Constitutive scaffolding of multiple Wnt enhanceosome components by Legless/BCL9
Figures
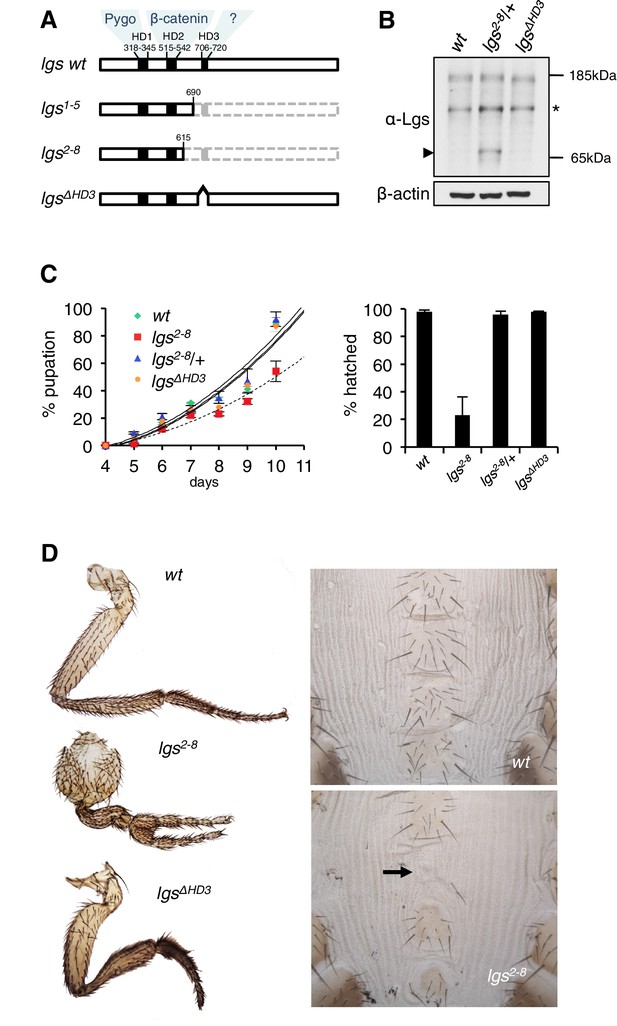
The C-terminus of Legless is required for Wg-dependent patterns in flies.
(A) Cartoon of lgs mutants, with domain boundaries indicated (grey, deleted sequences). (B) Western blot of lysates from lgs mutant embryos (genotypes indicated above panels), probed with antibodies as indicated, confirming stability of the lgs2-8 truncation product (~65 kDa, arrowhead; an unspecific cross-reactivity of this α-Lgs antiserum is marked by asterisk). (C) Developmental rates and survival of wt and lgs homozygous mutant larvae as indicated; first-instar larvae were picked (n = 25), and % pupation (left) or hatching of flies (right) was scored daily; error bars, SEM of four independent experiments. (D) Posterior leg and abdominal phenotypes of wt and lgs mutant flies, as indicated; representative examples are shown; arrow, missing sternite.
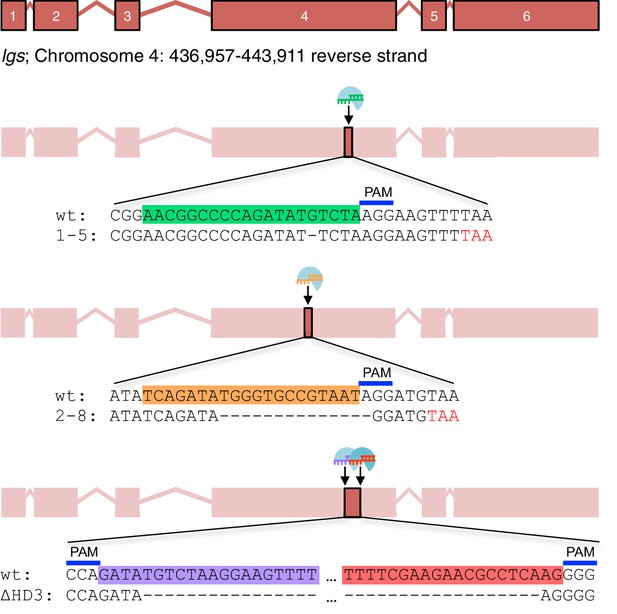
CRISPR/Cas9-based gene editing strategies for Drosophila legless.
Cartoon oflg sexon boundaries and targeting sites in exon 4; sgRNA target sequences and PAM sites are highlighted in the wt sequence, with corresponding mutant sequences underneath;red, premature stop codons generated in1-5and2-8alleles.
