A non canonical subtilase attenuates the transcriptional activation of defence responses in Arabidopsis thaliana
Figures
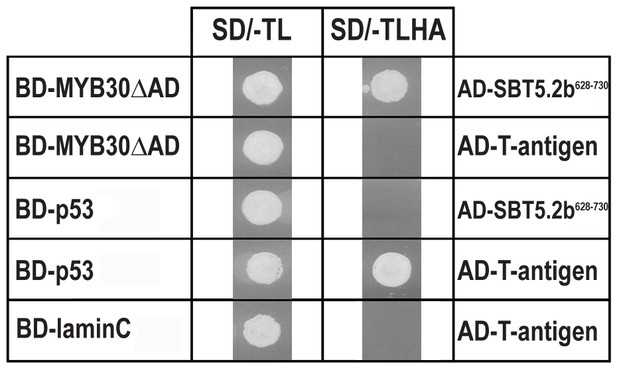
Specific interaction between MYB30 and SBT5.2 in yeast.
Yeasts are shown after growth for five days on low stringency (left; SD/-TL) or high stringency (right; SD/-TLHA) media. Co-expression of MYB30 deleted from its C-terminal transcription activation domain (MYB30△AD) and the isolated cDNA clone encoding the last 103 amino acids of SBT5.2 (SBT5.2628-730) resulted in yeast growth on selective medium. In a control experiment, yeast cells expressing MYB30△AD or SBT5.2628-730 with controls provided by Clontech (T-antigen or P53, respectively) were not able to grow on selective medium. BD, GAL4 DNA-binding domain; AD, GAL4 activation domain.
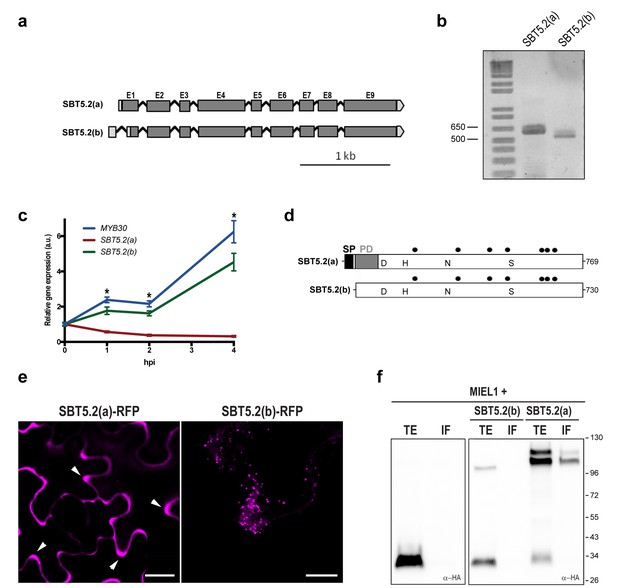
SBT5.2 is alternatively spliced.
(a) Genomic structure of SBT5.2 alternatively spliced variants. Exons are shown as dark gray boxes (E1-E9), introns as black lines between exons and 5’ and 3’ UTRs are shown in light gray. (b) RT-PCR analysis of SBT5.2(a) and SBT5.2(b) transcripts in four-week old Col-0 Arabidopsis leaves. (c) The relative expression of SBT5.2(a), SBT5.2(b) and MYB30 at the indicated timepoints after inoculation of Col-0 plants with Pst AvrRpm1 (5 × 107 cfu/ml). Expression values were normalized using SAND family gene as internal standard and related to the value of each gene at time 0, which is set at 1. The SEM values were calculated from 4 independent experiments (4 replicates/experiment). The asterisks indicate statistically significant values for the three tested genes according to a Student’s t-test (p<0.005) and with respect to gene expression values at time 0. (d) Schematic representation of SBT5.2(a) and SBT5.2(b) protein sequences. The signal peptide (SP) and the pro-domain (PD) in the SBT5.2(a) isoform are shown as black and grey boxes, respectively. Catalytically conserved Asp, His, Asn, and Ser residues are indicated. Putative N-glycosylation sites (PGSs) are indicated by black dots. (e) Confocal images of epidermal N. benthamiana cells 36 hr after Agrobacterium-mediated transient expression of the indicated constructs. Accumulation of SBT5.2(a)-RFP in apoplastic spaces is indicated by arrowheads. Bars = 10 µm. (f) Western blot analysis of total protein extracts (TE) and intercellular fluids (IF) from N. benthamiana leaves expressing the intracellular protein MIEL1 alone (left) or co-expressed with HA-tagged SBT proteins (right), as indicated. Molecular mass markers in kilodaltons are indicated on the right.
