Extensive cargo identification reveals distinct biological roles of the 12 importin pathways
Figures
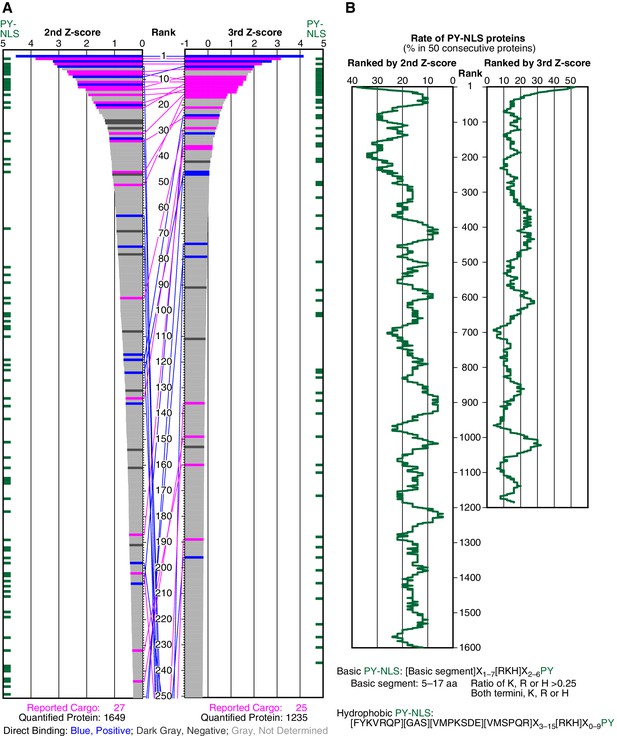
SILAC-Tp effectively sorts Trn-1 cargoes.
(A) Z-scores in the Trn-1 2nd- and 3rd-Z-rankings. The second (left) and third (right) Z-scores are presented for the top 250 proteins in the Trn-1 2nd- and 3rd-Z-rankings, respectively. The total number of ranked (quantified) proteins and the number of previously reported cargoes included in the ranking are indicated at the bottom. The magenta bars represent previously reported cargoes. The blue and dark gray bars represent the proteins that did and did not bind directly to Trn-1, respectively, in the bead halo assays (Supplementary file 2). Identical proteins marked by the colors are connected by lines. Proteins that carry PY-NLS motifs are indicated by green bars. (B) Distribution of PY-NLS motif-containing proteins in the rankings. The percentage of the proteins carrying PY-NLS motifs in 50 consecutively aligned proteins is presented along with the 2nd- and 3rd-Z-rankings (left and right, respectively). For example, the top 50 proteins in the 2nd-Z-ranking include 19 (38%) PY-NLS motif-containing proteins, and thus the value at position 1 is 38%. Two types of PY-NLS motifs, basic and hydrophobic, are defined as presented at the bottom.
-
Figure 1—source data 1
Statistical analysis of reported cargoes in the Trn-1 2nd- and 3rd-Z-ranking.
(A, B) Reported cargo rates, recall, and p-values of the Trn-1 2nd-Z-percentile ranking. Reported cargoes and proteins that have not been reported as cargoes (unreported candidate cargoes) in the top 1% to 20% ranks were counted in 1% rank increments, and the reported cargo rates (a lower bound on precision), recall (sensitivity), and p-values were calculated using two definitions of negative examples: (i) assuming any protein not reported as a cargo is negative (A), or (ii) assuming only those annotated to have non-nuclear localization in Uniprot to be negative (B). p-values were calculated by Fisher's exact test. The row marked in cyan corresponds to the 2nd-Z-15% cargoes. (C, D) Reported cargo rates, recall, and p-values of the Trn-1 3rd-Z-percentile ranking. Reported cargo rates, recall, and p-values of the top 1% to 20% ranks were calculated similarly using two definitions of negative examples. The row marked in magenta corresponds to the 3rd-Z-4% cargoes. See also Figure 1—figure supplement 3.
- https://doi.org/10.7554/eLife.21184.003
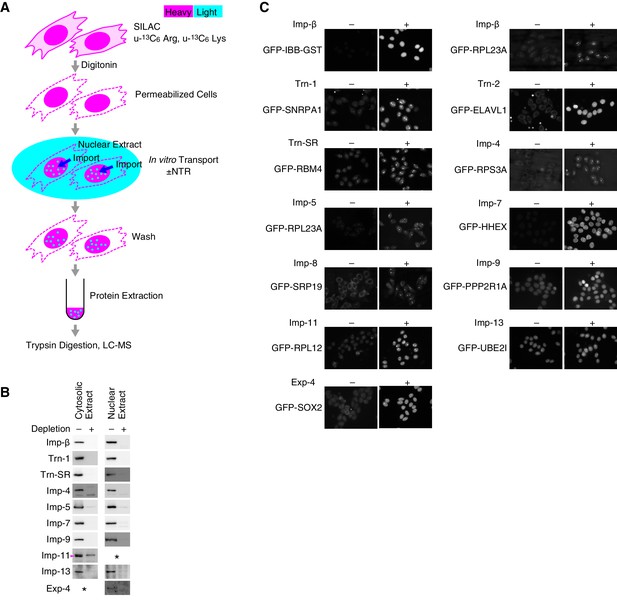
SILAC-Tp experimental system.
(A) Methods outline. For details, see the Materials and methods. This figure was originally published by Kimura et al. (2013a). (B) Depletion of the Imp-β family NTRs from the cytosolic and nuclear extracts. The cytosolic and nuclear extracts before and after the NTR depletion were analyzed by Western blotting with antibodies specific for each NTR. Asterisk, no band was detected. For Imp-11, the lower band indicated by the arrowhead was removed. For the antibodies, see Supplementary file 12B. (C) In vitro transport of specific cargoes. The reported cargoes were expressed as GFP-fusion proteins and purified. Then, the cargoes were added to the in vitro transport system with (+) or without (–) each specific NTR. The import was observed by fluorescence microscopy. IBB, Imp-β-binding domain of Imp-α. For the protein accessions, see Supplementary file 12B.
© 2013 The American Society for Biochemistry and Molecular Biology. All Rights Reserved. Figure 1—figure supplement 1A reproduced from Kimura et al. (2013).
