Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance
Figures
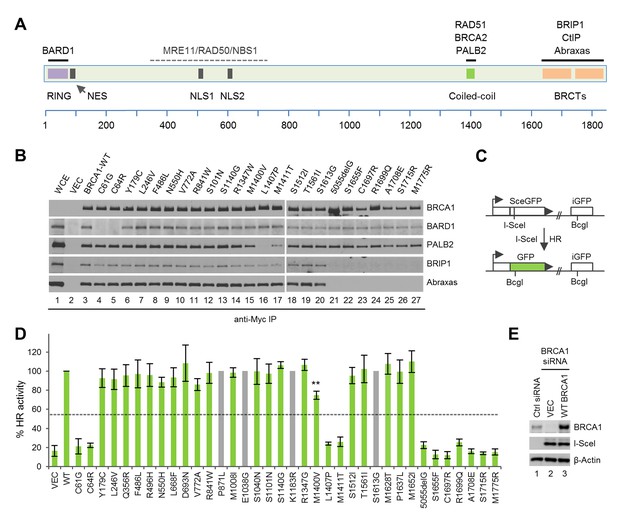
Sequence alterations generated in BRCA1 and their effects on protein-protein interactions and HR.
(A) Domain structure of BRCA1 and the binding sites for its interacting partners. NES, nuclear export signal; NLS, nuclear localization signal. (B) Effects of the BRCA1 variants on the binding of BARD1, PALB2, BRIP1 and Abraxas. The 3xMyc-tagged BRCA1 proteins were transiently expressed in 293T cells and IPed with an anti-Myc antibody. WCE, whole cell extract. (C) Schematic of the HR reporter assay. The DR-GFP reporter contains two defective copies of the GFP gene, one disrupted by an I-SceI site and the other lacking a promoter. I-SceI cutting of the first copy generates a DSB, and repair by HR with the second copy as a template leads to restoration of a functional GFP gene. (D) HR activities of the variants relative to the wt BRCA1 protein. Data shown are the means from two to seven independent experiments for each variant or mutant. Error bars represent standard deviations (SDs). The grey bars indicate variants that are among the top 20 missense variants but are already present in BRCA1 cDNAs obtained from three independent sources. The calculated cutoff threshold is indicated by horizontal lines. **p<0.01. See Figure 1—source data 1 for details. (E) Levels of BRCA1 protein following knockdown and re-expression. Cells treated with a control siRNA (NSC1) were used as a control for the endogenous protein abundance.
-
Figure 1—source data 1
HR activities of the BRCA1 variants and mutants analzyed in this study.
See texts and materials and methods for details.
- https://doi.org/10.7554/eLife.21350.004
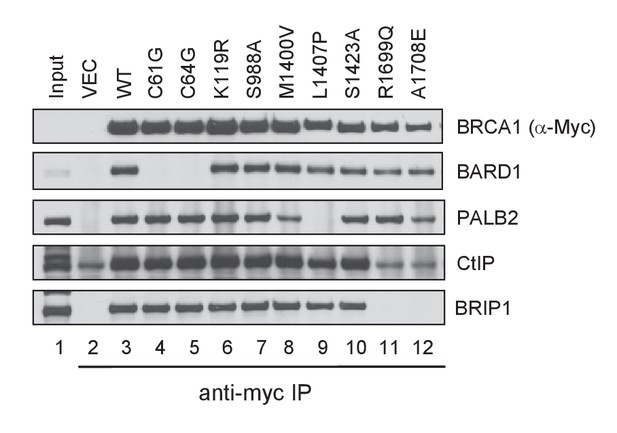
Capacity of the BRCA1 variants to bind BARD1, PALB2, CtIP and BRIP1.
The 3xMyc-tagged BRCA1 proteins were transiently expressed in 293T cells and IPed with an anti-Myc antibody. Note that the amount of CtIP co-IPed with BRCT mutants R1699Q and A1708E was the same as the background level in the vector lane.