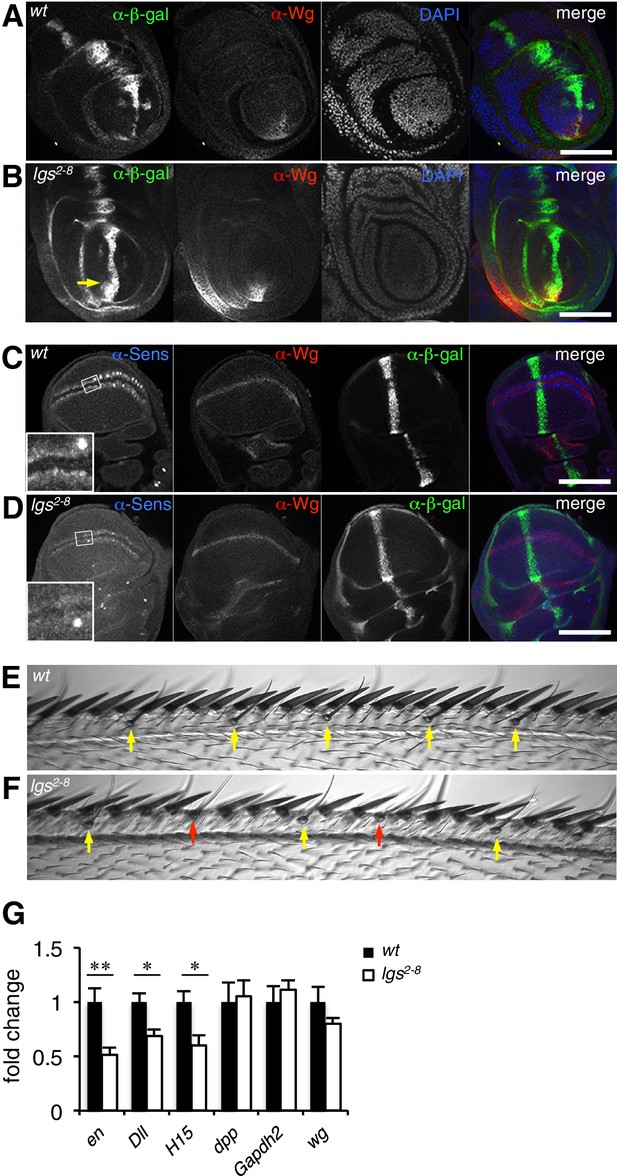
The C-terminus of Legless is required for nuclear Wg responses in imaginal discs.
(A, B) Third leg discs from third instar (A) wt or (B) lgs2-8/lgs2-8 larvae, fixed and stained with antibodies as indicated in panels (merges on the right), showing derepression of dpp.lacZ in the ventral compartment (arrow); space bar, 50 µm. (C, D) Corresponding wing discs, showing attenuated Sens expression along the prospective wing margin (see also insets); space bar, 100 µm. (E, F) Anterior margin segments of escaper flies, focused on stout margin bristles; yellow arrows, chemosensory bristles; red arrows, missing stout bristles causing gaps that are occupied by ectopic chemosensory bristles. (G) RT-qPCR assays of wing discs dissected from climbing wt and lgs2-8/lgs2-8 third instar larvae, as indicated; values were normalized relative to RpL32 (internal control), and are shown as mean ± SEM relative to wt (set to 1); * = p<0.05, ** = p<0.01.
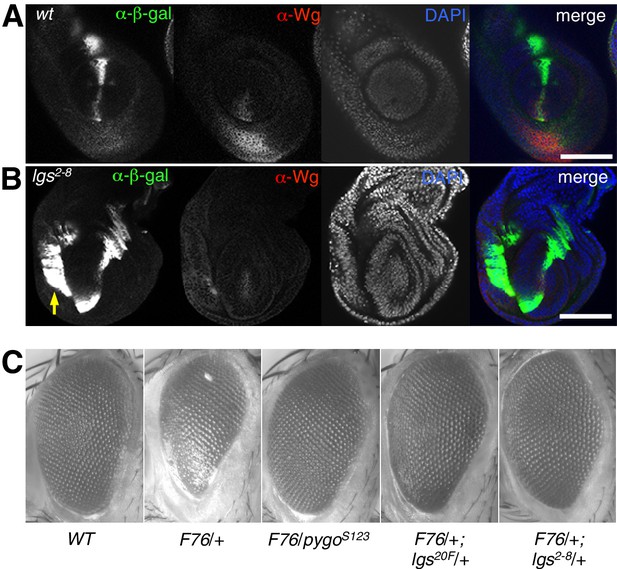
Additional analysis of lgs2-8 duringfly development.