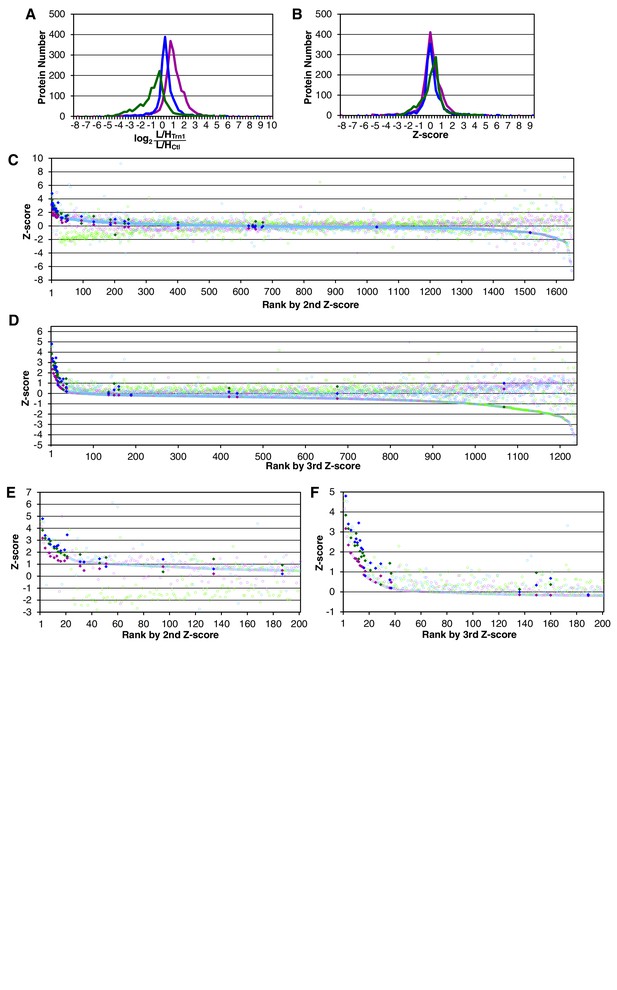
Trn-1 cargoes are effectively sorted by the second or third Z-scores in three replicates of SILAC-Tp.
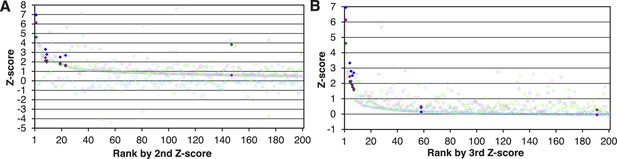
(A) Frequency distributions of the log2[(L/H+NTR)/(L/HCtl)] = log2(+NTR/Ctl) in three replicates of SILAC-Tp with Trn-1. (B) Frequency distributions of the Z-scores of log2[(L/H+NTR)/(L/HCtl)] in three replicates of SILAC-Tp with Trn-1. The Z-score of each protein was calculated as Z=(X-μ)/σ, where X is log2[(L/H+NTR)/(L/HCtl)] of a protein, μ is the mean of X in one replicate, and σ is the standard deviation of X. (C) Distribution of the three Z-scores in the Trn-1 2nd-Z-ranking. The Trn-1 2nd-Z-ranking consists of 1649 proteins. Both of the two or all three of the Z-scores are plotted against the rank by the second Z-score. The Z-scores in one replicate are presented as circles with the same color (blue, green, or violet), and the diamonds drawn in dark blue, dark green, and dark violet represent the Z-scores of reported cargoes in the respective replicates. (D) Distribution of the three Z-scores in the Trn-1 3rd-Z-ranking. The Trn-1 3rd-Z-ranking consists of 1235 proteins. All three of the Z-scores are plotted against the rank by the third Z-score. The marker colors are the same as those in (C). (E) Z-scores of the top 200 proteins in the Trn-1 2nd-Z-ranking. Close up of the top 200 in (C). (F) Z-scores of the top 200 proteins in the Trn-1 3rd-Z-ranking. Close up of the top 200 in (D).
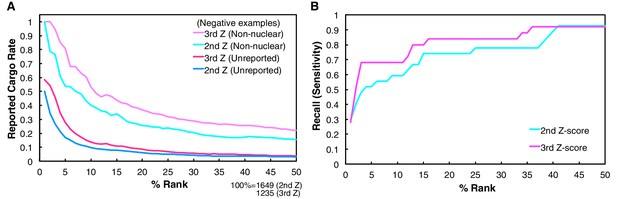
Reported cargo rates and recall of the Trn-1 2nd- and 3rd-Z-ranking.
(A) Rates of reported cargoes in the Trn-1 2nd- and 3rd-Z-percentile rankings. The reported cargo rates in the top 1% to 50% ranks are shown under the two definitions of negative examples: unreported, proteins not reported as cargoes; and non-nuclear, proteins annotated with non-nuclear localization. (B) Recall by ranks of the Trn-1 2nd- and 3rd-Z-percentile rankings. The recall of reported cargoes in the top 1% to 50% ranks are indicated. See also Figure 1—source data 1.
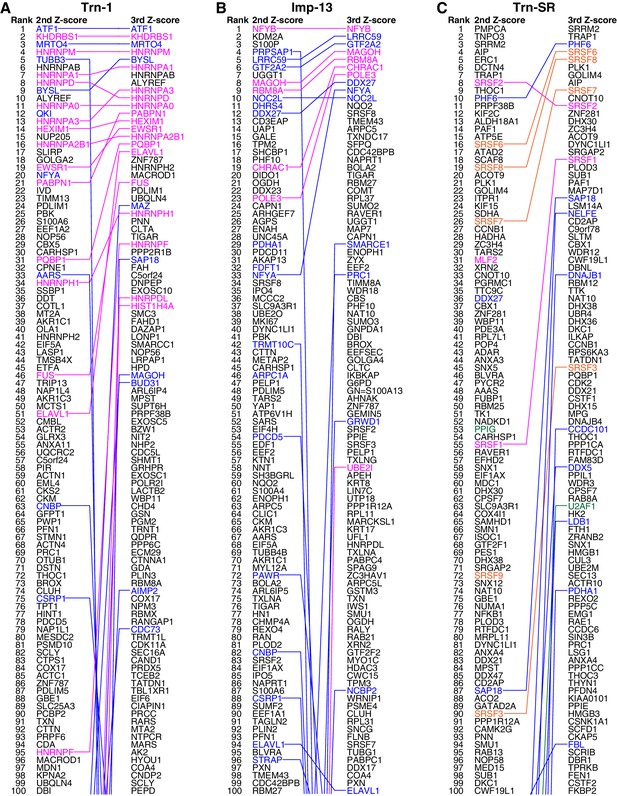
Trn-1, Imp-13, and Trn-SR cargo rankings.
(A–C) The top 100 proteins in the Trn-1 (A), Imp-13 (B), and Trn-SR (C) 2nd- and 3rd-Z-rankings (left and right, respectively). Magenta, reported cargoes; blue, proteins bound directly to the NTR in the bead halo assays (Supplementary file 2); orange in (C), SR-rich SFs that have not been reported; and green in (C), other RS (SR)-domain proteins. Identical proteins marked by the colors are connected by lines.