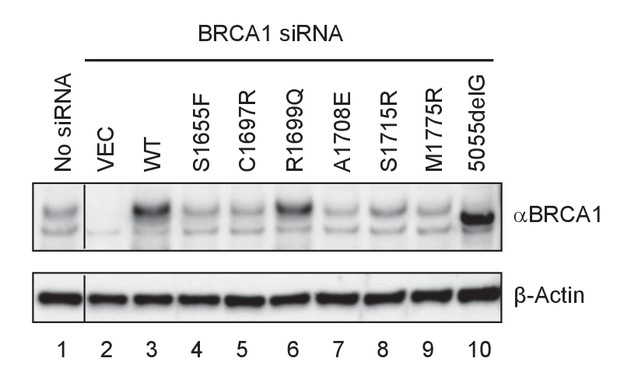
Expression levels of BRCA1 BRCT missense and truncating mutants in U2OS/DR-GFP cells first depleted of the endogenous BRCA1 and then transfected with cDNA expressing the mutants.
Cells untreated with any siRNA were used as a control for the endogenous protein abundance (spliced from the same gel).
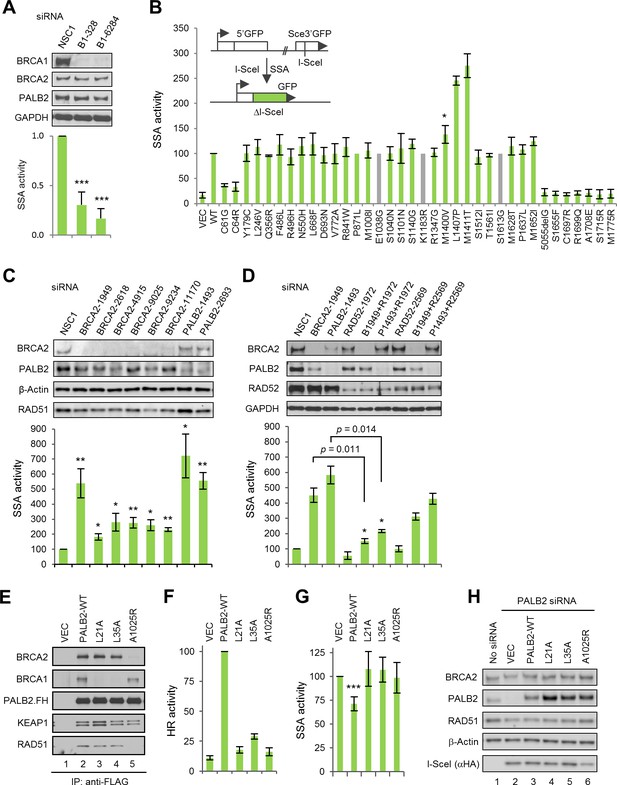
The BRCA1-PALB2 interaction suppresses SSA.
(A) Effect of BRCA1 depletion on SSA. Data shown are the means of the four data points obtained from two independent experiments each performed in duplicates. Error bars represent SDs. ***p<0.001. (B) SSA activities of the BRCA1 variants relative to the wt protein. A schematic of the SA-GFP reporter assay is shown at the upper left corner. Data shown are the means ± SDs from two to five independent experiments for each variant or mutant. *p<0.05. See Figure 2—source data 1 for details. (C) Effects of BRCA2 and PALB2 depletion on SSA. Data shown are the means ± standard errors of mean (SEMs) from four independent experiments. *p<0.05; **p<0.01. (D) Requirement of RAD52 for SSA upregulation following BRCA2 and PALB2 depletion. Data shown are the means ± SEMs from three independent experiments. *p<0.05; **p<0.01. (E) BRCA1 and BRCA2-binding defects of PALB2-L21A, L35A and A1025R mutants. The FLAG-HA-tagged mutants were transiently expressed in 293T cells and IPed with anti-FLAG M2 agarose beads. (F–G) HR (F) and SSA (G) activities of PALB2-L21A, L35A and A1025R mutants. Data shown are the means ± SDs from three to five independent experiments for each mutant. ***p<0.001. (G) Levels of PALB2 protein following knockdown and re-expression in U2OS/DR-GFP cells. Cells untreated with any siRNA were used as a control for the endogenous protein abundance.
-
Figure 2—source data 1
SSA activities of the BRCA1 variants and mutants analyzed in this study.
See texts and materials and methods for details.