(A, B) Leg discs as in main Figure 2 but giving rise to first or second leg, showing similar derepression of dpp. LacZ in the ventral compartments (arrow) of lgs2-8/lgs2-8 homozygous larvae as seen in their third leg discs; space bar, 50 mm. (C) Stereo images of eyes from wt or heterozygous mutant females also expressing activated Armadillo (F76) (Freeman and Bienz, 2001), as indicated in the panels, showing comparable suppression of the rough eye phenotype by lgs2-8/+ as by heterozygosity of the strongest known lgs allele (lgs20F; Kramps et al., 2002), or by apygo null allele (pygoS123; Thompson et al., 2002).
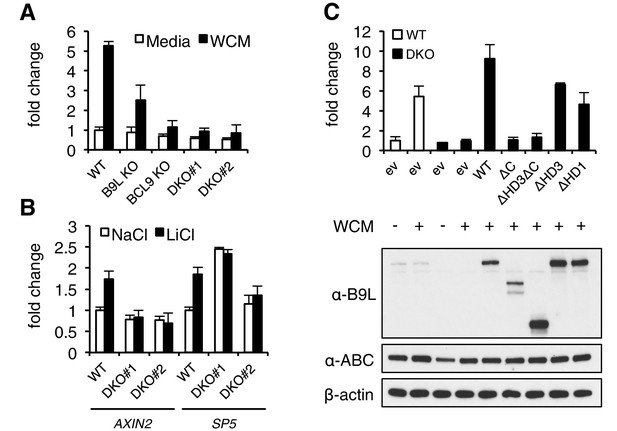
The C-terminus of BCL9/B9L is required for transcriptional Wnt responses in human cells.
(A) SuperTOP assays in wt or KO HEK293T cells lacking BCL9 and/or B9L, as indicated, ±6 hr of Wnt stimulation (WCM, Wnt3a-conditioned-media); mean ± SEM (n = 3 independent experiments). (B) RT-qPCR assays in wt or DKO HEK293T cells, ±6 hr of 20 mM NaCl or LiCl as indicated, revealing relative transcript levels of AXIN2 or SP5; values were normalized to TBP (internal control), and are shown as mean ± SEM relative to unstimulated wt controls (set to 1); note that the uninduced levels of SP5 were unusually high in one of the two DKO clones, but neither of the DKO clones showed Wnt-inducible SP5. (C) SuperTOP assays in wt or DKO HEK293T cells as in (A), 24 hr after transfection with wt and mutant B9L as indicated (below, corresponding Western blots; α-ABC, active unphosphorylated β-catenin); ev, empty vector control.
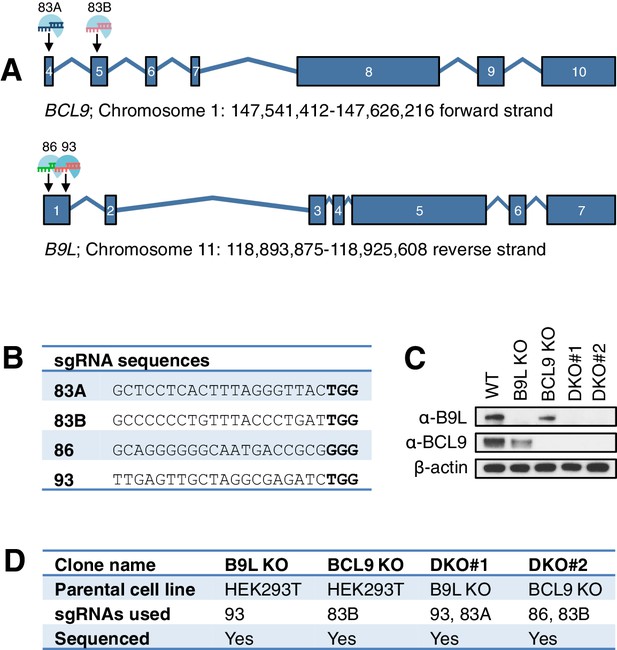
CRISPR/Cas9-based gene editing strategies for BCL9 and B9L in HEK293T cells.
(A) Cartoon of BCL9 and B9L exon boundaries (with non-coding 5’-and-3’-UTR exons omitted); sgRNA targeting sites are marked for both genes. (B) sgRNA sequences, with PAM sites in bold. (C) Western blot of lysates from individual HEK293T clones with BCL9 or B9L single KO, or with BCL9/B9L DKO, probed with antibodies as indicated on the left. (D) Summary of BCL9/B9L KO and DKO lines generated in this study.