Imp-13 cargoes are effectively sorted by the second or third Z-scores in three replicates of SILAC-Tp.
(A) Distribution of the three Z-scores in the Imp-13 2nd-Z-ranking. The Imp-13 2nd-Z-ranking consists of 2060 proteins. Both of the two or all three of the Z-scores are plotted against the rank by the second Z-score. The Z-scores in one replicate are presented as circles with the same color (blue, green, or violet), and the diamonds drawn in dark blue, dark green, and dark violet represent the Z-scores of reported cargoes in the respective replicates. Z-scores of the top 200 proteins are presented. (B) Distribution of the three Z-scores in the Imp-13 3rd-Z-ranking. The Imp-13 3rd-Z-ranking consists of 1671 proteins. All three of the Z-scores are plotted against the rank by the third Z-score. The marker colors are the same as those in (A). Z-scores of the top 200 proteins are presented.
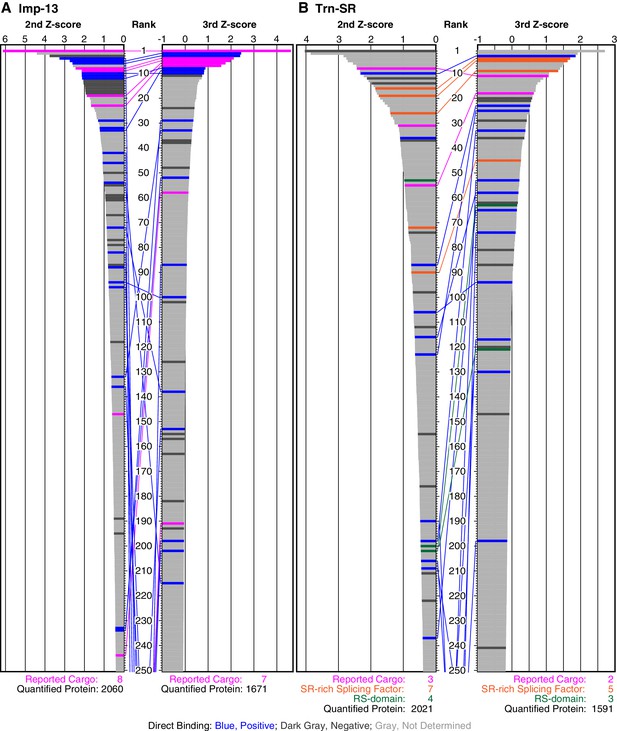
SILAC-Tp effectively sorts Imp-13 and Trn-SR cargoes.
(A) Z-scores in the Imp-13 2nd- and 3rd-Z-rankings. The second (left) and third (right) Z-scores are presented for the top 250 proteins in the Imp-13 2nd- and 3rd-Z-rankings, respectively. The total number of the ranked (quantified) proteins and the number of the previously reported cargoes included in the ranking are indicated at the bottom. The magenta bars represent the reported cargoes. The blue and dark gray bars represent proteins that did and did not bind directly to Imp-13, respectively, in the bead halo assays (Supplementary file 2). Identical proteins marked by the colors are connected by lines. (B) Z-scores in the Trn-SR 2nd- and 3rd-Z-rankings. The Z-scores in the Trn-SR 2nd- and 3rd-Z-rankings are presented as in (A). The orange and green bars represent SR-rich splicing factors that have not been reported and other RS-domain proteins, respectively.
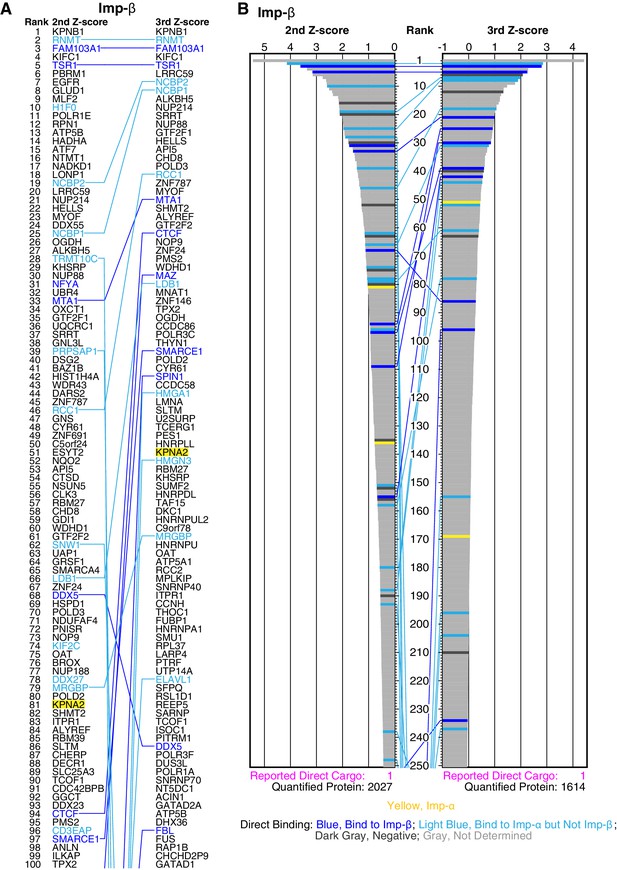
Imp-β cargo ranking and Z-scores in the 2nd- and 3rd-Z-rankings.
(A) The top 100 proteins in the Imp-β 2nd- and 3rd-Z-rankings (left and right, respectively). Blue indicates proteins bound directly to Imp-β or both Imp-α and -β, and light blue indicates proteins bound directly to Imp-α but not Imp-β in the bead halo assays (Supplementary file 2). Yellow highlight indicates Imp-α. Identical proteins marked by the colors are connected by lines. (B) Z-scores in the Imp-β 2nd- and 3rd-Z-rankings. The second (left) and third (right) Z-scores are presented for the top 250 proteins in the Imp-β 2nd- and 3rd-Z-rankings, respectively. The total number of the ranked proteins and the number of previously reported Imp-β direct cargoes that are included in the ranking are indicated at the bottom. Bar colors: blue, proteins bound directly to Imp-β or both Imp-α and -β in the bead halo assays; light blue, proteins bound directly to Imp-α but not Imp-β; dark gray; proteins that did not bind to either Imp-α or Imp-β; and yellow, Imp-α.