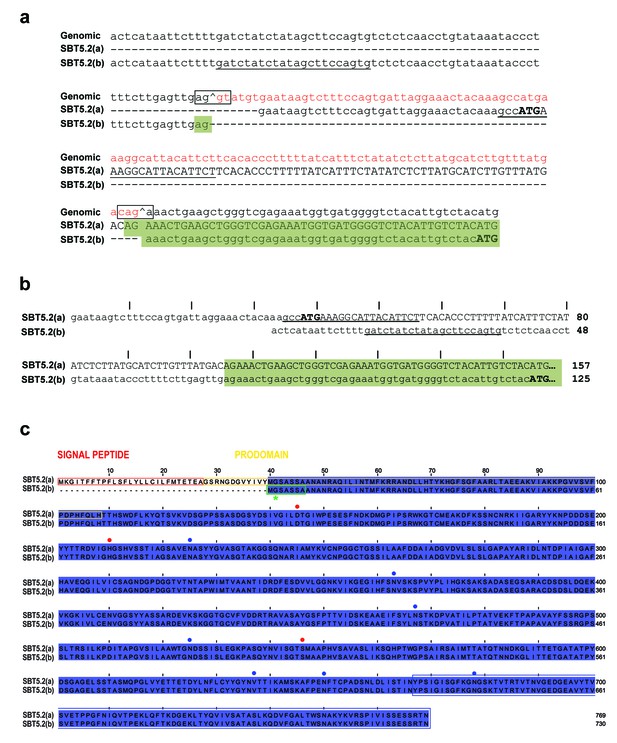
Alternative splicing of SBT5.2 gives raise to two distinct variants.
(a) Alignment of the two SBT5.2 cDNA sequences with the genomic sequence. The consensus splice sequences flanking the intron that is retained in SBT5.2(a) and shown in red in the genomic sequence are boxed. In the genomic sequence, ^ indicates 5’ and 3’ splice sites. In SBT5.2(a), the retained intron contains an ATG (in bold) encoding the first coding SBT5.2(a) Met residue. Splicing of this intron gives rise to the SBT5.2(b) isoform. Lower case in SBT5.2(a) and SBT5.2(b) cDNA sequences indicates untranslated 5’UTRs whereas upper case indicates coding sequences. Sequences used to specifically amplify SBT5.2(a) or SBT5.2(b) are underlined: for amplification of SBT5.2(a) the primer was situated in the intron [that is absent in SBT5.2(b)], whereas a primer in a sequence of the SBT5.2(b) 5’UTR [that is absent in SBT5.2(a)] was used for SBT5.2(b) amplification. Identical sequences are highlighted by a green box. From the ATG (in bold) in SBT5.2(b) both nucleotide sequences are identical and, thus, not shown. (b) Nucleotide sequences of 5’ends of SBT5.2(a) and SBT5.2(b) obtained by 5’RACE amplification. The ATG encoding the first coding Met residue in each protein is shown in bold. Identical sequences are highlighted by a green box. From the ATG in SBT5.2(b) both nucleotide sequences are identical and, thus, not shown. Sequences used to specifically amplify SBT5.2(a) or SBT5.2(b) are underlined. Every tenth residue is marked by |. (c) Sequence alignment of SBT5.2(a) and SBT5.2(b) proteins. Identical amino acids are highlighted in blue. The signal peptide and the prodomain in SBT5.2(a) are boxed in red and yellow, respectively. Catalytical conserved residues are indicated by red dots. Putative N-glycosylation sites (PGSs) are indicated by blue dots. The 103 C-terminal amino acids encoded by the partial SBT5.2 cDNA clone identified in the yeast two-hybrid screen as interacting with MYB30ΔAD is boxed in blue. A putative myristoylation domain in SBT5.2(b) is boxed in green with an asterisk indicating the putative myristoylated glycine residue. Every tenth residue is marked by |.
