The LIM protein complex establishes a retinal circuitry of visual adaptation by regulating Pax6 α-enhancer activity
Figures
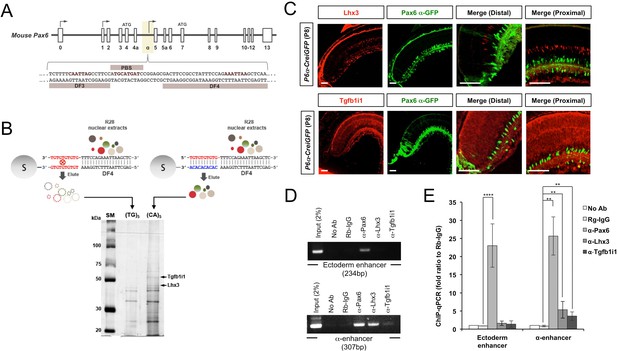
Identification of Lhx3 and Tgfb1i1 as Pax6 α-enhancer binding proteins.
(A) (Top) The genomic structure of the mouse Pax6 gene. Exons are shown as boxes, and arrows denote transcription initiation sites. (Bottom) The DF3, PBS, and DF4 sequences in the retina-specific α-enhancer are indicated with their core homeodomain (HD) and paired domain (PD) binding sites colored red. (B) Nuclear extracts from R28 rat retinal precursor cells were incubated with DF4 dsDNA oligomers with single-stranded 5’-(GT)5-3’ overhangs. DF4 oligomer-protein complexes were then added to Sepharose 6B columns conjugated with single-stranded DNA (ssDNA) of 5’-(CA)5-3’, which is complementary to the single-stranded overhang sequence of the oligomer, or 5’-(TG)5-3’ non-specific binding control. Proteins bound to the ssDNA column were eluted for SDS-PAGE and detected by silver staining. Protein bands specifically enriched in the (CA)5 column were then eluted from the gel and digested for mass spectrometric identification. This analysis identified the two bands marked by arrows as Lhx3 and Tgfb1i1. (C) Lhx3 and Tgfb1i1 expression in post-natal day 8 (P8) Pax6 α-enhancer::Cre-IRES-GFP (P6α-CreiGFP) mouse retinas stained with rabbit anti-Lhx3 (top) and anti-Tgfb1i1 (bottom) antibodies (red). These were also co-stained with a chick anti-GFP antibody (green). Scale bars, 100 μm. (D) DNA fragments bound to Pax6, Lhx3, and Tgfb1i1 in P7 mouse retinas were isolated by chromatin immunoprecipitation (ChIP) using rabbit polyclonal antibodies against each protein. The relative enrichment of each protein on the ectoderm enhancer and the α-enhancer of Pax6 gene was determined by PCR amplification of each enhancer sequence from the ChIP DNA fragments. (E) qPCR threshold cycle (Ct) values for each ChIP sample were compared to those of a protein-A bead only sample to obtain relative expression (2-ΔCt). The graph shows the ratio of 2-ΔCt values for each sample to those of a pre-immune rabbit IgG (Rb-IgG) ChIP sample. Error bars indicate standard deviations (STD, n = 5).
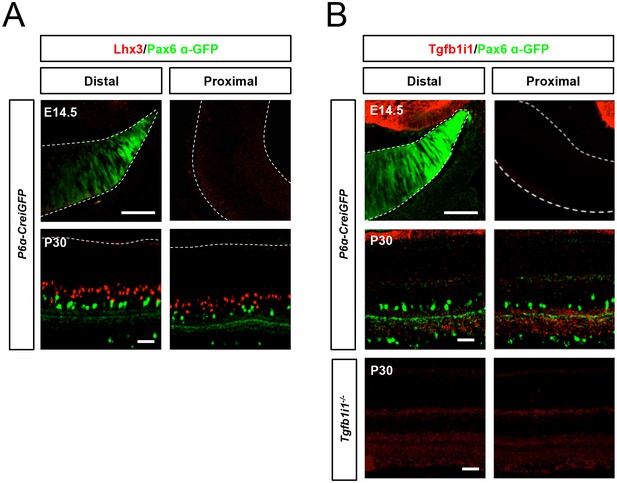
Lhx3 and Tgfb1i1 expression in embryonic and mature mouse retinas.
E14.5 and P30 P6α-CreiGFP mouse retinas stained with anti-Lhx3 (A) and anti-Tgfb1i1 (B) antibodies. Lhx3 is absent in E14.5 mouse retinas but expressed in bipolar cell subsets in post-natal (P8, Figure 1C) and adult (P30) mouse retinas. Tgfb1i1 is absent in E14.5 and P30 mouse retinas, but is expressed in P8 mouse retina (Figure 1C). The specificity of anti-Tgfb1i1 antibody was confirmed by staining P30 Tgfb1i1-ko mouse retinas (bottom). Scale bars, 50 μm.
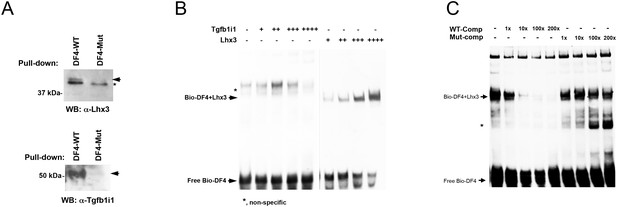
Binding abilities of Lhx3 and Tgfb1i1 to Pax6 α-enhancer sequence.
(A) P7 retinal nuclear extracts were incubated with either the wild-type DF4 (DF4-WT) dsDNA oligomers used in Figure 1B or mutant DF4 dsDNA oligomers (DF4-Mut) in which the homeobox core binding sequence ATTA was replaced with CGGC. Proteins captured by the (CA)5 ssDNA column were eluted for SDS-PAGE and Western blot (WB) analyses detecting Lhx3 and Tgfb1i1. Arrows indicate specific bands and the asterisk marks a non-specific band. (B) To evaluate direct binding of Lhx3 and Tgfb1i1 to DF4 sequence in the Pax6 α-enhancer, we performed an EMSA with biotin-labeled DF4 dsDNA oligomers (Bio-DF4) pre-incubated with in vitro translated Lhx3 and Tgfb1i1. (C) Lhx3 binding to the conserved homeodomain binding sequence in DF4 was measured by adding unlabeled competitor DNA (DF4 (WT-Comp) or mutated DF4 (Mut-Comp, ATTA to CGGC)) at 1-, 10-, 100-, and 200-fold the concentration of the Bio-DF4 probe. The asterisk marks non-specific bands.