- https://doi.org/10.7554/eLife.21350.010
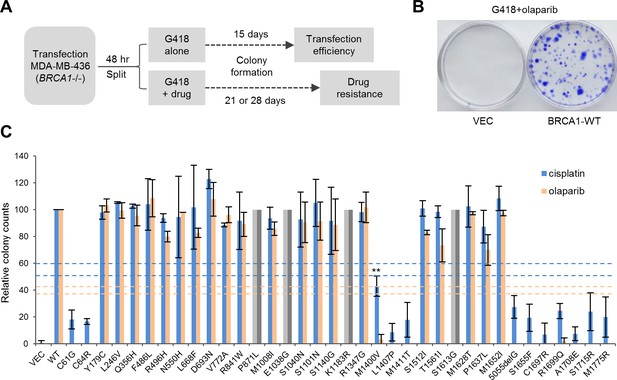
Abilities of the wt and mutant or variant BRCA1 proteins to confer cisplatin and olaparib resistance.
(A) Schematic of the colony formation assay. The BRCA1 mutant MDA-MB-436 breast cancer cells were transfected with BRCA1 expression plasmids, reseeded and then selected with G418 alone or G418 with cisplatin or olaparib. Cells were stained with crystal violet 3–4 weeks after selection. (B) Representative crystal violet-stained plates. (C) Relative activities of wt and mutant or variant BRCA1 proteins to support colony formation in the presence of cisplatin or olaparib. Data shown are the means ± SDs from two to three independent experiments for each variant or mutant. **p<0.01. Horizontal lines represent the cutoff thresholds for cisplatin (blue) and olaparib (orange). See Figure 3—source data 1 for details.
-
Figure 3—source data 1
Abilities of the BRCA1 variants and mutants to support olaparib and cisplatin resistance in MDA-MB-436 cells.
See texts and materials and methods for details.
- https://doi.org/10.7554/eLife.21350.012
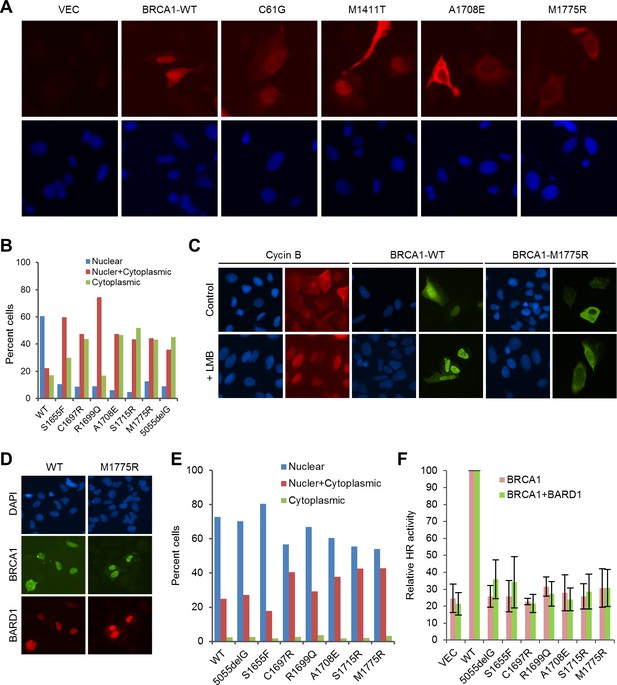
Rescue of the localization defect but not HR activity of BRCA1 BRCT missense mutants by BARD1.