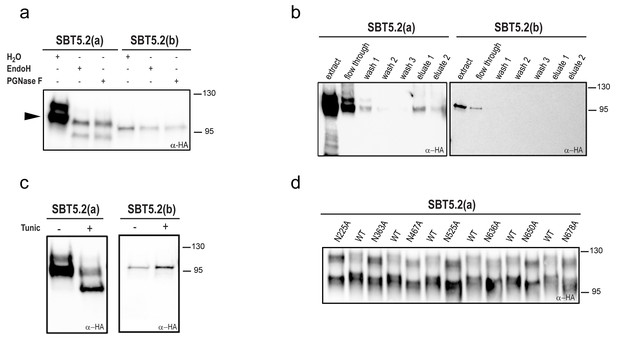
SBT5.2(a), but not SBT5.2(b), is glycosylated in planta.
(a) SBT5.2(a), but not SBT5.2(b), is deglycosylated by PNGase F and Endo H. Protein extracts containing HA-tagged SBT5.2(a) and SBT5.2(b) transiently expressed in N. benthamiana were treated (+) or not (−) with PNGase F or Endo H as indicated. The arrowhead on the left indicates the fully processed form of SBT5.2(a) whose mobility is not affected by the enzymatic treatment. (b) HA-tagged SBT5.2(a), but not SBT5.2(b) can be affinity purified using a concanavalin A resin. (c) Glycosylation of HA-tagged SBT5.2(a), but not SBT5.2(b), is blocked by tunicamycin treatment (+) after transient expression in N. benthamiana. (d) Electrophoretic mobility of individual HA-tagged PGS SBT5.2(a) mutants. Mutated N to A residues are indicated. WT: wild-type SBT5.2(a) proteins were interspersed to facilitate detection of the mobility shifts. In all cases, Western blot analyses were performed using anti-HA antibodies.
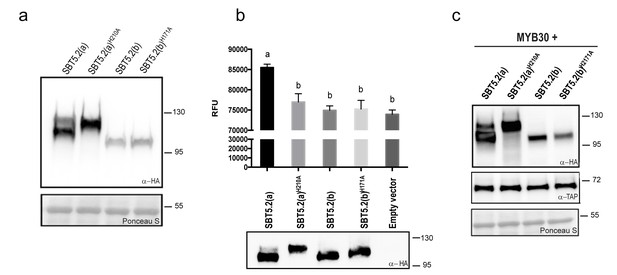
SBT5.2(a), but not SBT5.2(b), shows serine protease activity.
(a) Western blot analysis shows expression of HA-tagged SBT5.2(a), SBT5.2(b) and their catalytic mutant versions in N. benthamiana, as indicated. Ponceau S staining confirms equal loading. Molecular mass markers in kilodaltons are indicated on the right. (b) Fluorimetric assay to detect protease activity following incubation of Arabidopsis protoplasts expressing the indicated proteins with fluorescein isothiocyanate (FITC)-conjugated casein (top). RFU: relative fluorescence units. Error bars indicate SEM. Lowercase letters indicate significant differences as determined by Bonferroni-corrected p-values (p<0.001) obtained after ANOVA and subsequent LSD post-hoc test. All proteins were detected by Western blot (bottom). (c) TAP-tagged MYB30 was expressed in N. benthamiana alone or with SBT5.2(a), SBT5.2(b) and their catalytic mutant versions, as indicated. Western blot analysis shows the expression of TAP-tagged MYB30 and HA-tagged SBT proteins. Ponceau S staining confirms equal loading. Molecular mass markers in kilodaltons are indicated on the right.
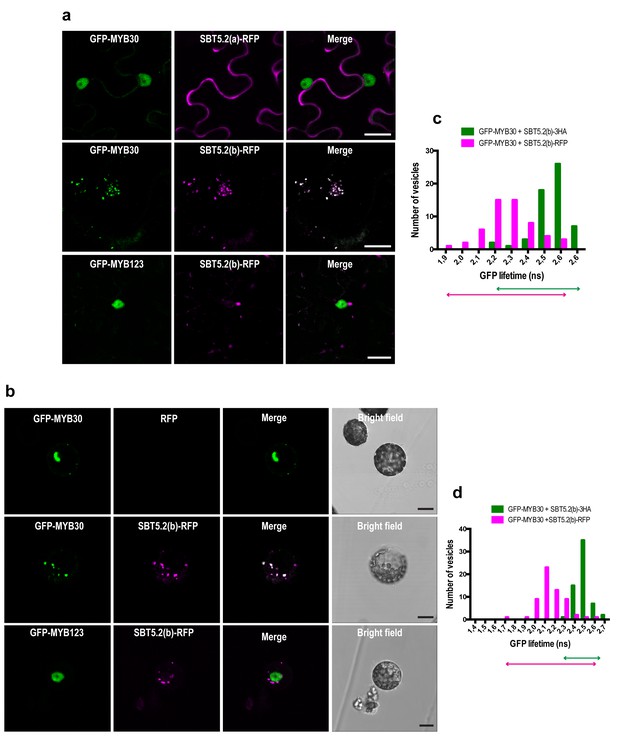
SBT5.2(b) mediates retention of MYB30 in intracellular vesicles.