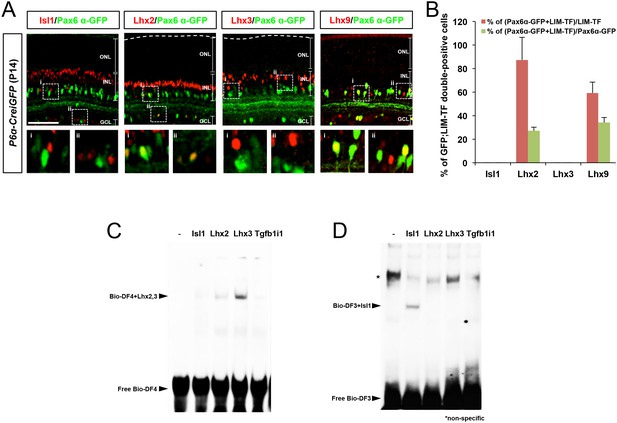
Relationship between LIM domain transcription factor expression and Pax6 α-enhancer activity in mouse retina.
(A) P14 P6α-CreiGFP mouse retinas stained with rabbit antibodies recognizing LIM domain transcription factors (LIM-TF), Isl1, Lhx2, Lhx3, and Lhx9, and a mouse antibody recognizing GFP, which represents Pax6 α-enhancer activity. Images in the bottom row are magnified versions of the dotted areas in the top row. Scale bars, 100 μm. (B) Population of Pax6 α-GFP-positive cells co-expressing each LIM domain transcription factor in total LIM-TF-expressing cells (red bars) or in total GFP-expressing cells (green bars) were obtained and shown in a graph. Error bars represent standard deviations (STD; n = 4, three litters). (C and D) EMSA performed with biotin-labeled dsDNA probes for the Pax6 α-enhancer DF4 (Bio-DF4; C) or DF3 (Bio-DF3; D) sequences. Unbound free DNA probes and LIM domain protein-bound DNA probes are indicated by arrows. An asterisk indicates a non-specific protein-bound probe band.
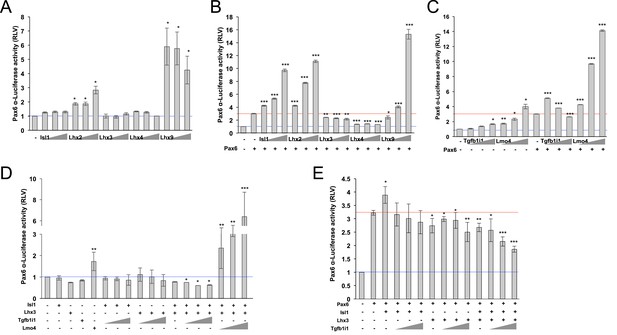
Lhx3 and Isl1 inhibit Pax6 α-enhancer activity in a Tgfb1i1-sensitive manner.
(A) The effects of LIM domain transcription factors on Pax6 α-enhancer activity were measured with a Pax6 α-enhancer luciferase reporter in HEK293T cells. These cells were co-transfected with DNA constructs encoding cDNAs of the indicated genes as well as the Pax6 α-luciferase reporter (0.2 μg). The triangles denote increasing doses of the indicated constructs (0.5 μg, 1 μg, and 2 μg). The relative luciferase activity of each sample was normalized to co-expressed β-galactosidase activity. (B) The effects of LIM domain transcription factors on Pax6-induced activation of the α-enhancer were also examined in the cells transfected with same DNA constructs used in (A) plus Pax6 construct (0.5 μg). (C) Regulatory effects of Tgfb1i1 and Lmo4 on Pax6 α-enhancer activity were also examined in the transfected cells as described in (A) and (B). (D and E) Cooperative effects of Isl1, Lhx3, and Tgfb1i1 on Pax6 α-enhancer activity were examined with the indicated combinations. (A – E) The blue lines indicated relative luciferase activity in samples expressing only the luciferase reporter, while red lines indicate activity of samples expressing the reporter with Pax6. The values on the Y-axes are averages. Error bars indicate STD (n > 5); *p<0.05, **p<0.01, ***p<0.001.
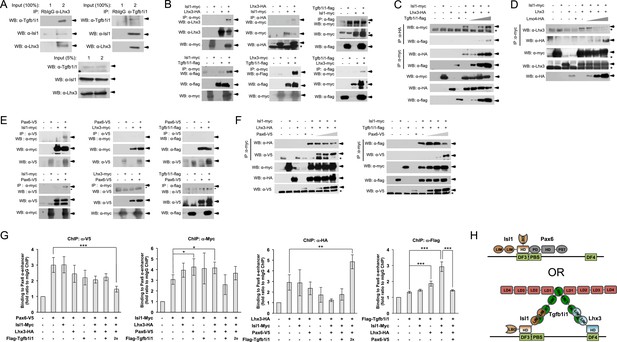
Pax6 and Tgfb1i1 antagonistically regulate Isl1-Lhx3 complex formation.
(A) Interactions between endogenous Isl1, Lhx3, and Tgfb1i1 in P7 mouse retinas measured by reciprocal co-immunoprecipitation (co-IP) and subsequent Western blotting (WB) with the indicated antibodies. P7 mouse retinal cell lysates were divided into two input tubes (1 and 2) in prior to the co-IP with indicated antibodies and subsequent WB detection of co-immunoprecipitated proteins. 5% of input cell lysates were used to compare the relative amount of the proteins in the retinal cell lysates used for co-IP. (B) Interactions between epitope-tagged Lhx3 and Isl1, Lhx3 and Tgfb1i1, and Isl1 and Tgfb1i1 in HEK293T cells were determined by co-IP and WB. The successful expression of each transfected cDNA was determined by WB for each protein in cell lysates (50 μg/lane; 5% of the co-IP input) with the corresponding epitope-tag antibodies. Arrows indicate specific WB bands, and asterisks indicate non-specific bands. (C and D) The effects of Tgfb1i1 and Lmo4 on Isl1-Lhx3 complex formation in HEK293T cells. Triangles denote increasing amounts of each DNA construct (1 μg, 2 μg, and 4 μg). Interaction between Pax6 and LIM domain proteins (E) and effect of Pax6 on LIM domain protein complex formation (F) in HEK293T cells were also examined by co-IP and WB analyses. (G) Reciprocal effects of LIM domain proteins and Pax6 on the binding to human PAX6 α-enhancer sequence in the transfected HEK293T cells were measured by qPCR amplification of α-enhancer sequences in DNA fragments isolated by ChIP with the indicated epitope tag-specific antibodies. Relative enrichment of each protein on the α-enhancer was determined by comparing the qPCR value of the transfected samples with those produced by antibodies bind non-specifically to the enhancer element in untransfected HEK293T cells. Error bars indicate STD (n > 5); *p<0.05, **p<0.01, ***p<0.001. (H) Schematic model depicting the binding of the Pax6-Isl1 and Isl1-Tgfb1i1-Lhx3 complexes to the Pax6 α-enhancer element. HD, homeodomain; LBD, LIM-binding domain; LD, leucine-rich domain; PD, paired domain.