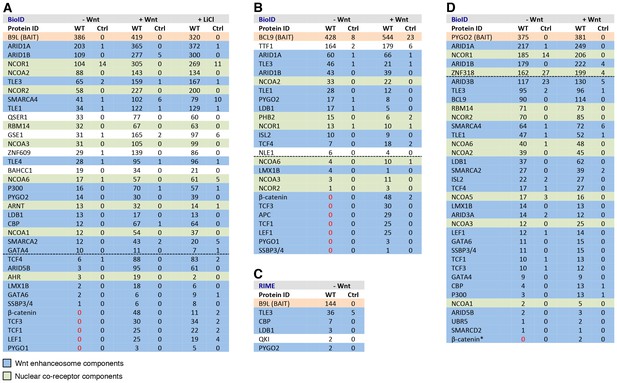
BCL9/B9L and PYGO2 are constitutively associated with the Wnt enhanceosome, and nuclear co-receptor complexes.
(A, B) List of BioID hits for (A) B9L-BirA* and (B) BCL9-BirA*±10–12 hr of WCM; names above the dotted line refer to the top hits, while names below this line refer to hits selected on relevance to Wnt (blue) or nuclear co-receptors (green); only specific hits with a > 5 spectral count ratio relative to the BirA* control are shown; numbers represent unweighted spectral counts (>95% probability). (C) RIME hits for FLAG-B9L-BirA*; only specific hits with a >5 spectral count ratio relative to the control are shown. (D) List of BioID hits for PYGO2-BirA*, as in (A, B); *, identified with lower confidence (>55% probability).
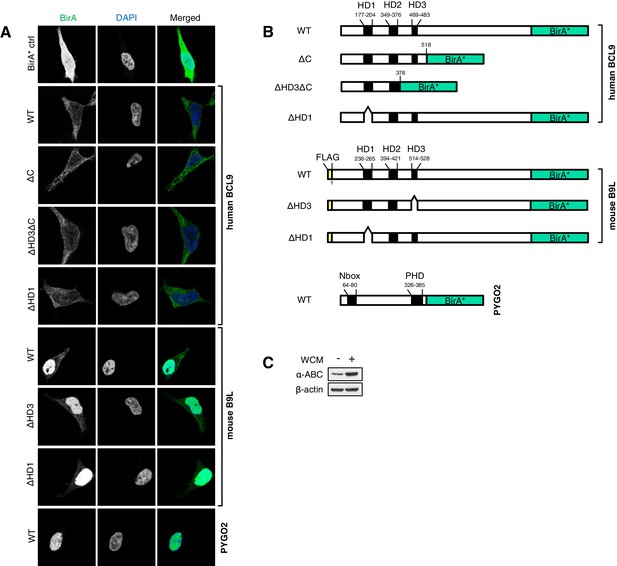
Stably transfected BCL9/B9L cell lines for BioID, and summary of wt and mutant BirA* baits.
(A) Immunofluorescence of stable BioID T-REx-293 cell lines expressing wt or mutant BCL9-BirA*, B9L- BirA* or BirA* alone, showing subcellular distribution of the various baits; nuclei are labeled by DAPI staining. (B) Cartoons of wt and mutant BCL9-BirA*, B9L-BirA* and PYGO2-BirA*. (C) Western blot probed with a-ABC antibody, to confirm Wnt induction of the stimulated PYGO2-BirA* cell lysates prior to BioID pull-downs and analysis.
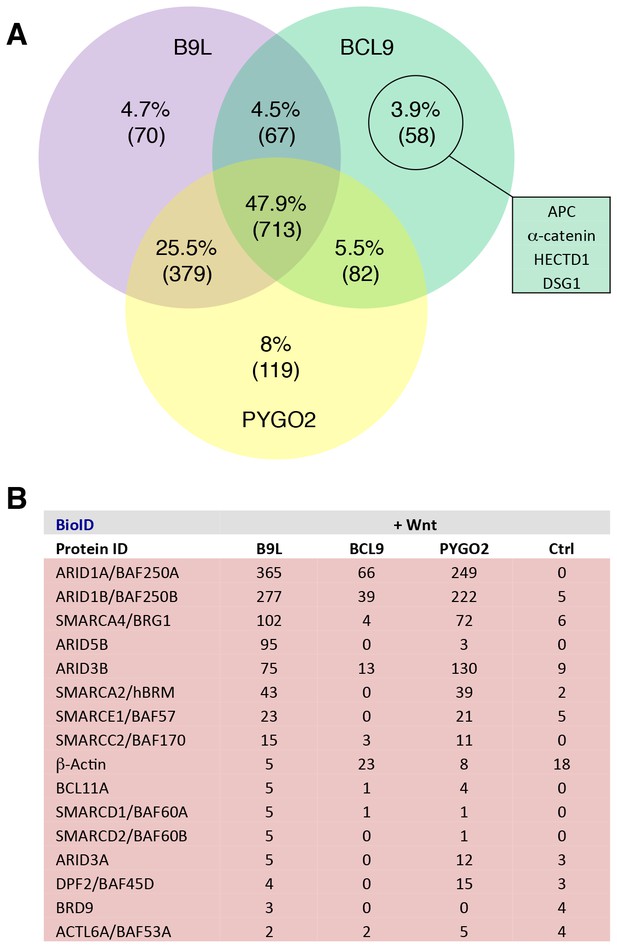
Additional analysis of BioID hits.