(A) Immunofluorescence staining of wt BRCA1 and representative RING, CC and BRCT mutants. 3xMyc-tagged BRCA1 expression constructs were transiently transfected into U2OS/DR-GFP cells first depleted of endogenous BRCA1, cells were fixed 48 hr after transfection, and the proteins were stained with an anti-Myc antibody. (B) Quantification of the subcellular distribution of wt BRCA1 and a panel of BRCT mutants. cDNA constructs were transfected into U2OS cells and the tagged BRCA1 proteins were stained as in (A). (C) Effect of Leptomycin B (LMB) on the localization of wt BRCA1 and BRCA1-M1775R. The BRCA1 proteins were expressed and stained as in (A). LMB (50 ng/ml) was added 48 hr after transfection, and cells were incubated with LMB for 12 hr prior to fixation. Staining of endogenous cyclin B in untransfected U2OS cells was used as a positive control. (D–E) Rescue of the localization defect of BRCA1 BRCT mutants by BARD1. 3xMyc-tagged BRCA1 and FLAG-HA-tagged BARD1 were transiently co-expressed in U2OS cells, cells were fixed 48 hr after transfection, and the proteins were stained with Myc and HA antibodies, respectively. Panel D shows the staining patterns of wt and two representative mutants of BRCA1 and the co-expressed BARD1. Panel E shows the quantification of the results. (F) No rescue of the HR defect of the BRCA1 BRCT mutants by BARD1 co-expression. U2OS/DR-GFP cells were depleted of the endogenous BRCA1 for 48 hr and then co-transfected with BRCA1 and BARD1 expression constructs. GFP-positive cells were scored another 52 hr later. The values were normalized against that of wt BRCA1, which was set as 1. Data shown are means ± SDs from three independent experiments.
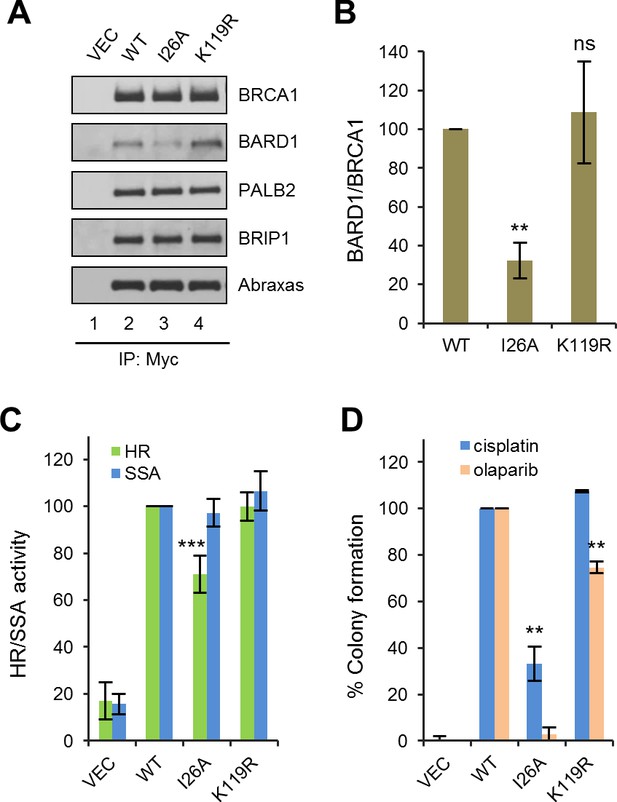
Effects of BRCA1 sumoylation and E3 ligase activity on HR and drug resistance.
(A) Effect of I26A and K119R mutations on BRCA1 binding to BARD1 and other interacting partners. The proteins were transiently expressed in 293T cells and IPed with anti-Myc. (B) Quantification of the BARD1-binding capacity of the BRCA1 mutants. Data shown are the means ± SDs of the ratios of BARD1 and BRCA1 band intensities from four independent experiments. **p<0.01. (C) HR and SSA activities of the BRCA1 mutants relative to that of the wt protein. Data shown are the means ± SDs from two to six independent experiments for each mutant. ***p<0.001. See Figure 1—source data 1 and Figure 2—source data 1 for details. (D) Abilities of the BRCA1 mutants to confer resistance to cisplatin and olaparib. Data shown are the means ± SDs from three independent experiments for both mutants. **p<001. See Figure 3—source data 1 for details.