-
Figure 3—source data 1
Protein-protein interaction between LIM proteins.
- https://doi.org/10.7554/eLife.21303.009
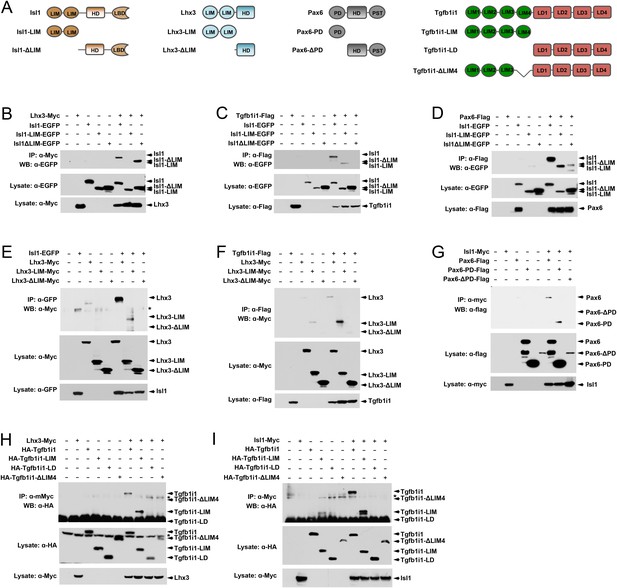
Pax6 and Tgfb1i1 antagonistically regulate Isl1-Lhx3 complex formation.
(A) Schematics for the full-length and deletion mutants of Isl1, Lhx3, Pax6, and Tgfb1i1 used in these experiments. HD, homeodomain; LIM, LIM domain; LBD, LIM binding domain; LD, leucin-rich domain; PD, paired domain; PST, transactivation domain enriched in proline, sereine, and threonine. (B – I) 293 T cells (~106) were transfected with DNA constructs (10 μg total) encoding the indicated protein fragments. Cell lysates collected at 48 hr post-transfection were incubated with antibodies against the epitope tags to immunoprecipitate each protein and its binding partners. Co-immunoprecipitated proteins were then analyzed by SDS-PAGE and subsequent WB with the indicated antibodies. In parallel, the cell lysates (containing 50 μg protein) were also analyzed by SDS-PAGE and WB with the indicated antibodies to examine relative levels of the overexpressed proteins in the transfected cells.
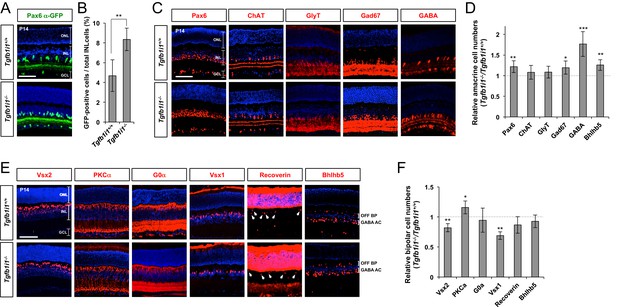
Elevated GABAergic amacrine cell number in Tgfb1i1−/−mouse retinas.
(A) Pax6 α-enhancer-active cells in P14 Tgfb1i1+/+ and Tgfb1i1−/−littermate mouse retinas were visualized by immunodetection of GFP expressed from the P6α-CreiGFP transgene. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (B) GFP-positive cell population in 250 μm x 250 μm retinal area. (C) P14 Tgfb1i1+/+ and Tgfb1i1−/−littermate mouse retinas stained with antibodies detecting amacrine cell subtype-specific markers. Pax6, pan-amacrine cells; ChAT, cholinergic amacrine cells; GlyT1, glycinergic amacrine cells; GABA, Gad67 and Bhlhb5 (in the bottom half of INL in the images in E), GABAergic amacrine cells. (D) Fold-changes of amacrine cell numbers in P14 Tgfb1i1−/− retinas compared to Tgfb1i1+/+ littermate retinas. (E) P14 Tgfb1i1+/+ and Tgfb1i1−/−mouse retinas stained for bipolar cell-specific markers. Vsx2, pan-bipolar cell marker; PKCα, rod bipolar cells; G0α, rod and ON-cone bipolar cells; Vsx1, OFF bipolar cells; Recoverin, photoreceptors (in the ONL) and type-2 OFF bipolar cells (in the INL); Bhlhb5 (in the top half of INL), type-2 OFF bipolar cells. (F) Fold-changes in marker-positive cell numbers in Tgfb1i1−/− retinas compared to Tgfb1i1+/+ littermate retinas. Values on the Y-axes of B, D, and F are averages. Error bars indicate STD (n = 4, three litters); *p<0.05; **p<0.01; ***p<0.001. Scale bars in the pictures, 100 μm.
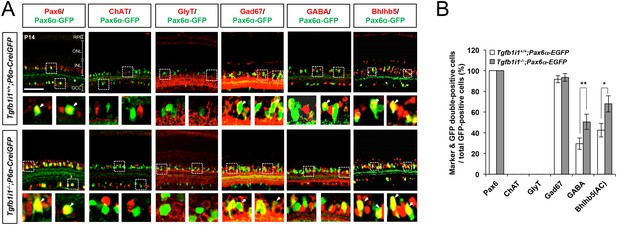
Elevation of Pax6 α-enhancer-active GABAergic amacrine cells in Tgfb1i1−/−mouse retinas.