(A) Venn diagrams, showing the number of hits shared between the BCL9, B9L and PYGO2 BioID baits with >95% probability (excluding the BirA*-only control);right, a selection of high-confidence BCL9-specific hits not found with the other two (nuclear) baits. (B) Components of the SWI/SNF complex found by all three baits.
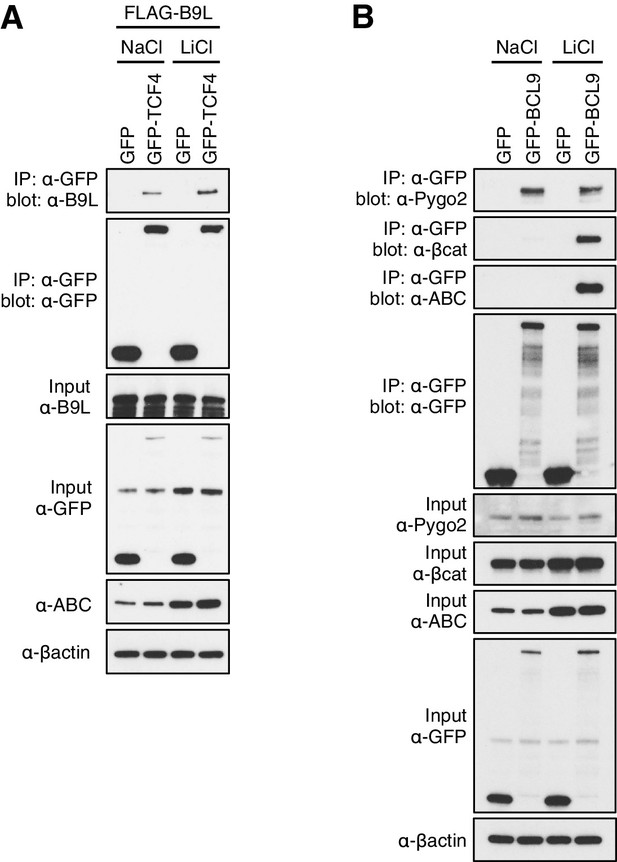
Constitutive association between B9L and TCF prior to Wnt stimulation.
CoIP assays in transiently transfected HEK293T cell, with Western blots showing (A) Wnt-independent association between GFP-TCF and FLAG-B9L, or (B) Wnt-dependent association between GFP-BCL9 with endogenous b-catenin or ABC (and Wnt-independent association with endogenous Pygo, as internal control). Transfected cells were exposed to LiCl, or NaCl as control, as described in the main Materials and methods.
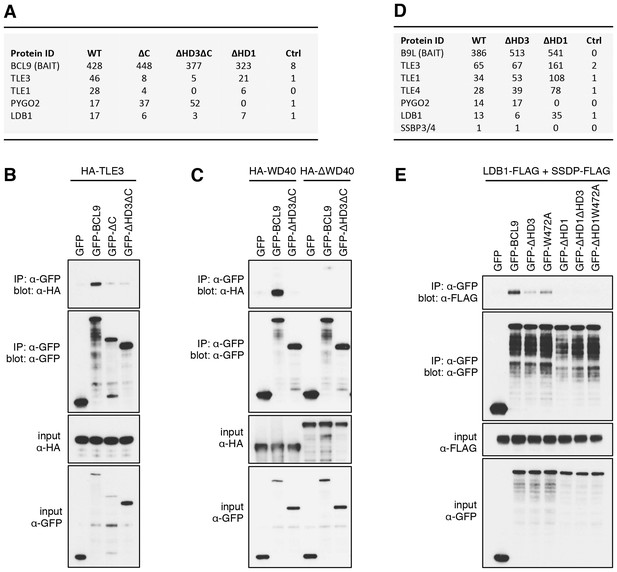
The Legless/BCL9 C-terminus binds to core Wnt enhanceosome components.
(A) Top BioID hits showing differential association with wt versus mutant BCL9-BirA* (unweighted spectral counts > 95% probability). (B, C) Western blots of coIPs of wt or mutant GFP-BCL9 with (B) HA-TLE3 or (C) HA-tagged truncations, after co-expression in HEK293T cells (lysed 48 hr after transfection), probed with antibodies as indicated on the left. (D) Top differential BioID hits of B9L-BirA* as in (A). (E) CoIP assays between co-expressed wt or mutant GFP-BCL9, LDB1-FLAG and SSDP-FLAG, as in (B); the band in the top panel corresponds to LDB1-FLAG.