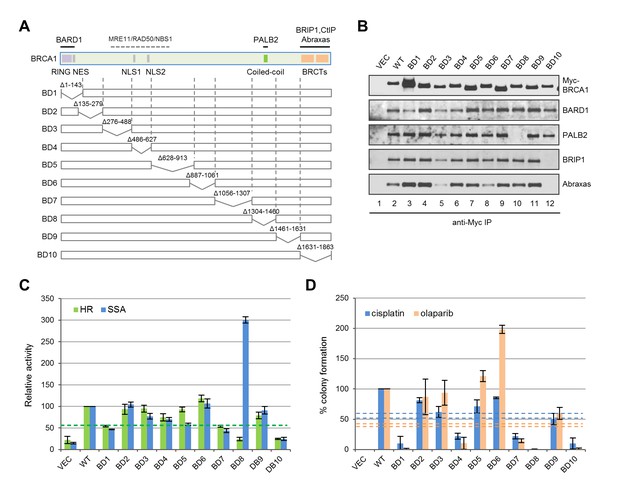
Functionalities of BRCA1 deletion mutants in HR, SSA and drug resistance.
(A) Schematic of wt BRCA1 and 10 overlapping deletions generated for this study. (B) Capacity of the deletion mutants in binding key interacting partners. The proteins were transiently expressed in 293T cells and IPed with anti-Myc. (C) HR and SSA activities of the deletion mutants. See Figure 1—source data 1 and Figure 2—source data 1 for details. (D) Levels of cisplatin and olaparib resistance conferred by the deletion mutants. Values presented are means ± SDs from two to four independent experiments for each mutant. See Figure 3—source data 1 for details.
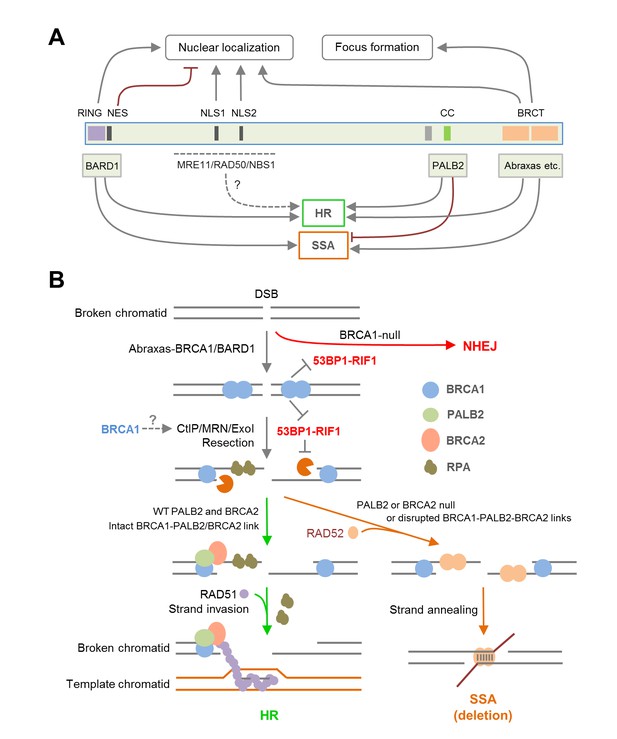
Summary models of the regulation and function of BRCA1 in HR and SSA.
(A) Roles of BRCA1 structural elements and binding partners on its nuclear localization and HDR (HR and SSA) activities. In brief, the NLSs and BRCT domain both promote BRCA1 nuclear entry, whereas the NES mediates BRCA1 export from the nucleus. BARD1 bound to the RING domain shields the NES of BRCA1 thereby promoting its nuclear retention. The RING and BRCT domains are required for both HR and SSA, whereas the CC domain promotes HR but inhibits or prevents SSA through its binding to PALB2. (B) A model of the BRCA1-PALB2-BRCA2 axis in the regulation of HR and SSA. Following DSB formation, the BRCA1/BARD1 complex was recruited to DNA damage sites by Abraxas and perhaps another yet to be defined factor(s). The presence of BRCA1 at damage sites promotes resection and inhibits non-homologous end joining (NHEJ), at least in part, by counteracting the resection-inhibiting activity of the 53BP1-RIF1 complex. At the same time, BRCA1 helps recruit the PALB2/BRCA2 complex, which displaces RPA from the resected ssDNA ends and loads RAD51 to initiate HR. When PALB2 or BRCA2 is lost or when the direct interactions in the BRCA1-PALB2-BRCA2 axis are disrupted, RAD52 binds to resected ends and mediates SSA when homologous sequences are available, leading to genomic deletions. It is unclear whether PALB2 or BRCA2 can directly suppress the binding of RAD52 to ssDNA or inhibiting its strand annealing activity.