(A) P14 Tgfb1i1+/+;P6α-CreiGFP and Tgfb1i1−/−;P6α-CreiGFP littermate mouse retinas co-stained with antibodies against amacrine cell subtype markers and GFP. Pax6, pan-amacrine cell marker; ChAT, cholinergic; GlyT1, glycinergic; Gad67, GABAergic; GABA, GABAergic subsets; Bhlhb5, GABAergic subsets (bottom of the INL). Outset images in the bottom row are magnified versions of the dotted box areas in the top row. Scale bar, 100 μm. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (B) Populations of GFP-positive cells co-expressing amacrine cell subset markers are shown in a graph. Values on the Y-axis are averages. Error bars indicate STD (n = 4, three litters). *p<0.05; **p<0.01.
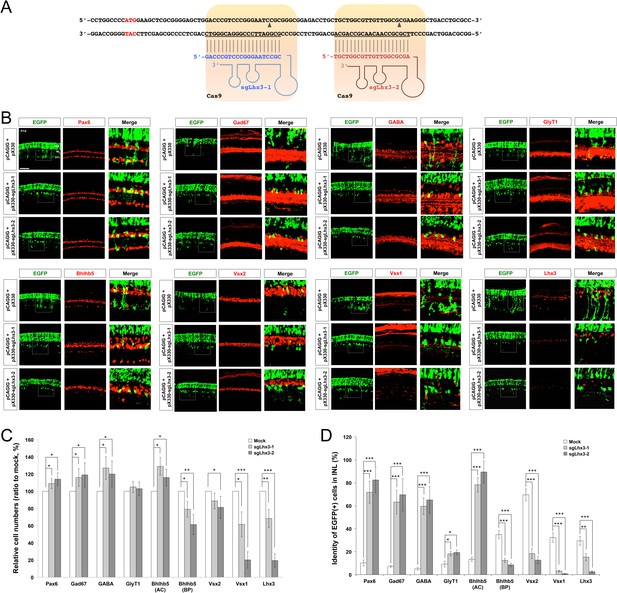
Deletion of Lhx3 in the post-natal mouse retina.
(A) To delete Lhx3 in the post-natal mouse retina, we designed two independent sgRNAs complementary to the sequences near the translation initiation site in the exon2 (highlighted in red), following the suggestion of the CRISPR Design server (http://crispr.mit.edu). The sequences were cloned into pX330 (pX330-U6-Chimeric_BB-CBh-hSpCas9) DNA construct, which express the cloned sgRNA and Cas9 endonuclease. (B) P14 mouse retinas, which were electroporated with the indicated pX330 DNA constructs at P0, were stained for the detection of various amacrine cell markers, including Pax6 (pan-amacrine), Gad67 (GABAergic), GABA (GABAergic subsets), Bhlhb5 (GABAergic subsets, bottom half of the INL), and GlyT1 (glycinergic), and bipolar cell markers, including Vsx2 (pan-bipolar), G0α (ON bipolar), Vsx1 (OFF bipolar), and Bhlhb5 (type-2 OFF bipolar, top half of the INL), as well as for EGFP, which is expressed from co-electroporated pCAGIG DNA construct. Thus, EGFP-positive retinal cells are expected to express sgRNA and Cas9 from the indicated pX330 DNA constructs. Successful loss of Lhx3 in the mouse retinas was examined by immunostaing of Lhx3. Scale bar, 100 μm. (C) Ratio of marker-positive cells to total INL cells of each sample was then compared with that of pX330+pCAGIG (Mock) sample. (D) The population of EGFP-positive cells co-expressing each amacrine or bipolar cell type-specific marker in total EGFP-positive INL cells were obtained and shown in a graph. Scores on the Y-axis of the graphs in C and D are averages (n = 6, two independent batches). Error bars indicate STD (n = 6, two independent batches); *p<0.05; **p<0.01; ***p<0.001.
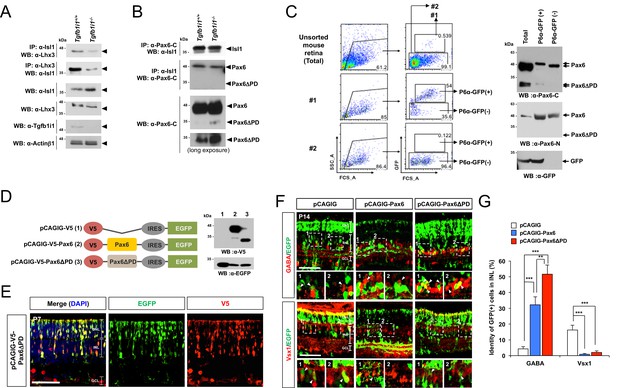
Pax6 α-enhancer-induced Pax6ΔPD isoform supports GABAergic amacrine cell fate.
(A) Reciprocal co-IP and WB analyses with the indicated antibodies reveal a reduced interaction between Isl1 and Lhx3 in P7 Tgfb1i1−/−mouse retinas compared with littermate Tgfb1i1+/+retinas (top two WB images). Tgfb1i1−/−retinal lysates show 1.6-fold higher Isl1 level than Tgfb1i1+/+retinal lysates and no significant change in the levels of Lhx3 and Actinβ1 (bottom four WB images). (B) No significant difference in the assembly of Isl1 and Pax6 was observed in P7 Tgfb1i1+/+and Tgfb1i1−/−littermate mouse retinas (top two WB images). Tgfb1i1−/− retinas show higher expression of the Pax6ΔPD isoform than Tgfb1i1+/+retinas and no change in full-length Pax6 (bottom two WB images). (C) Pax6 α-enhancer-active cells were isolated from P14 P6α-CreiGFP retinas by repeated FACS (see the Materials and Methods). Lysates of GFP(+) and GFP(-) retinal cells were then analyzed by SDS-PAGE and WB with a rabbit anti-Pax6 antibody. Successful purification of the cells was confirmed by WB detection of GFP in each fraction. (D) Diagram of pCAGIG DNA constructs encoding V5-tagged Pax6 (pCAGIG-V5-Pax6) and Pax6ΔPD (pCAGIG-V5-Pax6ΔPD). These constructs express EGFP from an IRES linked to the V5-Pax6 or V5-Pax6ΔPD cDNAs. This allowed for the confirmation of successful expression of the cDNAs in in P7 mouse retinas electroporated with the indicated pCAGIG DNA constructs at P0 by WB detection of EGFP and V5. (E) Co-expression of V5-Pax6ΔPD and EGFP in P7 mouse retinas was also determined by immunostaining with mouse anti-V5 (red) and chick anti-GFP (green) antibodies. (F) The identities of EGFP-positive retinal cells co-expressing Pax6 or Pax6ΔPD in P14 mouse retinas were determined by staining with antibodies against various amacrine and bipolar cell-specific proteins. The images are mouse retinal sections stained with anti-GABA (top) and anti-Vsx1 (bottom) antibodies. Arrowheads indicate cells positive to both of EGFP and the markers. Additional immunostaining results are provided in Figure 5—figure supplement 2. (G) EGFP-positive cells co-expressing each cell type-specific marker are shown as a percentage of total EGFP-positive INL cells. Values on the Y-axis are averages. Error bars indicate STD (n = 5); **p<0.01; ***p<0.001.