(a) Confocal images of epidermal N. benthamiana cells 36 hr after Agrobacterium-mediated transient expression of the indicated constructs. (b) Confocal images of Arabidopsis protoplasts 16 hr after transformation of the indicated constructs. (c,d) GFP lifetime distribution of GFP-MYB30 in N. benthamiana cells (c) or Arabidopsis protoplasts (d) expressing SBT5.2(b). Histograms show the number of vesicles according to GFP-MYB30 lifetime classes in the presence of SBT5.2(b)-HA (green bars) or SBT5.2(b)-RFP (magenta bars). The degree of overlap of GFP lifetime distribution is represented with magenta (SBT5.2(b)-RFP) and green (SBT5.2(b)-HA) arrows. Bars = 10 µm.
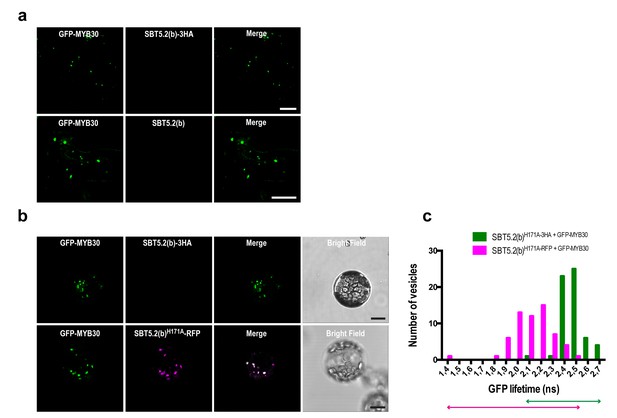
SBT5.2(b)-mediated retention of MYB30 outside the nucleus is independent of C-terminal tagging of the subtilase and of SBT5.2(b) catalytic triad.
(a) Confocal images of epidermal cells of N. benthamiana leaves 36 hr after Agrobacterium-mediated transient expression of GFP-tagged MYB30 and HA-tagged (top) or untagged (down) versions of SBT5.2(b) Bars = 10 µm. (b) Confocal images of Arabidopsis protoplasts 16 hr after transformation with the indicated constructs. Bars = 10 µm. (c) GFP lifetime distribution of GFP-MYB30 in protoplasts expressing SBT5.2(b)H171A. Histograms show the number of vesicles according to GFP-MYB30 lifetime classes in the presence of SBT5.2(b)H171A-HA (green bars) or SBT5.2(b)H171A-RFP (magenta bars). The degree of overlap of GFP lifetime distribution is represented with magenta (+SBT5.2(b)H171A-RFP) and green (SBT5.2(b)H171A-HA) arrows.
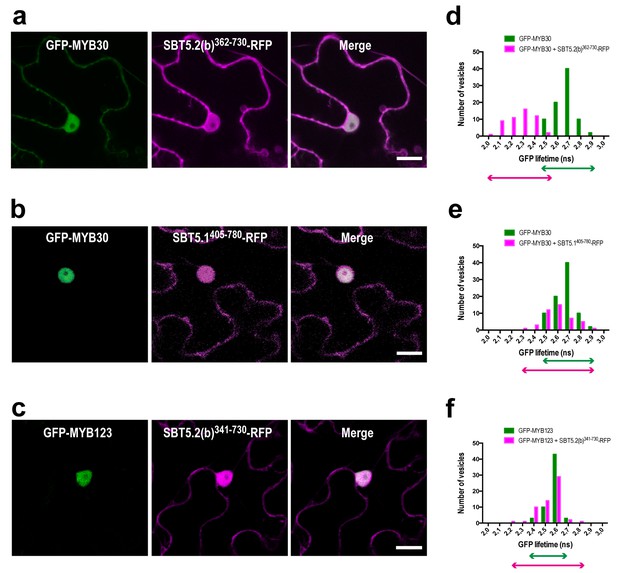
MYB30 interacts with SBT5.2(b) through the C-terminus of the subtilase.