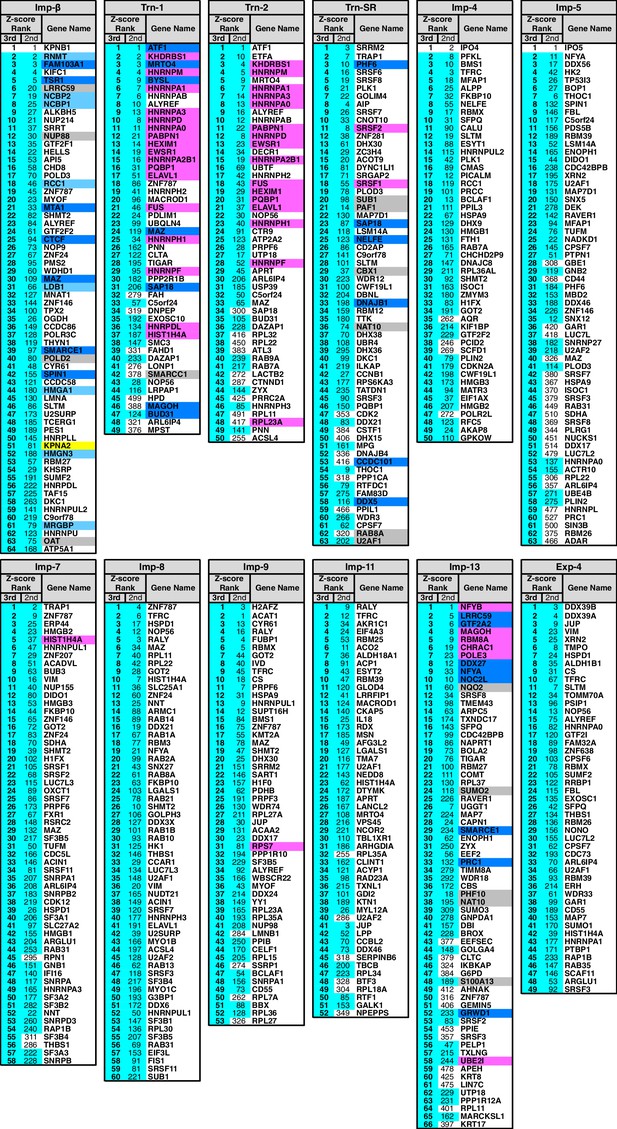
3rd-Z-4% cargoes of the 12 NTRs.
The 3rd-Z-4% cargoes of each NTR are listed by the gene names in the 3rd-Z-rank orders. The ranks by the second Z-scores are also shown. The 3rd-Z-4% and 2nd-Z-15% cargoes are indicated by cyan in the rank columns. Colors in the gene name columns: magenta, reported cargoes; blue, cargoes bound directly to the NTR in the bead halo assays (Supplementary file 2); light blue, cargoes bond directly to Imp-α but not to Imp-β; gray, proteins that did not bind to the NTRs; and yellow, Imp-α. For the 2nd-Z-15% cargoes, see Supplementary file 3.
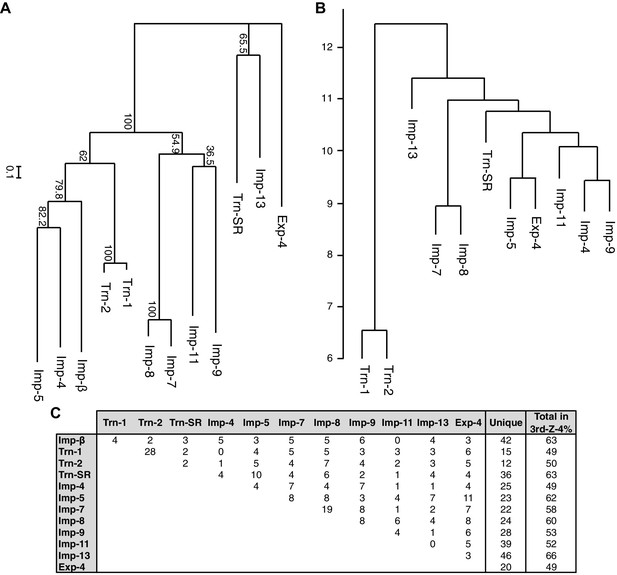
Phylogenetic tree and cargo profile hierarchical clustering of the Imp-β family import receptors.
(A) Phylogenetic tree of the 12 Imp-β family import receptors with the bootstrap values. Scale bar indicates substitutions per site. (B) A hierarchical clustering dendrogram of the same NTRs (except Imp-β) based on the similarities of their 3rd-Z-4% cargo profiles. Imp-β was excluded because Imp-α connects to Imp-β and many of the identified cargoes. The scale indicates the intercluster distance. (C) The numbers of 3rd-Z-4% cargoes shared by two NTRs. For the 2nd-Z-15% cargoes, see Supplementary file 5A.
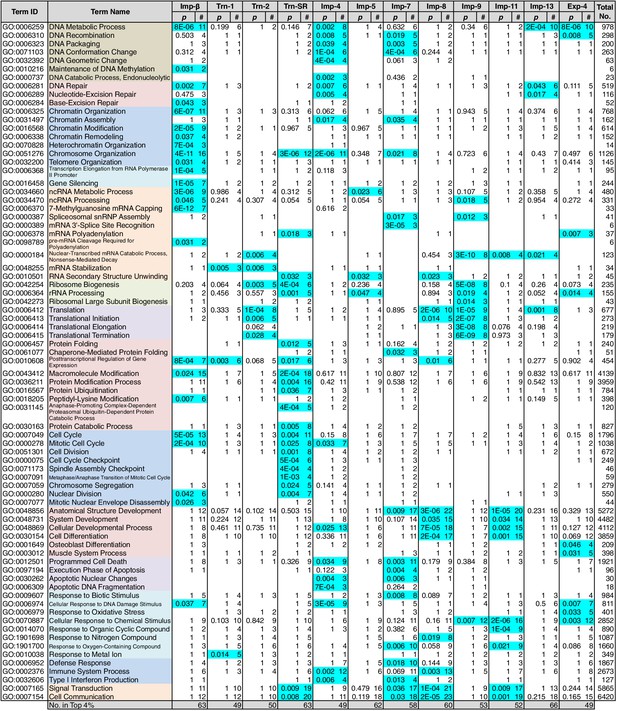
GO term (Biological Process) enrichments of the 3rd-Z-4% cargoes.
The 3rd-Z-4% cargoes were analyzed for GO term (term type, Biological Process) enrichment. The significantly enriched terms (p<0.05, cyan) in the 3rd-Z-4% cargoes of four or fewer NTRs were selected, and a representative term for each group of highly similar is presented with their p-values and the numbers (#) of cargoes annotated with them. Total No. denotes the number of proteins annotated with each term in the database. Related terms are bundled in the same color. This table was extracted from Supplementary file 6B. All the GO terms annotated to the 3rd-Z-4% cargoes are listed in Supplementary file 7. The correspondence between each 3rd-Z-4% cargo and GO term is summarized in Supplementary file 9. For the 2nd-Z-15% cargoes, see Supplementary files 6A, 8, and 10.