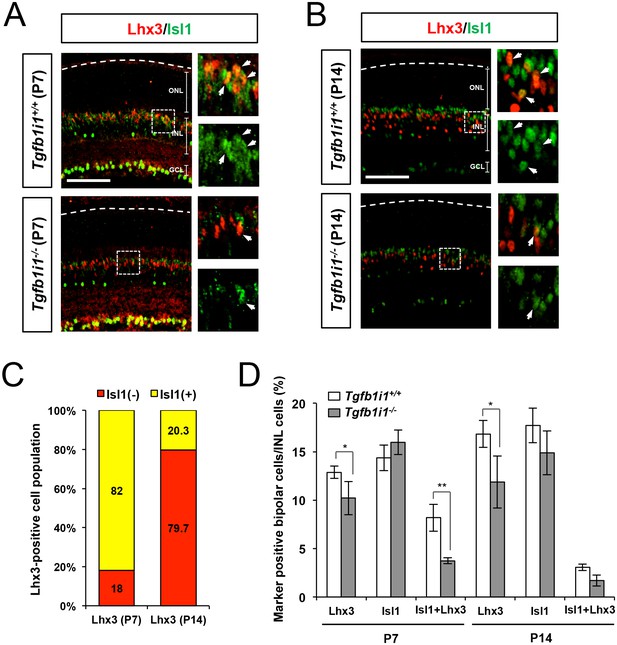
Distribution of Isl1- and Lhx3-expressing cells in Tgfb1i1+/+ and Tgfb1i1−/− mouse retinas.
P7 (A) and P14 (B) Tgfb1i1+/+ and Tgfb1i1−/− littermate mouse retinas stained with a guinea pig anti-Isl1 antibody (green) and a rabbit anti-Lhx3 antibody (red). Images in the right columns are magnified versions of the dotted areas in the left columns. Scale bars, 100 μm. Isl1(-) (red) and Isl1(+) (yellow) cells among Lhx3(+) cells are shown in the graph in (C) and populations expressing each marker in total INL cells are shown in the graph in (D). Y-axis values in the graphs are averages and error bars indicate STD (n = 4, three independent litters). *p<0.05; **p<0.01.
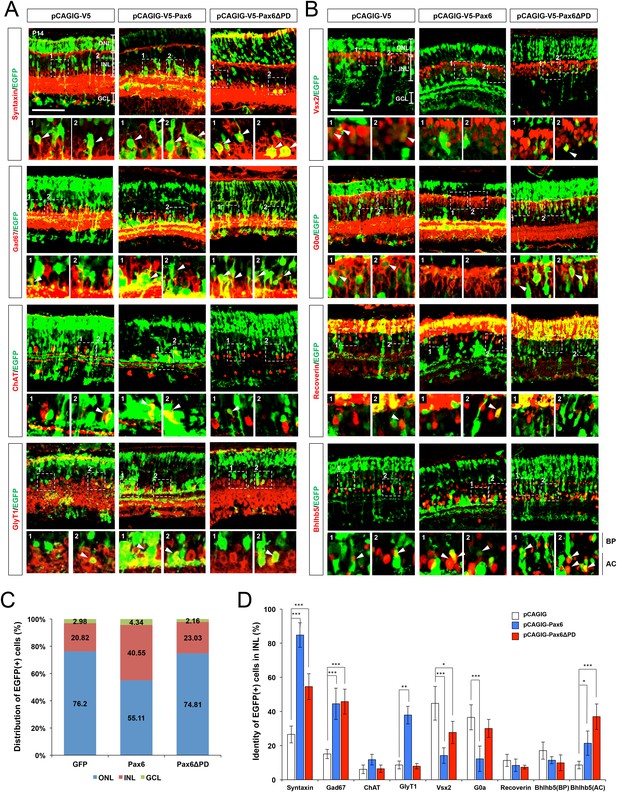
Ectopic expression of Pax6 isoforms in the post-natal mouse retinas.
(A) P14 mouse retinas, which had been electroporated with the indicated DNA constructs at P0, were stained for the detection of various amacrine cell markers, including Syntaxin (pan-amacrine), Gad67 (GABAergic), GABA (GABAergic subsets; results are in Figure 5G), Bhlhb5 (GABAergic subsets, bottom half of the INL), ChAT (cholinergic), and GlyT1 (glycinergic). EGFP cDNA is linked to the Pax6 cDNAs via IRES, thus those two cDNAs are transcribed in a single mRNA. Thus, the cells expressing EGFP together with the amacrine cell markers can be counted to investigate the effects of overexpressed Pax6 isoforms on retinal cell fate determination. Scale bar, 100 μm. (B) The retinas were also stained for the detection of bipolar cell markers Vsx2 (pan-bipolar), G0α (ON bipolar), Vsx1 (OFF bipolar; results are in Figure 5G), Recoverin (type-2 OFF bipolar), and Bhlhb5 (type-2 OFF bipolar, top half of the INL). Scale bar, 100 μm. (C) Retinal layer distribution of EGFP-positive cells in the indicated electroporated mouse retinas. (D) EGFP-positive cells co-expressing each amacrine or bipolar cell type-specific marker are shown as a percentage of total EGFP-positive INL cells. Scores on the Y-axis in the graphs in (C) and (D) are averages. Error bars indicate STD (n = 6, four independent batches); *p<0.05; **p<0.01; ***p<0.001.