(a–c) Confocal images of epidermal cells of N. benthamiana leaves 36 hr after Agrobacterium-mediated transient expression of the indicated constructs. Bars = 10 µm. (d–f) GFP lifetime distribution of GFP-MYB30, or GFP-MYB123, in nuclei of N. benthamiana cells expressing the C-terminus of SBT5.2(b) [SBT5.2(b)341-730], or SBT5.1 [SBT5.1405-780], as indicated. Histograms show the number of nuclei according to GFP-MYB30, or GFP-MYB123, lifetime classes when the MYB protein is expressed alone (green bars) or in the presence of RFP-tagged SBT5.2(b)341-730, or SBT5.1405-780, (magenta bars), as indicated. The degree of overlap of GFP lifetime distribution is represented with magenta (+RFP-tagged SBT constructs) and green (GFP-tagged MYB constructs) arrows.
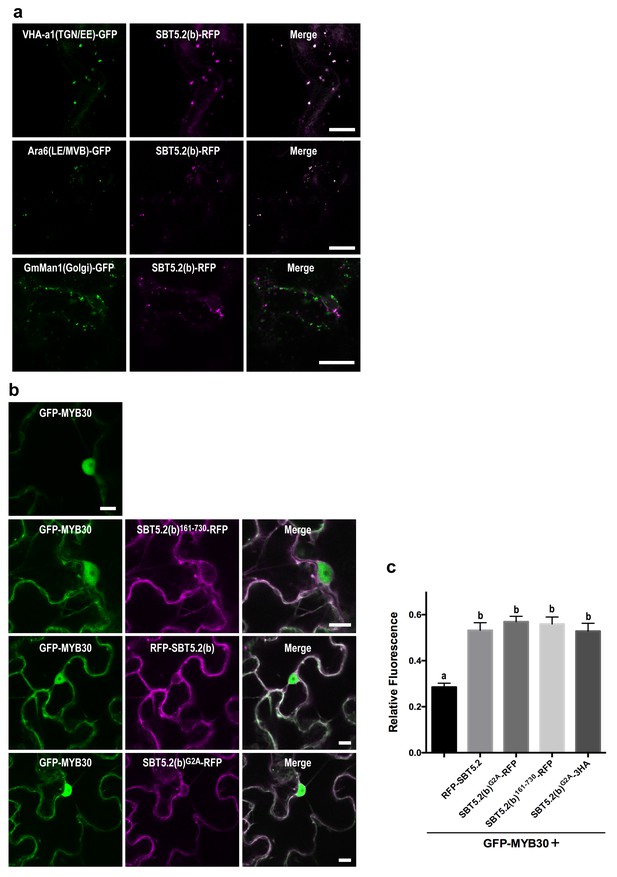
An N-terminal myristoylation site in SBT5.2(b) is required for its localization to endosomes and MYB30 nuclear exclusion.
(a,b) Confocal images of epidermal N. benthamiana cells 36 hr after Agrobacterium-mediated transient expression of the indicated constructs. Bars = 10 µm. (a) SBT5.2(b) localizes to endosomal vesicles. TGN/EE: trans-Golgi network/early endosomes; LE/MVB: late endosomes/multivesicular bodies. (b) A free N-terminal myristoylation site in SBT5.2(b) mediates its localization to endosomes and retention of MYB30 in endosomal vesicles. (c) Relative fluorescence values of cytoplasmic MYB30, expressed alone or with the indicated SBT5.2(b) versions, represented as ratios between cytoplasmic and nuclear fluorescence values in individual cells. Mean and SEM values were calculated from two independent experiments in which fifteen fluorescence measurements were taken per experiment and construct combination. Lowercase letters indicate statistically significant differences as determined by Bonferroni-corrected p-values (p<0.001) obtained after ANOVA and subsequent LSD post-hoc test.
