsRed positive females do not survive beyond the L1 stage (Figure 3) and the male parents (transgenics and their non-transgenic brothers) were equally productive (detailed calculations are shown in Figure 4—source data 1). Statistical analysis was performed using one-sample proportion tests for both lines (Z=5.0 and 8.1, respectively; p<0.001 in both cases, Figure 4—source data 1). The significantly larger transgenic male population in comparison to the expected value suggests that the transgenic Guy1 containing males are reproductively more competitive than non-transgenic males under these laboratory conditions.
-
Figure 4—source data 1
Assay for male reproductive competitiveness of nGuy1-1 and nGuy1-2 lines 1.
- https://doi.org/10.7554/eLife.19281.010
Tables
Transient injection of the nGuy1, but not the Guy1m, plasmid in early embryos confers strong male bias in Anopheles stephensi*.
| Plasmid | Male | Female |
|---|---|---|
| nGuy1, replicate 1 | 25 | 1 |
| nGuy1, replicate 2 | 19 | 2 |
| Guy1m, replicate 1 | 24 | 34 |
| Guy1m, replicate 2 | 10 | 6 |
-
Notes:
-
*An EGFP reporter plasmid under the control of the Drosophila melanogaster actin 5C promoter was co-injected with either the nGuy1 or Guy1m plasmid to ensure effective embryonic injections as indicated by an EGFP signal in the larvae. The adults that developed from EGFP positive larvae were sexed according to antennae morphology.
Sex ratios of transgenic and non-transgenic progeny of five transgenic lines*.
| Line | Transgenic male | Transgenic female | Negative male | Negative female |
|---|---|---|---|---|
| nGuy1-1 † | 71 | 0 | 58 | 56 |
| nGuy1-2 † | 129 | 0 | 91 | 83 |
| Guy1m † | 69 | 62 | 26 | 34 |
| bGuy1C-1 ‡ | 78 | 0 | 75 | 66 |
| bGuy1N-1 ‡ | 77 | 0 | 66 | 65 |
-
Notes:
-
*Crosses were done between the heterozygous transgenic males and wildtype females. Screening and sexing was initially done at the L3 instar stage. The sexed negative and positive larvae were reared separately to adulthood and sex was further confirmed on the basis of antennae morphology. The total numbers of each sex for the transgenic and non-transgenic groups are listed.
-
†The numbers for these lines are from generation 7 (G7). Note that Guy1m has a point mutation that abolished the Guy1 open reading frame. There are more transgenic individuals than non-transgenic individuals in the Guy1m line because both transgenic females and wild type females were mated with transgenic males to maintain the line.
-
‡Two bGuy1C lines and nine bGuy1N lines were obtained and only one of each is shown in this Table. The numbers are from generation 4 (G4).
Primers and probes used in this study.
| GUY1_Full-F1 | CCTGAAATGATGCTCTGGAAA |
| GUY1_Full-R1 | GAAACGTTTTTCCAACATGTGA |
| GUY1_Full+AscI-F1 | CACTGGCGCGCCCCTGAAATGATGCTCTGGAAA |
| GUY1_Full+PacI-R1 | TCATGCAATTAATTGAAACGTTTTTCCAACATGTGA |
| RPS4-F2 | GAGTCCATCAAAGGAGAAAGTCTAC |
| RPS4-R2 | TAGCTGGCGCATCAGGTAC |
| sYG2_F2 | TGCCGGACATGACATTTG |
| sYG2_R2 | TCAATGCGAACAGAAGGCTAA |
| DsRed_F2 | CCCCGTAATGCAGAAGAAGA |
| GUY1_R2 | GATCCGTTAAAAATTGACACCA |
| GUY1_F3 | TTTACTCGTCAAAGCTGCCA |
| GUY1_R3 | GATCCGTTAAAAATTGACACCA |
| DsRed_F4 | CCCCGTAATGCAGAAGAAGA |
| DsRed_R4 | GGTGATGTCCAGCTTGGAGT |
| DsRed_P4 | FAM-TACATGGCCAAGAAGCCCGT-BHQ1 |
| AutoRef_ddP_F4 | ATCACCACTCGTCGTCCGTT |
| AutoRef_ddP_R4 | CGAACGAACTCGATTGACCC |
| AutoRef_ddP_P4 | HEX-GCAAACACCACAACAGCAGC-BHQ1 |
| GUY1_ddP_F4 | GTCAAAGCTGCCACGGATCT |
| GUY1_ddP_R4 | TCCAATGTCACAGCAGAGTGTTT |
| GUY1_ddP_P4 | FAM-TCACAAAGTAGGCGATACAAAAACA-BHQ1 |
| iPCR_PBR_F5 | TACGCATGATTATCTTTAACGTA |
| iPCR_PBR_R5 | TGGCTCTTCAGTACTGTCAT |
| iPCR_PBRnest_F5 | GTCACAATATGATTATCTTTCTA |
| iPCR_PBRnest_R5 | CACTTCATTTGGCAAAATAT |
| Guy1m_F6 | TTAGATAACAGAAAGCGTACACT |
| Guy1m_R6 | ACTTGATTTATCATTCCAAGTCA |
| bGuy1C-F7 | GTTTATTGCAGCTTATAATGGTTAC |
| bGuy1C-R7 | TCTGACTTGGAATGATAAATCAAG |
| bGuy1N_F7 | GAGAAAATAGCTGAATTGAAGG |
| bGuy1N_R7 | AACACACAAAGTGGTCTTATGC |
| RPS4_F7 | CACGAGGATGGATGTTGGAC |
| RPS4_R7 | ATCAGGCGGAAGTATTCACC |
-
Notes: Primers are presented in seven sets which alternates in white and grey backgrounds. (1) Amplification of the full length Guy1 gene sequence followed by adding a 5’ adaptor with an AscI site and a 3’ adaptor with a PacI site. (2) Primers used for genotyping embryos. RPS4, ribosomal protein subunit 4, was used as the positive control, sYG2 was used as a Y chromosome marker, and the transgene was amplified using a DsRed and a Guy1 primer, which is only present in transgenic individuals. (3) Primer sets used for RT-PCR of Guy1 on single embryos. (4) Digital droplet PCR primer sets used to determine Guy1 copy number and number of insertions in transgenic lines. (5) Inverse PCR primer sets used to determine the site of integration near the piggyBac right hand flanking sequences. (6) RT-PCR primers to amplify a 130 bp fragment of Guy1 to allow differentiation of Guy1m transcript from endogenous Guy1 transcript by MseI digestion. (7) RT-PCR primers used to amplify cDNA made from bGuy1C and bGuy1N transcripts. A set of RPS4 primers different from set 2 was used.
Additional files
-
Supplementary file 1
Number of male and female transgenics (DsRed positive) in the nGuy1 and Guy1m lines.
- https://doi.org/10.7554/eLife.19281.012
-
Supplementary file 2
Number of transgenic (DsRed positive) males in the bGuy1C and bGuy1N lines.
- https://doi.org/10.7554/eLife.19281.013
-
Supplementary file 3
Supplemental sequences.
- https://doi.org/10.7554/eLife.19281.014