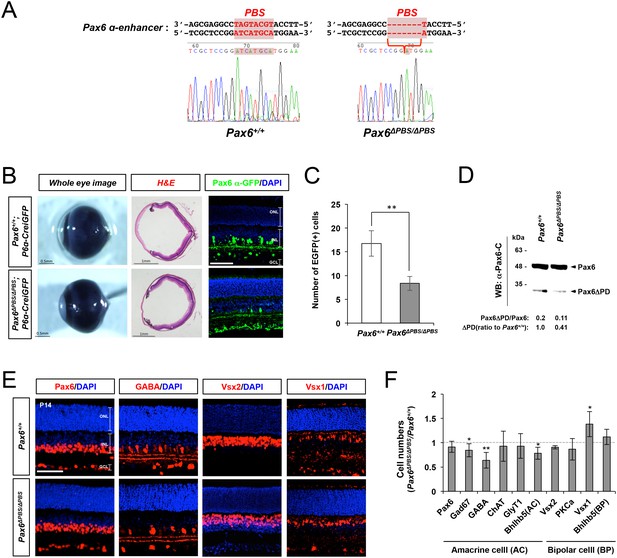
Pax6-dependent Pax6 α-enhancer activation is positively correlated with GABAergic amacrine cell number.
(A) Genomic DNA was isolated from the tails of Pax6+/+(left) and Pax6ΔPBS/ΔPBS (right) mice for sequencing the Pax6 α-enhancer region. The Pax6 binding sequence (PBS) in the α-enhancer is colored red. The Pax6ΔPBS allele is missing six nucleotides (5’-TGCATG-3’) in the PBS. (B) Whole eye images of P30 Pax6+/+;P6α-CreiGFP and Pax6ΔPBS/ΔPBS;P6α-CreiGFP littermate mice (left) and the mouse eye sections stained with H&E (center) or an anti-GFP antibody (right). Scale bar in the rightmost column is 100 μm. (C) Pax6 α-GFP-positive cells in P30 Pax6+/+and Pax6ΔPBS/ΔPBS retinas (250 μm x 250 μm). Error bars indicate STD (n = 4, two independent litters). (D) Full-length Pax6 and Pax6ΔPD in P14 Pax6+/+ and Pax6ΔPBS/ΔPBS retinal cell lysates were detected by WB with anti-Pax6 antibody and WB band intensities were compared to show the relative values below the WB image. (E) Distributions of pan-amacrine cell marker Pax6, GABAergic amacrine cell subset marker GABA, pan-bipolar cell marker Vsx2, and OFF bipolar cell marker Vsx1 in P14 Pax6+/+and Pax6ΔPBS/ΔPBS littermate retinas were visualized with immunostaining with antibodies recognizing respective markers. Scale bars, 100 μm. Additional images of amacrine and bipolar cell subtypes are shown in Figure 6—figure supplement 2. (F) Quantification of relative numbers of amacrine and bipolar cell subsets in mouse retinas. Error bars indicate STD (n = 5, three independent litters). *p<0.05; **p<0.01.
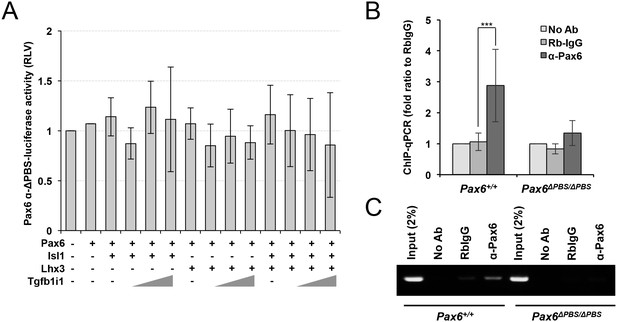
Impaired response of the Pax6ΔPBS α-enhancer to Pax6.
(A) Luciferase expression at downstream of a Pax6 α-enhancer mutant lacking its PBS (Pax6-αΔPBS) was measured by detecting chemiluminescence emitted from the lysates of HEK293T cells combinatorially expressing Pax6, Lhx3, Isl1, and Tgfb1i1 (n = 4). Bindings of Pax6, Isl1, Lhx3, and Tgfb1i1 to the Pax6 α-enhancer sequence in P30 Pax6+/+ and Pax6ΔPBS/ΔPBS mouse retinas were assessed by qPCR (B, n = 4) and PCR (C) amplification of DNA fragments isolated by ChIP with a rabbit IgG recognizing each respective protein. Error bars indicate STD; ***p<0.001.
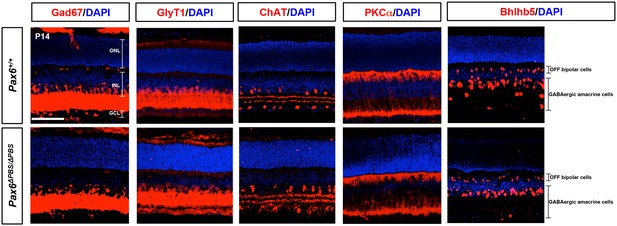
Distribution of amacrine and bipolar cell subsets in Pax6+/+ and Pax6ΔPBS/ΔPBS mouse retina.
P14 Pax6+/+ and Pax6ΔPBS/ΔPBS littermate retinas co-stained with amacrine cell and bipolar cell subtype marker-specific antibodies. Gad67, GABAergic amacrine cells; ChAT, cholinergic amacrine cells; GlyT1, glycinergic amacrine cells; PKCα, rod bipolar cells; Bhlhb5, OFF bipolar cells and GABAergic amacrine cells. Scale bar, 100 μm. Quantification results are shown in Figure 6F.
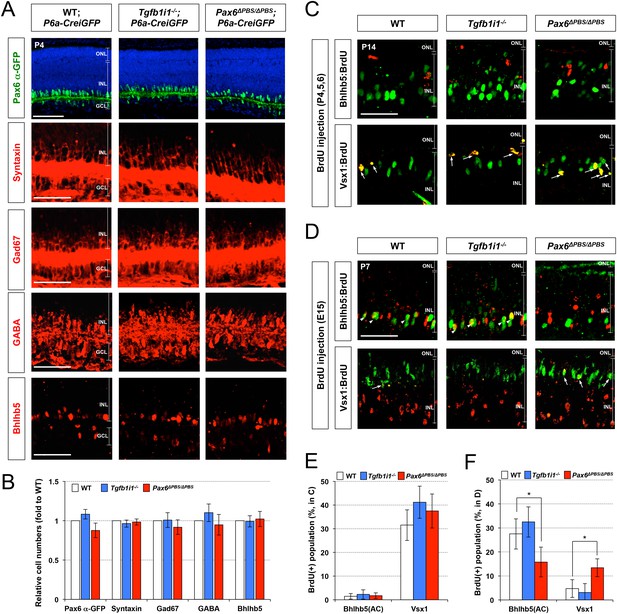
Fate determination of GABAergic amacrine cells and OFF bipolar cells in the post-natal mouse retinas.