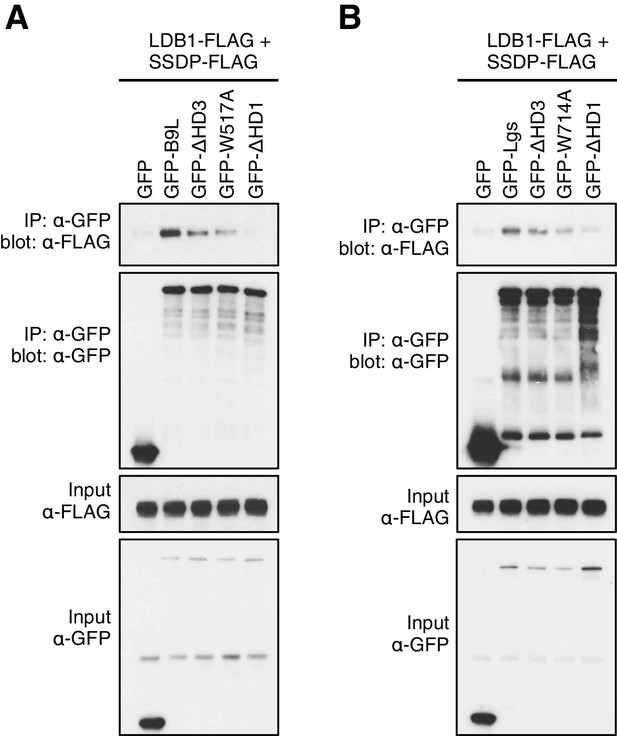
HD3-dependent interaction between LDB1 and Legless/B9L.
CoIP assays between co-expressed wt or mutant (A) GFP-B9L or (B) GFP-Lgs, and LDB1-FLAG and SSDP-FLAG, as inmain Figure 5E.
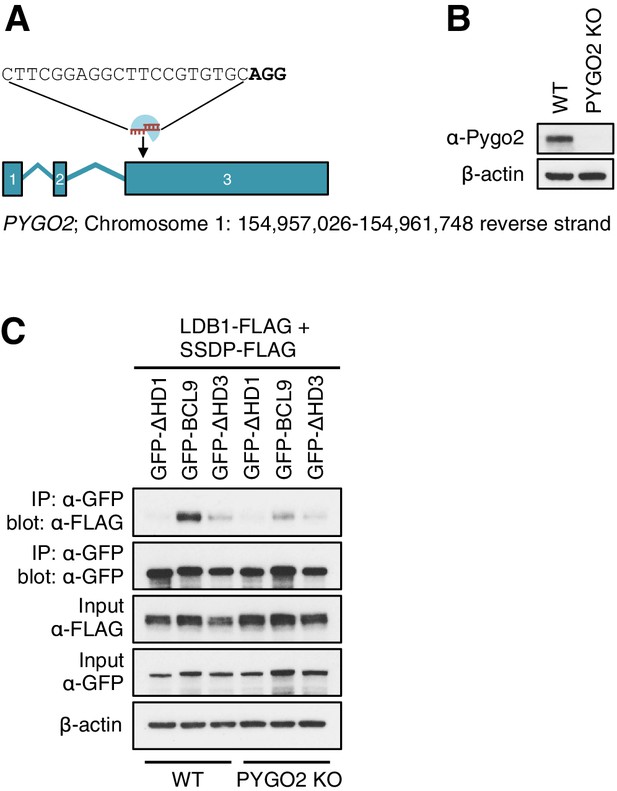
CRISPR/Cas9-based gene editing strategy for PYGO2 in HEK293T cells.
(A) Cartoon of PYGO2 exon boundaries and targeting site in exon 3; top, sgRNA sequence, with PAM site in bold; underneath, chromosomal location. (B) Western blot of lysates from individual HEK293T clones with PYGO2KO, probed with antibodies as indicated on the left. (C) CoIP assays between co-expressed wt or mutant GFP-BCL9, and LDB1-FLAG plus SSDP-FLAG in wt or PYGO2KO cells, confirming that the association of this BCL9 paralog with the ChiLS core complex is partly mediated by its binding to Pygo via HD1.
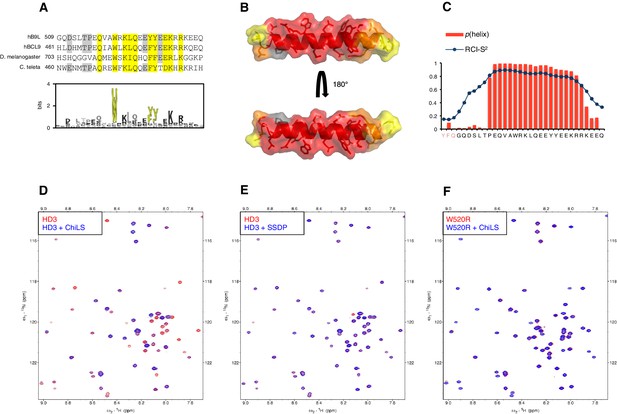
HD3 binds directly to ChiLS.