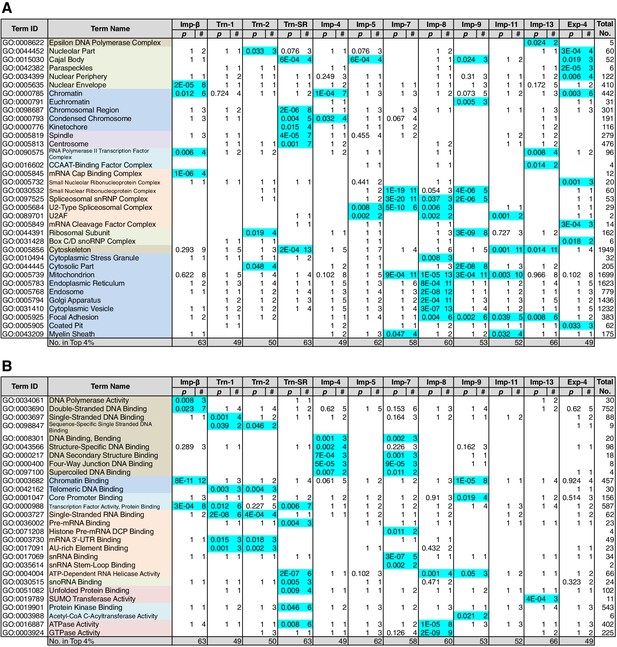
GO term (Cellular Component and Molecular Function) enrichments of the 3rd-Z-4% cargoes.
The 3rd-Z-4% cargoes were analyzed, and the results are presented in a format similar to that of Figure 5. (A) Term type, Cellular Component. (B) Term type, Molecular Function. These tables were extracted from Supplementary file 6B. All the GO terms annotated to the 3rd-Z-4% cargoes are listed in Supplementary file 7. The correspondence between each 3rd-Z-4% cargo and GO term is summarized in Supplementary file 9. For the 2nd-Z-15% cargoes, see Supplementary file 6A, 8, and 10.
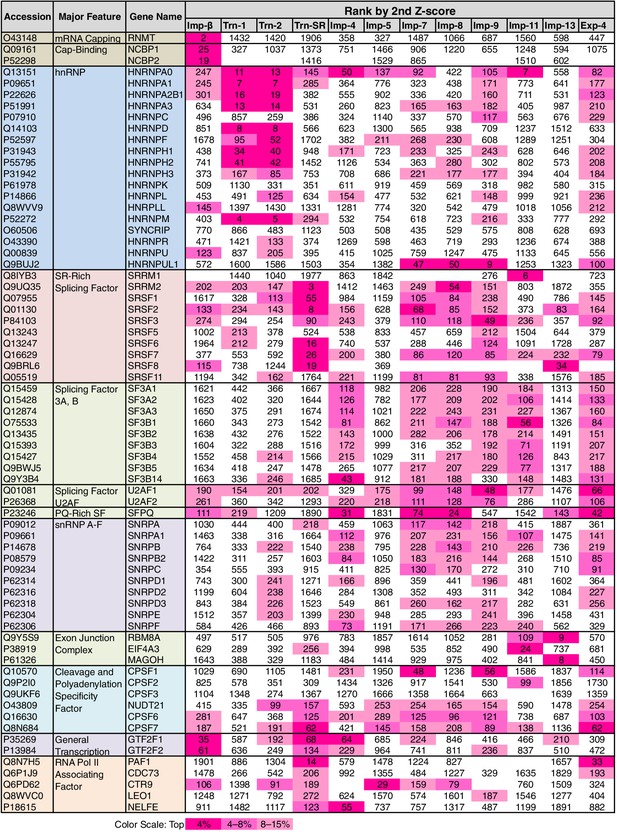
mRNA processing factors in the 2nd-Z-rankings.
The ranks of the mRNA processing factors in the 2nd-Z-rankings of the 12 NTRs are presented. The color scale is set by percentile rank as indicated. The 2nd-Z-rankings of the 12 NTRs include 275 proteins in total that are annotated with mRNA processing in GO. Of these, 69 were selected and are presented. For other factors and the 3rd-Z-rankings, see Supplementary file 11A.
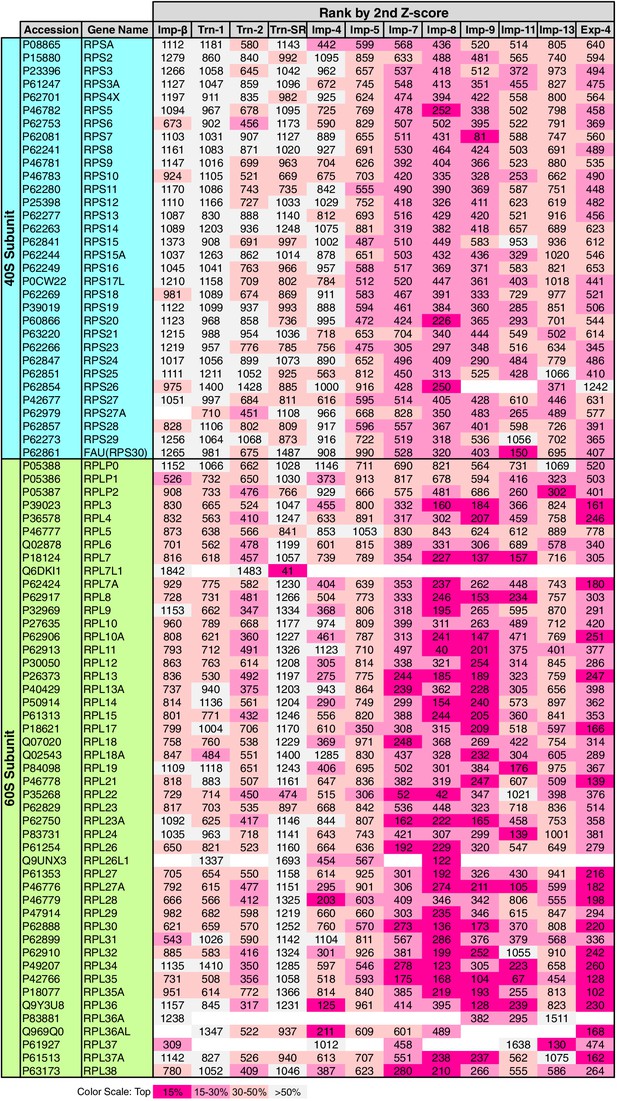
Ribosomal proteins in the 2nd-Z-rankings.