(A) The effects of deletions of Tgfb1i1 (Tgfb1i1−/−) and PBS sequence of Pax6 α-enhancer (Pax6ΔPBS/ΔPBS) on GABAergic amacrine cell development were investigated by immunostaining of various GABAergic amacrine cell markers, including Gad67, GABA, and Bhlhb5. Distribution of entire amacrine cells was examined by immunostaining of pan-amacrine cell marker Syntaxin. The effects of the gene deletions on Pax6 α-enhancer activity was also determined by detecting cells expressing Pax6 α-GFP. Scale bars, 100 μm (top) and 50 μm (rest). (B) Relative numbers of marker-positive cells in P4 Tgfb1i1−/−and Pax6ΔPBS/ΔPBS mouse retinas are determined by comparing with those in their WT littermate mice. Error bars denote STD (n = 4, two independent litters). *p<0.05. (C) To identify the fate of cells were born in WT, Tgfb1i1−/−, and Pax6ΔPBS/ΔPBS between post-natal day 4 and 7 (P4 and P7) when bipolar cells and Müller glia are predominantly generated, the mice were repeatedly injected with BrdU (5 mg/kg) at P4, P5, and P6. Eye sections of the BrdU-injected mice were obtained at P14 for the immunodetection of Bhlhb5-positive GABAergic amacrine cells and Vsx1-positive OFF bipolar cells, which had exited cell cycle after incorporating BrdU between P4 and P7. Scale bar, 50 μm. (D) To trace the fates of cells produced in the embryonic retina when amacrine cells are generated, pregnant mice were injected with BrdU (5 mg/kg) at 15 dpc (E15) and the identities of cells had exited cell cycle after incorporating BrdU were examined at P7. Scale bar, 50 μm. (E and F) BrdU-labeled cell population in Bhlhb5-positive GABAergic amacrine cells, which locate the bottom half of INL, and that in Vsx1-positive OFF bipolar cell population in P14 mouse retinas as (C) and P7 mouse retina as (D) are quantified. Values in the Y-axis are average and error bars denote STD (n = 4, two independent litters). *p<0.05.
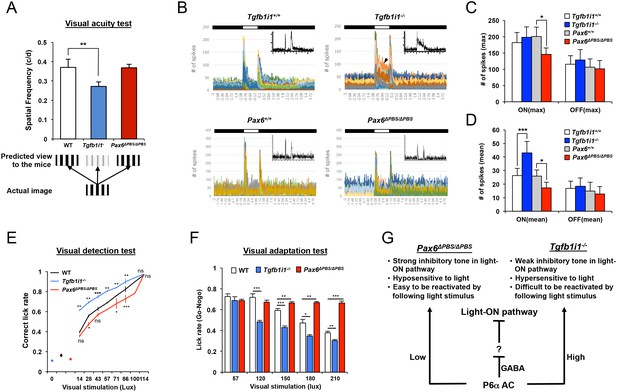
Pax6 α-enhancer-active amacrine cells are important for visual adaptation.
(A) Visual acuity was measured in P60 mice using the OptoMotry system as previously described (Prusky et al., 2004) (for details, see the Materials and Methods). Error bars indicate STD (n = 6). **p<0.01. (B) Peristimulus time histograms (PSTHs) for RGCs in P60 Tgfb1i1+/+ and Tgfb1i1−/− littermate and P60 Pax6+/+ and Pax6ΔPBS/ΔPBS littermate mouse retinas were obtained by multielectrode array (MEA) recordings. Maximum and mean numbers of spike were counted from each PSTH. Insets are representative PSTH patterns. Arrowhead indicates the sustained light-ON responses of RGCs. Maximum (max, C) and mean (D) numbers of spikes were counted from each PSTH. The numbers on the Y-axis are averages (WT, n = 526 (in four mice); Tgfb1i1−/−, n = 534 (in six mice); Pax6+/+, n = 175; Pax6ΔPBS/ΔPBS, n = 276). Error bars indicate STD. Statistical significance was determined using the D’Agostino and Pearson omnibus normality test followed by one-way ANOVAs and Sidak’s test for multiple comparisons. *p<0.05; ***p<0.001. (E) Visual detection in P60 Tgfb1i1−/−, Pax6ΔPBS/ΔPBS, and their WT littermate mice trained to lick water in response to light stimuli. The experimental scheme and task learning curves are provided in Figure 7—figure supplement 3A and B (for details, see the Materials and Methods). (F) The mice were also given water in association with a continuous light stimulus (2 s) but not with a continuous light stimulus (1 s) followed by a drifting grate stimulus (1 s) (see the experimental scheme and task learning curves in Figure 7—figure supplement 3C and D). Visual responses were quantified as ratios of hit rates (HitR, Go) to false alarm rates (FAR, Nogo). Error bars in (E) and (F) indicate STD. *p<0.05; **p<0.01; ***p<0.001 (Unpaired t-test). (G) Diagram depicting the modulation of retinal circuitry important for visual adaptation by Pax6 α-enhancer-active (P6α) GABAergic amacrine cells.
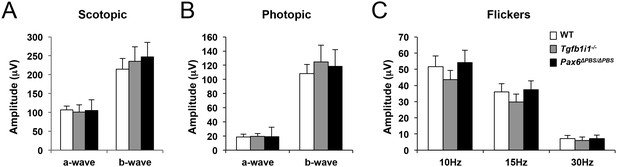
ERGs of mouse retinas.
P60 WT, Tgfb1i1−/−, and Pax6ΔPBS/ΔPBS mice were dark-adapted for 16 hr. Then, their scotopic ERG responses were assessed at a light intensity of 2.5 cds (left). Average amplitudes of scotopic ERG a-waves and b-waves measured from WT (white bars, n = 8), Tgfb1i1−/−(gray bars, n = 6), and Pax6ΔPBS/ΔPBS (black bars, n = 4) eyes. Photopic (center) and flicker (right) ERG responses of these mice were also measured after adaptation under room light (30 cd/m2).