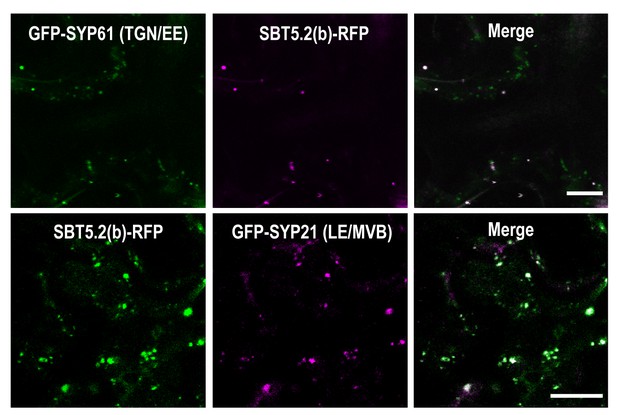
SBT5.2(b) localizes to endosomal vesicles.
Confocal images of epidermal cells of N. benthamiana leaves 36 hr after Agrobacterium-mediated transient expression of the indicated constructs. TGN/EE: trans-Golgi network/early endosomes; LE/MVB: late endosomes/multivesicular bodies. Bars = 10 µm.
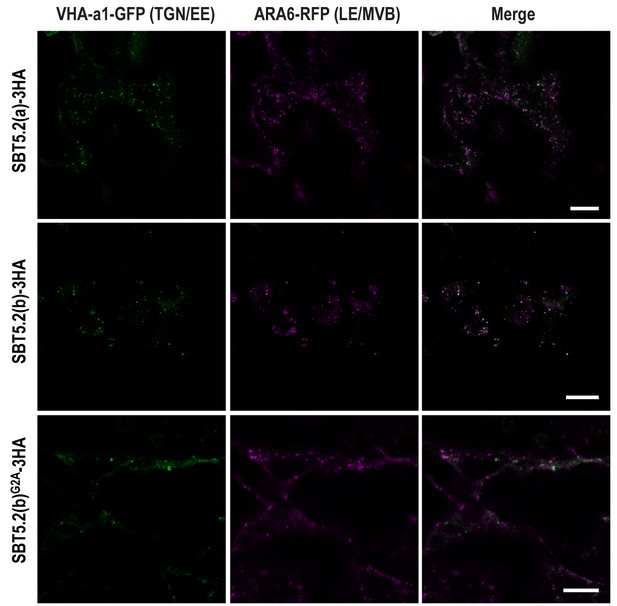
SBT5.2(b) leads to the formation of hybrid endosomal compartments in a myristoylation-dependent manner.
Confocal images of epidermal N. benthamiana cells 36 hr after Agrobacterium-mediated transient expression of the indicated constructs. TGN/EE: trans-Golgi network/early endosomes; LE/MVB: late endosomes/multivesicular bodies. Bars = 10 µm.
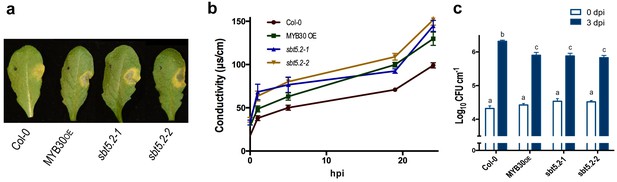
SBT5.2(b) is a negative regulator of resistance and HR responses in Arabidopsis in response to bacterial inoculation.
(a) Symptoms developed by the indicated Arabidopsis lines 60 hpi with Pst AvrRpm1 (2 × 106 cfu/ml). The pictures are representative of three independent experiments in which 4 plants of each line were infiltrated. (b) Quantification of cell death by measuring electrolyte leakage of the indicated Arabidopsis lines in a time course of 24 hr. Plants were inoculated with Pst AvrRpm1 (5 × 106 cfu/ml). Mean and SEM values were calculated from four independent experiments in which three plants were inoculated (four leaves/plant). (c) Growth of Pst AvrRpm1 in the indicated Arabidopsis lines. Bacterial growth 0 (white bars) and three days (blue bars) was measured after inoculation (5 × 105 cfu/ml). Mean bacterial densities were calculated from 6 independent experiments with 6 individual plants (4 leaves/plant). Statistical differences using multiple factor analysis of variance (ANOVA) (p<0.001) are indicated by letters.