The ranks of the ribosomal proteins in the 2nd-Z-rankings of the 12 NTRs are presented. The color scale is set by the percentile rank as indicated. For the 3rd-Z-rankings, see Supplementary file 11B.

NTRs in the nuclei of permeabilized cells (Western).
https://doi.org/10.7554/eLife.21184.029
Imp-α1 in depleted cytosolic extract (Western).
https://doi.org/10.7554/eLife.21184.030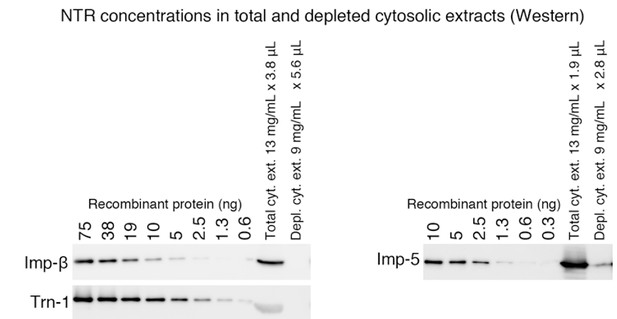
NTR concentrations in total and depleted cytosolic extracts (Western).
(Left panel) Reproduced from Kimura et al., 2013, Mol. Cell. Proteomics 12:145-157.
Additional files
-
Supplementary file 1
Results of SILAC-Tp.
Each sheet contains the result of SILAC-Tp with one of the 12 NTRs. The proteins that exhibited values at least once in the three replicates (Experiments 1–3) are listed with the values. The second and third Z-scores and the ranks according to those scores are also presented. The 2nd-Z-15% and 3rd-Z-4% cargoes are indicated in cyan. The LC-MS/MS quantitation data for each replicate (Experiments 1–3) are also included. Light/Heavy, the median of quantified L/H values; Light/Heavy count, the number of quantified values; Light/Heavy variability, coefficient-of-variation for log-normal distributed data. aReport: The proteins listed by Chook and Süel (2011) are regarded as reported cargoes, and the references are provided in the Legend sheet. For Imp-β, only direct cargoes are listed. For Trn-2, the reported Trn-1 cargoes are listed. bDirect Binding: The results of the bead halo assays (Supplementary file 2) are summarized. ++ or +, positive; ± or –, negative. cGO Nucleus: Annotated with ‘nucleus’ in Gene Ontology (term type, cellular component). The rows can be sorted into preferable orders with Excel. To access the mass spectra, chromatograms, or raw data, see Supplementary file 12A. Statistics: For each experiment, the number of proteins assigned with values (proteins assigned with Z-scores), the mean and standard deviation (S.D.) of are listed. where of each protein, is the mean of X in one experiment, and is the S.D. of .
- https://doi.org/10.7554/eLife.21184.017
-
Supplementary file 2
NTR–cargo direct binding.
The direct binding of the candidate cargoes to the NTRs was analyzed by bead halo assay. From well-characterized proteins that have not been reported as cargoes, (i) proteins ranked high (within the top 15% in the 2nd-Z-rankings or 4% in the 3rd-Z-rankings), around presumptive cutoffs (within about top 15–25% in the 2nd-Z-rankings), or lower and (ii) highly ranked proteins that are suspected as indirect cargoes or false positives based on their well-known features, e.g., PMPCA, GALE, UAP1, NQO2, EEF1A2, RAB2A, RAB8A, S100A4, S100A6, S100A13, and S100P, were selected and analyzed. Proteins in (i) verify the cargo identification and cutoff setting, and proteins in (ii) serve for finding indirect cargoes and false positives. The negative rate of these bead halo assays should be higher than the true overall false positive rate of the SILAC-Tp, because proteins in (ii) were selected preferentially. GST or GST-NTR was attached to glutathione-Sepharose beads, mixed with an extract of E. coli expressing GFP or a GFP-fusion protein, and observed by fluorescence microscopy. Q69L-Ran, which inhibits the NTR–cargo functional binding, was added as appropriate. The contrast of the bead fluorescence between the GST and GST-NTR indicates the binding, and the inhibition of this binding by Q69L-Ran certifies the specificity of the binding; ++ or +, positive; ± or –, negative. Summary of the results (p2–5): The results are summarized in both 2nd- and 3rd-Z-rank order. The 2nd-Z-15% and 3rd-Z-4% cargoes are indicated by cyan, and positive binding (++ or +) is indicated by blue. Trn-1 (p6–8): The GFP-fusion proteins were divided into five groups (A–E) according to the expression levels. Because GFP binds weakly to Trn-1, the concentrations of GFP (control) and GFP-fusion proteins were equalized within each group, and the binding was observed in the same conditions. The images are comparable within a group. Trn-2 (p9): GFP weakly binds to Trn-2, and the concentrations of GFP and GFP-fusion proteins were equalized. The images are comparable. Proteins whose ranks differed substantially between the Trn-1 and Trn-2 Z-ranking were assayed. Imp-13 (p10–13): GFP does not bind to Imp-13, and GFP was added to the control mixture at the highest concentration. Three images (GST, GST-Imp-13, and GST-Imp-13 + Q69L-Ran) for each GFP-fusion protein were acquired under identical conditions, and the background intensities and dynamic ranges were equalized. Trn-SR (p14–16): GFP weakly binds to Trn-SR, and the procedures were similar to those used for Trn-1. The GFP-fusion proteins were divided into four groups (A–D), and the images are comparable within a group. Imp-α/β (p17–20): GFP does not bind to Imp-α or -β, and the procedures were similar to those used for Imp-13. GST-Imp-α2 lacks the N-terminal Imp-β-binding domain. Western blotting (p21–22): The GFP-fusion proteins in the E. coli extracts were relatively quantified by Western blotting using an anti-GFP antibody (Roche). The extracts containing the amounts of protein (ng) indicated at the bottoms were loaded. The arrowheads indicate the expected full-length products. The GFP-moieties including those of the partial products were quantified by chemiluminescence. GFP was used as the standard. The Western blots were replicated more than three times. Accessions and sequences (p23–27): The cDNAs were cloned from a HeLa cDNA library by PCR. The accession numbers of the proteins are listed. If the sequence of a used protein is different from that in the database, the deleted, substituting, or inserted amino acids are indicated by the colors. The sequences that matched perfectly are not presented.
- https://doi.org/10.7554/eLife.21184.018
-
Supplementary file 3
The 2nd-Z-15% cargoes of the 12 NTRs.