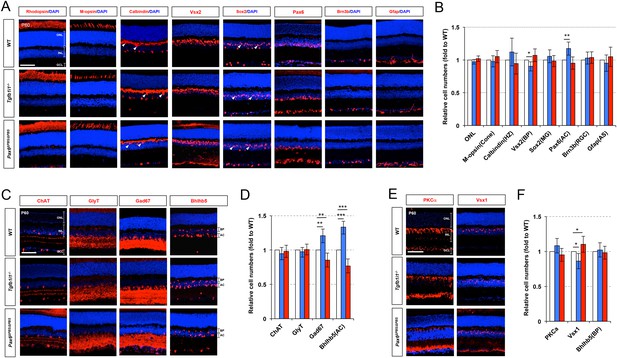
Cell composition of P60 WT, Tgfb1i1−/−, and Pax6ΔPBS/ΔPBS mouse retinas.
(A) Composition of P60 WT, Tgfb1i1−/−, and Pax6ΔPBS/ΔPBS mouse retinas were determined by examining cell type-specific markers. Rhodopsin, rod photoreceptors; M-opsin, M-cone photoreceptors; Calbindin, horizontal cells (HZ; arrowheads); Vsx2, bipolar cells (BP); Sox2, Müller glia (MG; arrowheads); Pax6, amacrine cells (AC); Brn3b, retinal ganglion cells (RGCs); glial fibrillary acidic protein (Gfap), astrocytes (AS). Scale bar, 100 μm. (B) Relative numbers of marker-positive cells in P60 Tgfb1i1−/−and Pax6ΔPBS/ΔPBS mouse retinas were determined by comparing with those in their WT littermate mice. Error bars denote STD (n = 4, two independent litters). *p<0.05; **p<0.01. (C) Distribution of amacrine cell subtypes in P60 WT, Tgfb1i1−/−, and Pax6ΔPBS/ΔPBS mice were determined by examining cell type-specific markers. ChAT, cholinergic; GlyT1, glycinergic; Gad67 and Bhlhb5 (AC in the bottom half of INL), GABAergic. Scale bar, 100 μm. (D) Relative numbers of marker-positive cells in P60 Tgfb1i1−/−and Pax6ΔPBS/ΔPBS mouse retinas were determined by comparing with those in their WT littermate mice. Error bars denote STD (n = 4, two independent litters). **p<0.01; ***p<0.001. (E) Distribution of bipolar cell subtypes in P60 WT, Tgfb1i1−/−, and Pax6ΔPBS/ΔPBS mice were determined by examining cell type-specific markers. PKCα, rod bipolar cell; Vsx1 and Bhlhb5 (BP in the top of INL in (C)), OFF bipolar cells. Scale bar, 100 μm. (F) Relative numbers of marker-positive cells in P60 Tgfb1i1−/−and Pax6ΔPBS/ΔPBS mouse retinas were determined by comparing with those in their WT littermate mice. Error bars denote STD (n = 4, two independent litters). *p<0.05; **p<0.01; ***p<0.001.
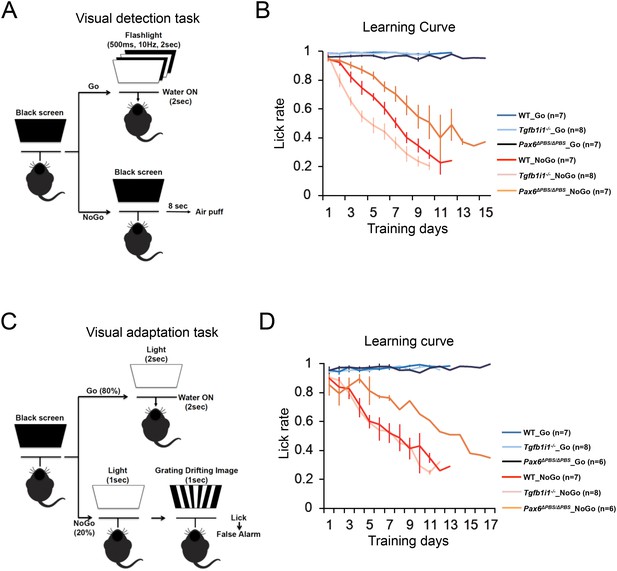
Experimental scheme assessing mouse visual responses.
(A) P60 mice were trained to associate water rewards with flashing light stimuli. Correct and incorrect lick rates were used to measure visual detection. (B) Lick rates during the learning period for mice responding to various intensity of light as shown in Figure 4E. (C) P60 mice were trained to associate water rewards only with a continuous (2 s) light stimulus and not a continuous (1 s) light followed by a drifting grate image (1 s). Correct and incorrect lick rates were used to measure visual discrimination of the drifting grate from various intensities of light stimulus. (D) Lick rates during the learning period.
Tables
Antibody used in this study.
Antigen | Species | Producer | Dilution |
|---|---|---|---|
Bhlhb5 | Goat | Santa Cruz | 1:100 |
Brn3b | Goat | Santa Cruz | 1:200 |
Calbindin | Mouse | Sigma | 1:200 |
Calretinin | Mouse | Millipore | 1:1000 |
ChAT | Goat | Millipore | 1:200 |
Isl1 | Rabbit | gift from Dr. Mi-Ryoung Song | 1:500 |
Isl1 | Guinea Pig | gift from Dr. Mi-Ryoung Song | 1:10,000 |
Gad67 | Mouse | Millipore | 1:500 |
GABA | Guinea Pig | Millipore | 1:300 |
GFAP | Rabbit | Abcam | 1:500 |
GFP | Chick | Abcam | 1:200 |
GFP | Rabbit | Santa Cruz | 1:500 |
GlyT1 | Rabbit | Abcam | 1:200 |
G0α | Mouse | Millipore | 1:300 |
G/R opsin | Rabbit | Millipore | 1:200 |
Lhx2 | Goat | Santa Cruz | 1:200 |
Lhx3 | Rabbit | Abcam | 1:1000 |
Lhx9 | Rabbit | Santa Cruz | 1:500 |
Pax6 | Rabbit | Abcam | 1:200 |
Pax6 | Rabbit | Covance | 1:300 |
PKCα | Mouse | Sigma | 1:200 |
Recoverin | Rabbit | Chemicon | 1:200 |
Rhodopsin | Mouse | Millipore | 1:500 |
Sox2 | Goat | Santa Cruz | 1:100 |
Sox9 | Rabbit | Santa Cruz | 1:200 |
Tgfb1i1(Hic-5) | Mouse | BD | 1:100 |
Tgfb1i1(Hic-5) | Rabbit | Abcam | 1:100 |
Vsx1 | Goat | Santa Cruz | 1:50 |
Vsx2(Chx10) | Mouse | Santa Cruz | 1:200 |
V5 | Mouse | Genway Biotech | 1:1000 |