(A) Top, sequence alignments of HD3; yellow, conserved residues; grey, semi-conserved residues. Bottom, position-specific alignment (HMMER; Finn et al., 2011) for B9L HD3; yellow, conserved tryptophan and phenylalanine/tyrosine doublet. (B) Predicted structure of HD3 (by I-TASSER), with heat-map indicating relative line broadenings upon incubation of 15N-HD3 with ChiLS (see D), ranging from 80% (red) to 40% (yellow); grey, proline (not detectable). (C) TALOS+ predictions of α-helicity of HD3, based on backbone secondary chemical shifts in Figure 6—figure supplement 1; for each position, the rigidity index (RCI-S2) and helical probability (p) are indicated (pink, N-terminal linker residues). (D–F) Overlays of HSQC spectra of 100 μM 15N-labeled (D, E) wt HD3 or (F) W520R mutant alone (red), and probed with 300 μM (D, F) MBP-Chip205-436-Lip-SSDP1-92 or (E) Lip-SSDP1-92 (blue); line broadening owing to ChiLS binding to wt HD3 (D) results in the disappearance of selected resonances in the blue (HD3 + ChiLS) spectrum, revealing the corresponding resonances from the red (HD3-only) spectrum.
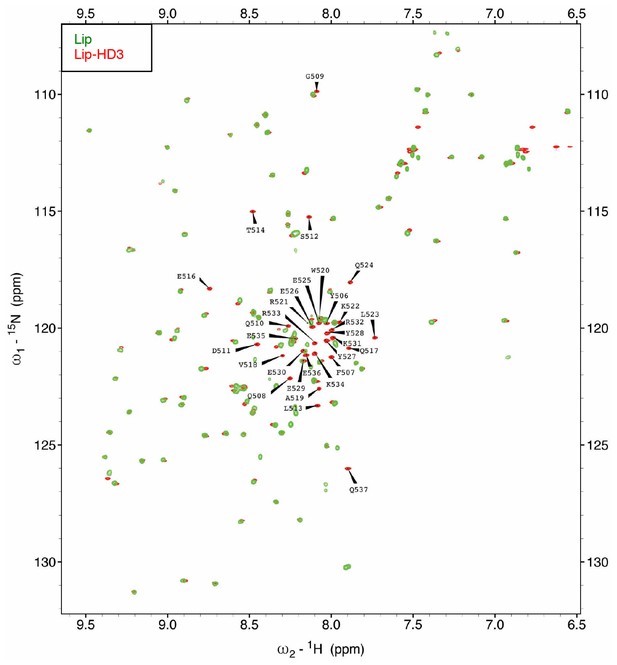
Assignment of the [1H-15N]-HSQC spectrum of HD3.
Assignments of 1H-15N correlations for the backbone amide resonances of [15N-13C]-Lip-HD3 residues, after overlay with [15N-13C]-Lip (to identify Lip residues).
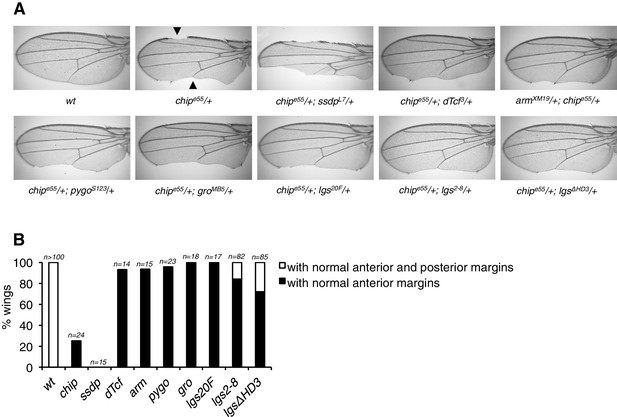
Modification of the chipp henotype by loss of HD3.
(A) Genetic interactions between chipe55/+ heterozygosity (causing wing margin defects, marked by arrowheads) and heterozygosity of Wnt pathway components; representative wings are shown. Note that this phenotype is exacerbated by lowering the dose of SSDP protein (as expected, given that Chip and SSDP form an obligatory complex; see Fiedler et al., 2015), but ameliorated by lowering the dose of associated proteins such as dTCF, Armadillo, Pygo, Legless or Groucho, possibly because normal levels of the latter sequester some of the limiting amounts of Chip (which is reduced in these heterozygotes). Note the strong suppression by the HD3 allele, possibly indicative of the direct binding of Chip to this domain. (B) Restoration of normal wing margins by heterozygosity of genes, as indicated (see also A); black, partial restoration (only anterior margin); white, restoration of both margins.