The 2nd-Z-15% cargoes of each NTR are listed by the gene names in the 2nd-Z-rank orders. The ranks by the third Z-scores are also shown. Cyan in the rank columns indicates the 2nd-Z-15% and 3rd-Z-4% cargoes. Colors in the gene name columns: magenta, reported cargoes; blue, cargoes bound directly to the NTR in the bead halo assays (Supplementary file 2); light blue, cargoes bound directly to Imp-α but not Imp-β; gray, proteins that did not bind to the NTRs; yellow, Imp-α; and green, reported export cargoes. For the 3rd-Z-4% cargoes, see Figure 3.
- https://doi.org/10.7554/eLife.21184.019
-
Supplementary file 4
Example of extracted ion chromatograms (EICs) of peptides.
EICs of GLUD1 peptides in the SILAC-Tp with Trn-1: As a general problem of high-throughput LC-MS/MS quantitation, quantitative values of proteins with fewer quantified peptides deviate among the replicates. Referring to EICs of the quantified peptides is useful to avoid misidentification of cargoes. For example, the L/H ratios of a Trn-1 2nd-Z-15% cargo GLUD1 (P00367, ranked 110th and 342nd by the second and third Z-score, respectively) deviated largely in the three replicates of SILAC-Tp with Trn-1 (Supplementary file 1). Panels (A–H) show EICs of the indicated peptide (trypsin targets, K and R, are written in lower cases) in the three (three Ctl and there +Trn) experiments (some peptides were not identified in all the experiments). Magenta letters indicate the quantified peptides and L/H rations. In panel (A), the elution time of the peptide TAMkYNLGLDLr differs largely between the expriment-1 Ctl and experiment-2 +Trn-1, the peak shape of the experiment-2 +Trn-1 is irregular, and the L/H ratio of it is much higher than those of other peptides in +Trn-1 experiments (B and E). Thus, there is concern about misidentification. Because the L/H count of GLUD1 in the experiment-2 +Trn-1 is two (TAMkYNLGLDLr and NLNHVSYGr, A and B) and the L/H ratio of a protein is defined as the median, the L/H ratio of GLUD1 in the expriment-2 +Trn-1 is affected by the L/H ratio of TAMkYNLGLDLr. Exclusion of the L/H ratio of TAMkYNLGLDLr in the experiment-2 +Trn-1 lowers the Z-score rank of GLUD1 significantly. In panel (C), the chromatogram of the peptide HGGTIPIVPTAEFQDr in the experiment-1 Ctl has an irregular peak, and the L/H ratio of it is much higher than those of other peptides in the Ctl experiments (A–H). Thus, overlap with other peptide or other failures may be possible. However, the L/H count of GLUD1 in the experiment-1 Ctl is four (TAMkYNLGLDLr, HGGTIPIVPTAEFQDr, ALASLMTYk, and GASIVEDkLVEDLr) and the value of HGGTIPIVPTAEFQDr does not affect the median. (The L/H ratio of TAMkYNLGLDLr, whose EIC differ between the experiment-1 Ctl and expriment-2 +Trn in (A) as mentioned above, may affect the L/H ratio of GLUD1 in the experiment-1 Ctl, but we assumed that it is reliable.) As above, the L/H ratios of proteins with low L/H counts (Supplementary file 1) may be affected by LC-MS/MS artifacts, and misidentification can be avoided by referring to the EICs. All the EICs and MS spectra in this work can be accessed by downloading the mass spectrometry data and Proteome Discoverer software (see the Materials and methods and Supplementary file 12A).
- https://doi.org/10.7554/eLife.21184.020
-
Supplementary file 5
Redundancy of NTRs: Cargoes shared by NTRs.
(A) The numbers of the 2nd-Z-15% cargoes shared by two NTRs. For the 3rd-Z-4% cargoes, see Figure 4. (B) Redundancy of the 3rd-Z-4% cargoes. Many proteins are included in the 3rd-Z-4% cargoes of multiple NTRs. The ranks of these cargoes in the 3rd-Z-rankings for all 12 NTRs are presented. (C) Relationships between the NTRs and the characteristics of their 3rd-Z-4% cargo proteins. The 3rd-Z-4% cargoes are grouped according to their characteristics (functions or biological processes that the proteins act in), and their ranks are presented as in (B). To make our points clear, typical terms for the protein characteristics and typical proteins related to the terms have been selected with reference to Gene Ontology (GO) and UniProt. Thus, the terms in this sheet are slightly different from those in the databases, fewer proteins than annotated in the databases are grouped, and the list is redundant. For the complete linkages between the GO terms and the 3rd-Z-4% cargoes, see Supplementary file 7. (D) Redundancy of the 2nd-Z-15% cargoes. The ranks of the 2nd-Z-15% cargoes are presented as in (B). (E) Relationships between the NTRs and the characteristics of their 2rd-Z-15% cargo proteins. The 2nd-Z-15% cargoes are grouped, and their ranks are presented in a manner similar to that in (C). For the complete linkages between the GO terms and the 2nd-Z-15% cargoes, see Supplementary file 8.
- https://doi.org/10.7554/eLife.21184.021
-
Supplementary file 6
GO term enrichments of the identified cargoes.
(A) Extraction of the GO term enrichments of the 2nd-Z-15% cargoes. The 2nd-Z-15% cargoes were analyzed for GO term enrichment in (C). The terms that were significantly enriched (p<0.05, cyan) in the 2nd-Z-15% cargoes of four or fewer NTRs were selected, and terms that represent many similar terms are presented. With the p-values, the numbers (#) of cargoes annotated with each of the terms are presented. Total No. represents the number of proteins annotated with each term in the database. Related terms are bundled in the same color. For the 3rd-Z-4% cargoes, see Figures 5 and 6. The correspondences between each 2nd-Z-15% cargo and GO term are summarized in Supplementary file 10. All the GO terms annotated to the 2nd-Z-15% cargoes are listed in Supplementary file 8. (B) Full table of the GO term enrichments of the 3rd-Z-4% cargoes. The 3rd-Z-4% cargoes were analyzed for GO term enrichment. For all combinations of GO terms and NTRs, the p-values for the term enrichments in the 3rd-Z-4% cargoes and the numbers (#) of cargoes annotated with the terms are presented. The numbers following ‘# in’ are the total numbers of 3rd-Z-4% cargoes. Total No. represents the number of proteins annotated with each term in the database. Cyan, p<0.05. Figures 5 and 6 were extracted from this table. This table was derived from Supplementary file 7, and see Supplementary file 7 to retrieve the protein accessions. (C) Full table of the GO term enrichments of the 2nd-Z-15% cargoes. The 2nd-Z-15% cargoes were analyzed and are presented in a manner similar to that in (B). (A) was extracted from this table. This table was derived from Supplementary file 8, and see Supplementary file 8 to retrieve the protein accessions.