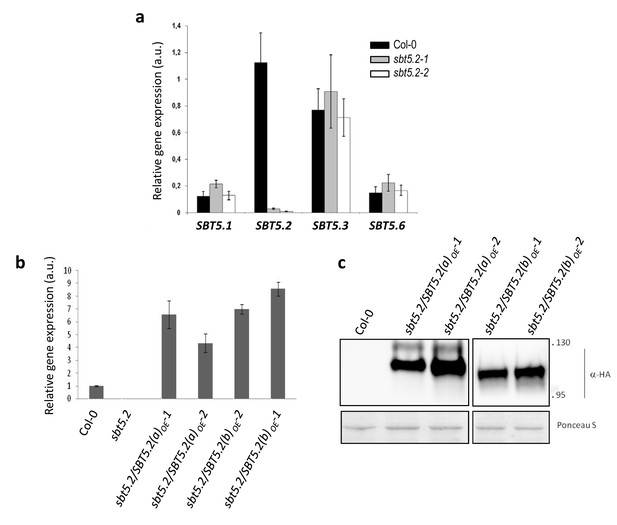
Characterization of sbt5.2 mutant lines before and after transformation with SBT5.2(a) and SBT5.2(b) overexpresing constructs.
(a) The expression of SBT5.2 is strongly repressed in sbt5.2 mutant lines. qRT-PCR analysis of the six members of the clade V of Arabidopsis subtilases in wild-type Col-0 (black bars), sbt5.2–1 (gray bars) and sbt5.2–2 (white bars) Arabidopsis leaves. Expression values were normalized using SAND family gene as internal standard. Mean and SEM values were calculated from 4 independent replicates. Expression of SBT 5.4 and SBT5.5 was not detected in any line. (b) The relative expression of SBT5.2 in Arabidopsis leaves in wild-type Col-0, SBT5.2(a)OE and SBT5.2(b)OE lines was determined by qRT-PCR. The expression values were normalized by using SAND family gene as internal standard, and related to the value of Col-0, which is set at 1. Mean and SEM values were calculated from four individual plants per line. (c) Western blot analysis showing SBT5.2(a)-HA or SBT5.2(b)-HA protein accumulation in Arabidopsis transgenic and Col-0 wild-type plants. Ponceau S staining confirms equal loading. Molecular mass markers in kiloDaltons are indicated on the right.
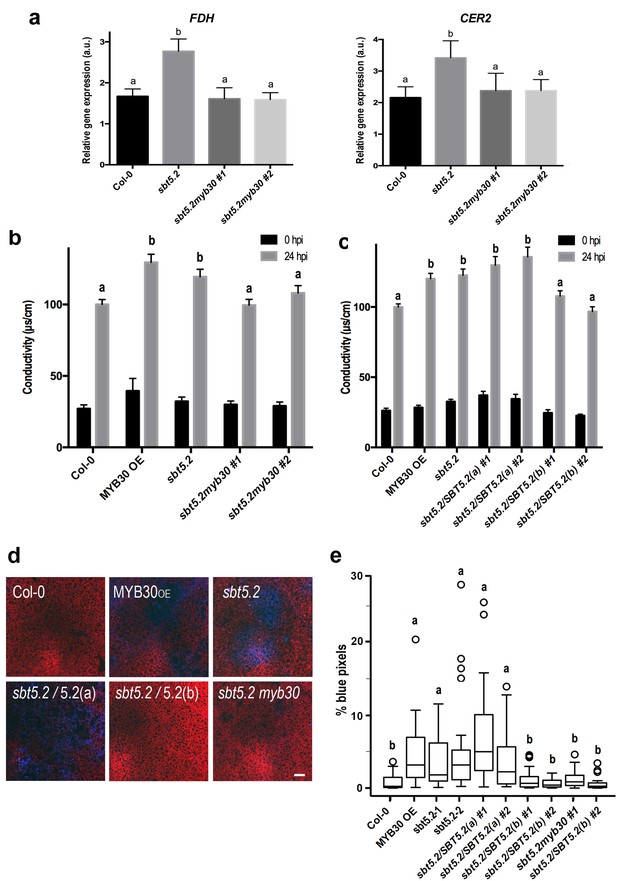
SBT5.2(b) attenuates MYB30-dependent transcriptional activation of VLCFA-related genes and hypersensitive cell death.