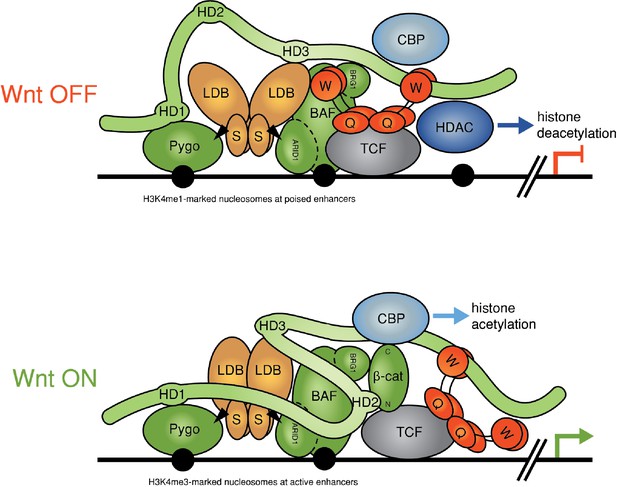
Refined model of the Wnt enhanceosome.
The Wnt enhanceosome complex associated with a Wnt-responsive enhancer in its OFF or ON state (for initial model, see Fiedler et al. [2015], illustrating multivalent constitutive interactions of Legless/BCL9 with Pygo (through HD1), ChiLS (through HD3) and Groucho/TLE (through its C-terminus). OFF, HD2 is poised to interact with Armadillo/β-catenin upon Wnt-induced stabilization; ON, rearrangement of Legless/BCL9 upon recruitment of Armadillo/β-catenin, resulting in the apposition of its C-terminus to TCF. CBP/p300 is associated with both states (possibly through direct binding to Legless/BCL9); its activity may be directed towards histones upon Armadillo/β-catenin binding, which antagonizes Groucho/TLE-dependent silencing and promotes the transcription of linked target genes. Likewise, the BAF complex is associated with both states (through the NPF motif of its subunit Osa/ARID1), earmarking the complex for feedback inhibition (see Figure 7—figure supplement 1). S, SSDP; Q, Q domain of TLE (tetramerizing, and binding to TCF); W, WD40 domain of TLE (binding to ChiLS); arrows, NPF-mediated interactions; green, positively-acting components; red, negatively-acting components; black circles, nucleosomes bearing H3K4me marks of poised or active enhancers (Kharchenko et al., 2011).
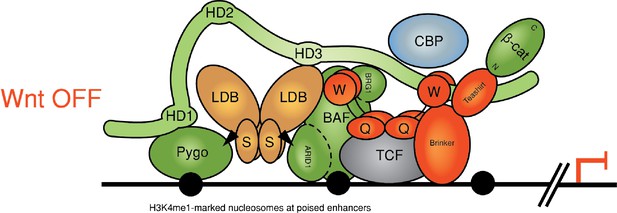
Reinstalling silencing on a Wnt-responsive enhancer.
Revised model for feedback inhibition of the Wnt enhanceosome in response to high signaling levels at the Wnt signaling source, which depends on the BAF complex and its Osa/ARID1 subunit (for initial model, see Fiedler et al., 2015). The homeodomain protein Brinker has been implicated in this process by studies of the Wg-responsive Ubx midgut enhancer in flies, in which the Osa response element maps to the primary Brinker binding site (Collins and Treisman, 2000). Brinker confers repression of this enhancer in response to high Wg signaling levels by binding to the WD40 domain of Groucho, but its repressive activity also requires recruitment of its co-repressor Teashirt, whose expression is locally induced by high levels of signaling emanating from the Wg source (Saller et al., 2002). Repression of thedppleg enhancer near the Wg source in the ventral leg disc also depends on Brinker (Theisen et al., 2007). Notably, Teashirt binds to Armadillo (Gallet et al., 1999) and could thus compete with its binding to dCBP, suggesting a mechanism by which the Brinker-Teashirt complex may reinstall silencing on a Wnt-responsive enhancer. Note that the BAF complex might also be responsible for silencing the enhanceosome after cessation of Wnt signaling.
Additional files
-
Supplementary file 1
Complete list of specific BioID hits obtained with B9L-BirA*.
- https://doi.org/10.7554/eLife.20882.021
-
Supplementary file 2
Complete list of specific BioID hits obtained with BCL9-BirA*.
- https://doi.org/10.7554/eLife.20882.022
-
Supplementary file 3
Complete list of specific BioID hits obtained with PYGO2-BirA*.
- https://doi.org/10.7554/eLife.20882.023