- https://doi.org/10.7554/eLife.21184.022
-
Supplementary file 7
The 3rd-Z-4% cargoes annotated with GO terms.
With respect to each NTR, the accessions of the 3rd-Z-4% cargoes annotated with each GO term are listed. Cyan, significant term enrichment (p<0.05) in the 3rd-Z-4% cargoes of the NTR. Total No. represents the number of proteins annotated with each term in the database. Supplementary files 6B and 9 were derived from this table. For the 2nd-Z-15% cargoes, see Supplementary file 8.
- https://doi.org/10.7554/eLife.21184.023
-
Supplementary file 8
The 2nd-Z-15% cargoes annotated with GO terms.
With respect to each NTR, the accessions of the 2nd-Z-15% cargoes annotated with each GO term are listed. Cyan, significant term enrichment (p<0.05) in the 2nd-Z-15% cargoes of the NTR. Total No. represents the number of proteins annotated with each term in the database. Supplementary files 6C and 10 were derived from this table. For the 3nd-Z-4% cargoes, see Supplementary file 7.
- https://doi.org/10.7554/eLife.21184.024
-
Supplementary file 9
Correspondences between the 3rd-Z-4% cargoes and GO terms.
Each sheet shows the correspondences between the 3rd-Z-4% cargoes of one NTR and selected GO terms. A term annotation to a cargo is indicated by ‘1’ in the corresponding cell. Reported cargoes are indicated by magenta in the gene name cells, and the results of the bead halo assays (Supplementary file 2) are also indicated by colors in the gene name cells: blue, cargoes directly bound to the NTR; light blue, cargoes directly bound to Imp-α but not Imp-β; gray, proteins that did not bind to the NTR. GO terms that represent many similar terms were selected from the terms enriched significantly (p<0.05) for the 3rd-Z-4% cargoes of each NTR, and broadly defined terms were deselected. Magenta and orange in the term ID cells indicate terms that are significantly enriched for the cargoes of four or fewer NTRs, and of them magenta indicates the terms presented in Figures 5 and 6. Related GO terms are bundled in the same color, and different colors are used to distinguish the columns easily. The NTRs added in the transport reactions (white in the rank cells) were not analyzed. This table was derived from Supplementary file 7. For 2nd-Z-15% cargoes, see Supplementary file 10.
- https://doi.org/10.7554/eLife.21184.025
-
Supplementary file 10
Correspondences between the 2nd-Z-15% cargoes and GO terms.
Each sheet shows the correspondences between the 2nd-Z-15% cargoes of one NTR and selected GO terms. A term annotation to a cargo is indicated by ‘1’ in the corresponding cell. Reported cargoes are indicated by magenta in the gene name cells, and the results of the bead halo assays (Supplementary file 2) are also indicated by colors in the gene name cells: blue, cargoes directly bound to the NTR; light blue, cargoes directly bond to Imp-α but not Imp-β; gray, proteins that did not bind to the NTR. GO terms that represent many similar terms were selected from the terms enriched significantly (p<0.05) for the 2nd-Z-15% cargoes of each NTR, and broadly defined terms were deselected. Magenta and orange in the term ID cells indicate terms that are significantly enriched for the cargoes of four or fewer NTRs, and of them magenta indicates the terms presented in Supplementary file 6A. Related GO terms are bundled in the same color, and different colors are used to distinguish the columns easily. The NTRs added in the transport reactions (white in the rank cells) were not analyzed. This table was derived from Supplementary file 8. For 3rd-Z-4% cargoes, see Supplementary file 9.
- https://doi.org/10.7554/eLife.21184.026
-
Supplementary file 11
mRNA processing factors, ribosomal proteins, and transcription factors.
(A) Ranks of mRNA processing factors. All of the 2nd-Z-15% and 3rd-Z-4% cargoes that are annotated with mRNA processing in Gene Ontology (GO) are listed with the ranks in the 2nd- and 3rd-Z-rankings. The color scale is set by percentile rank as indicated. Figure 7 was extracted from this table. (B) The 3rd-Z-rankings of ribosomal proteins. The ranks of the ribosomal proteins in the 3rd-Z-rankings of the 12 NTRs are presented. The color scale is set by percentile rank as indicated. For the 2nd-Z-rankings, see Figure 8. (C) Extracts of the GO term enrichments of the transcription factors found in the 2nd-Z-15% cargoes. Seventeen to 36 proteins in the 2nd-Z-15% cargoes of each NTR are annotated with ‘transcription factor activity, sequence-specific DNA binding' or ’transcription factor activity, protein binding’ in GO. The factors were analyzed for GO term enrichment (term type, biological process, BP) in (D). Typical terms that represent similar terms were extracted from (D). The p-value for the term enrichment and the number (#) of factors annotated with the term are presented. Total No. represents the number of proteins annotated with each term in the database. Cyan, p<0.05. Related terms are bundled in the same color. (D) Full table of the GO term enrichments of the transcription factors found in the 2nd-Z-15% cargoes. The 2nd-Z-15% cargoes that are annotated with ‘transcription factor activity, sequence-specific DNA binding" or ’transcription factor activity, protein binding’ in GO were analyzed for GO term enrichment (term type, BP). The analyzed transcription factors are listed at the bottom. For all of the combinations of GO terms and NTRs, the p-value for the term enrichment and the number (#) of transcription factors annotated with the term are presented. The numbers following ‘# in’ are the total numbers of transcription factors in the 2nd-Z-15% cargoes. Total No. represents the number of proteins annotated with each term in the database. Cyan, p<0.05. (C) was extracted from this table. (E) SRSRSR motif in the 3rd-Z-4% cargoes. The 3rd-Z-4% cargoes that contain an ‘SRSRSR’ hexa-peptide sequence were counted.
- https://doi.org/10.7554/eLife.21184.027
-
Supplementary file 12
MS data files, recombinant cargoes, and antibodies.
(A) MS data files. The mass spectrometry proteomics data (.msf and .raw files) have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org/) with the dataset identifier PXD004655. The results of protein and peptide identification and quantitation are summarized in Supplementary file 1, and the .msf and .raw data files corresponding to each experiment in Supplementary file 1 are listed in this table. The quantitation results can be seen by opening .msf files by Proteome Discoverer software. To see spectra and chromatograms, .msf files and corresponding .raw files must be in the same local directory. A demo version of Proteome Discoverer can be downloaded at the Thermo Scientific omics software portal site (https://portal.thermo-brims.com/). (B) GFP-fusion proteins used for in vitro transport and antibodies.
- https://doi.org/10.7554/eLife.21184.028