Tables
BIC database and ClinVar reports of patient-derived BRCA1 missense variants characterized in this study.
| HGVS cDNA | BIC designation | BIC entry count | BIC clinical importance | ClinVar individuals | ClinVar designation | Align-GVGD grade | IARC classification |
|---|---|---|---|---|---|---|---|
| c.4837A>G | S1613G | 248 | No | 1319 | Benign | C0 | 1 |
| c.181T>G | C61G | 239 | Yes | 323 | Pathogenic | C65 | 5 |
| c.2612C>T | P871L | 211 | No | 1348 | Benign | C0 | 1 |
| c.3113A>G | E1038G | 182 | No | 1237 | Benign | C0 | 1 |
| c.3548A>G | K1183R | 164 | No | 1215 | Benign | C0 | 1 |
| c.4039A>G | R1347G | 161 | Unknown | 712 | Benign | C0 | 1 |
| c.1067A>G | Q356R | 155 | Unknown | 315 | Benign | C0 | 1 |
| c.3024G>A | M1008I | 139 | Unknown | 146 | Benign | C0 | 1 |
| c.2521C>T | R841W | 119 | Unknown | 128 | Benign | C15 | 1 |
| c.4883T>C | M1628T | 96 | Unknown | 491 | Benign | C0 | 1 |
| c.1487G>A | R496H | 86 | Unknown | 90 | Benign | C0 | 1 |
| c.736T>G | L246V | 80 | Unknown | 83 | Benign | C0 | 1 |
| c.3119G>A | S1040N | 68 | Unknown | 141 | Benign | C0 | 1 |
| c.4910C>T | P1637L | 67 | Unknown | 76 | Benign | C0 | 1 |
| c.2077G>A | D693N | 65 | No | 229 | Benign | C0 | 1 |
| c.4956G>A | M1652I | 61 | Unknown | 94 | Benign | C0 | 1 |
| c.2315T>C | V772A | 60 | Unknown | 62 | Benign | C0 | 1 |
| c.4535G>T | S1512I | 58 | No | 72 | Benign | C0 | 1 |
| c.1456T>C | F486L | 56 | Unknown | 58 | Benign | C0 | 1 |
| c.536A>G | Y179C | 54 | Unknown | 59 | Benign | C35 | 1 |
| c.1648A>C | N550H | 54 | Unknown | 59 | Benign | C0 | 1 |
| c.5123C>A | A1708E | 46 | Yes | 80 | Pathogenic | C65 | 5 |
| c.5324T>G | M1775R | 31 | Unknown | 43 | Pathogenic | C45 | 5 |
| c.3418A>G | S1140G | 29 | Unknown | 40 | Benign | C0 | 1 |
| c.4682C>T | T1561I | 26 | Unknown | 33 | Conflicting | C0 | N/A |
| c.2002C>T | L668F | 25 | Unknown | 28 | Benign | C0 | 1 |
| c.3302G>A | S1101N | 14 | Unknown | 16 | Benign | C0 | 1 |
| c.190T>C | C64R | 12 | Unknown | 15 | Conflicting | C65 | N/A |
| c.5096G>A | R1699Q | 11 | Unknown | 22 | Conflicting | C35 | 5 |
| c.5145C>G | S1715R# | 5 | Unknown | 5 | Pathogenic | C65 | 5 |
| c.4964C>T | S1655F | 3 | Unknown | 4 | Conflicting | C25 | N/A |
| c.5098T>C | C1697R | 3 | Unknown | 7 | Conflicting | C65 | N/A |
| c.4198A>G | M1400V | 1 | Unknown | 1 | Uncertain | C0 | N/A |
| c.4220T>C | L1407P | 1 | Unknown | 1 | Uncertain | C65 | N/A |
| c.4232T>C | M1411T | 1 | Unknown | 1 | Uncertain | C65 | N/A |
| 5055delG | V1646Sfs | 5 | Yes | 6 | Pathogenic | - | - |
-
Align-GVGD grade: C0 to C65 denote increasing likelihood of a variant to cause damage (to protein function). IARC (ENIGMA) classification: 5, Definitely pathogenic; 4, Likely pathogenic; 3, Uncertain, 2, Likely not pathogenic or of little clinical significance; 1, Not pathogenic or of no clinical significance. # c.5145C>G, c.5145C>A and c.5143A>C have all been reported to generate BRCA1-S1715R. Data shown are up to date as of March 21, 2017.