(a) Expression analysis of the MYB30 target genes FDH and CER2 in the indicated Arabidopsis lines 1 hr after inoculation with Pst AvrRpm1 (5 × 107 cfu/ml). Expression values of the individual genes were normalized using SAND family as internal standard. Mean and SEM values were calculated from 3 independent experiments (4 replicates/experiment). Statistical significance according to a Student’s t-test (p<0.05) is indicated by letters. (b,c) Quantification of cell death by measuring electrolyte leakage of the indicated Arabidopsis lines before (black bars) and 24 hr after (gray bars) inoculation with Pst AvrRpm1 (5 × 106 cfu/ml). Mean and SEM values were calculated from four independent experiments (three plants/experiment and four leaves/plant) and related to the value displayed by wild-type Col-0 plants, which is set at 100%. Statistical differences using multiple factor analysis of variance (ANOVA) (p<0.01) are indicated by letters. (d) Representative pictures of accumulation of phenolic compounds (blue coloration indicative of cell death) in the indicated Arabidopsis lines detected by epifluorescence 24 hr after inoculation with Pst AvrRpm1 (2 × 105 cfu/ml). Bar = 100 µm. (e) Blue pixels in the indicated lines were quantified using Image-Pro Plus and are shown as the percentage of the total number of blue pixels in each image. Boxplots are as follows: box limits, values between first and third quartiles; middle bar, median. Whiskers cover 1.5 times the interquartile distance and circles represent extreme values. Lowercase letters indicate significant differences with respect to the sbt5.5–2 line as determined by Bonferroni-corrected p-values (p<0.01) obtained after ANOVA and subsequent LSD post-hoc test.
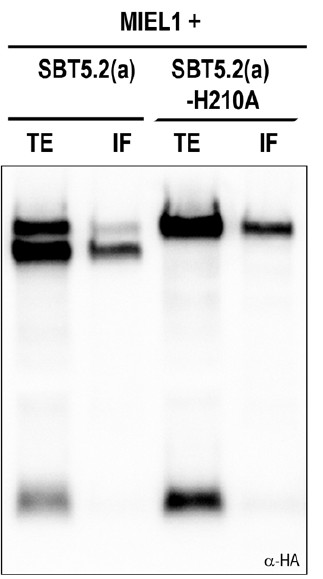
Western blot analysis of total protein extracts (TE) and intercellular fluids (IF) from N. benthamiana leaves co-expressing HA-tagged intracellular MIEL1 with wild-type and catalytically mutant SBT5.2(a) proteins.
https://doi.org/10.7554/eLife.19755.019Tables
FRET-FLIM analysis shows that MYB30 physically interacts with SBT5.2(b) at intracellular vesicles.
| Donor | Acceptor | Lifetime* | SD† | N‡ | E§ | p-value# |
|---|---|---|---|---|---|---|
| N. benthamiana | ||||||
| GFP-MYB30 | SBT5.2(b)-HA | 2.552 | 0.013 | 57 | - | |
| GFP-MYB30 | SBT5.2(b)-RFP | 2.274 | 0.019 | 54 | 10.86 | 5.8 × 10−21 |
| GFP-MYB30 | - | 2.669 | 0.009 | 82 | ||
| GFP-MYB30 | SBT5.2(b)362-730-RFP | 2.271 | 0.016 | 51 | 14.91 | 3.8 × 10−49 |
| GFP-MYB30 | SBT5.1(b)405-780-RFP | 2.592 | 0.018 | 44 | 2.86 | 4.2 × 10−05 |
| GFP-MYB123 | - | 2.570 | 0.013 | 59 | ||
| GFP-MYB123 | SBT5.2(b)362-730-RFP | 2.544 | 0.013 | 58 | 1.00 | 0.17 |
| A. thaliana | ||||||
| GFP-MYB30 | SBT5.2(b)-HA | 2.491 | 0.009 | 60 | - | |
| GFP-MYB30 | SBT5.2(b)-RFP | 2.151 | 0.018 | 60 | 13.65 | 1.2 × 10−33 |
| GFP-MYB30 | SBT5.2(b)H171A-HA | 2.473 | 0.012 | 60 | - | |
| GFP-MYB30 | SBT5.2(b)H171A-RFP | 2.115 | 0.023 | 60 | 14.49 | 7.8 × 10−26 |
-
* Mean lifetime in nanoseconds
-
† Standard deviation
-
‡ Total number of measured vesicles
-
§ Percentage of FRET efficiency (E = 1 - τDA/τD) calculated by comparing the lifetime of the donor in the presence of the acceptor (τDA) with its lifetime in the absence of the acceptor (τD).
-
# p value of the difference between the donor lifetimes in the presence and in the absence of the acceptor (t-test)
Additional files
-
Supplementary file 1
Oligonucleotide primers used in this study.
- https://doi.org/10.7554/eLife.19755.018