Comparison of results on HR activity and drug response of the BRCA1 variants analyzed in this study obtained from previous and this studies.
| Domain | HR activity (%) | Cisplatin response | Olaparib response | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sy et al., 2009 | Ransburgh et al. (2010) | Towler et al. (2013) | Bouwman et al. (2013) | Lu et al., 2015 | This study | Bouwman et al. (2013) | This study | Bouwman et al. (2013) | This study | ||
| Vector | N/A | ~10 | ~9 | ~20 | ~18 | 17.3 | S | S | S | S | |
| WT | 98 | 100 | 100 | 100 | 100 | 100 | R | R | R | R | |
| C61G | RING | - | ~17 | - | - | 23.6 | 21.2 | S | S | - | S |
| C64R | RING | - | - | - | - | - | 22.3 | - | S | - | S |
| Y179C | - | - | ~95 | - | 157.1 | 92.7 | - | R | - | R | |
| L246V | - | - | - | - | - | 91.5 | R | R | - | R | |
| Q356R | - | - | - | - | - | 95.5 | - | R | - | R | |
| F486L | - | - | - | - | 160 | 95.8 | - | R | - | R | |
| R496H | - | - | - | - | - | 95.9 | - | R | - | R | |
| N550H | - | - | - | - | 90.8 | 88.4 | - | R | - | R | |
| L668F | - | - | - | - | 96.8 | 93.6 | R | R | - | R | |
| D693N | - | - | - | - | - | 111.5 | R | R | - | R | |
| V772A | - | - | - | - | 110.4 | 84.5 | - | R | - | R | |
| R841W | - | - | - | - | - | 98.2 | - | R | - | R | |
| P871L | - | - | - | - | - | wt | - | wt | - | wt | |
| M1008I | - | - | - | - | - | 99.2 | R | R | - | R | |
| E1038G | - | - | - | - | - | wt | - | wt | - | wt | |
| S1040N | - | - | - | - | - | 98.0 | - | R | - | R | |
| S1101N | - | - | - | - | - | 97.3 | R | R | - | R | |
| S1140G | - | - | - | - | - | 106.2 | R | R | - | R | |
| K1183R | - | - | - | - | - | wt | - | wt | - | wt | |
| R1347G | - | - | - | - | - | 106.5 | - | R | - | R | |
| M1400V | CC | 56 | - | - | - | - | 74.8 | R | S | R | S |
| L1407P | CC | 24 | - | - | - | - | 24 | S | S | S | S |
| M1411T | CC | 25 | - | - | - | - | 26 | R | S | R | S |
| S1512I | - | - | - | - | - | 95.3 | - | R | - | R | |
| T1561I | - | - | - | - | 133.2 | 102.3 | - | R | - | R | |
| S1613G | - | - | - | - | - | wt | - | wt | - | wt | |
| M1628T | - | - | - | - | 107 | 107.6 | R | R | - | R | |
| P1637L | - | - | - | - | 98.8 | 99.5 | - | R | - | R | |
| 5055△G | BRCT | - | - | - | - | - | 22.5 | - | S | - | S |
| M1652I | BRCT | - | - | - | - | - | 106.2 | R | R | - | R |
| S1655F | BRCT | - | - | - | ~30 | - | 8.4 | S | S | - | S |
| C1697R | BRCT | - | - | - | - | - | 8.0 | - | S | - | S |
| R1699Q | BRCT | - | - | - | ~45 | - | 16.9 | S | S | S | S |
| A1708E | BRCT | - | - | - | - | - | 10.7 | - | S | - | S |
| S1715R | BRCT | - | - | - | - | - | 9.3 | - | S | - | S |
| M1775R | BRCT | 36 | - | ~6 | - | - | 10.2 | - | S | - | S |
-
R, resistant; S, sensitive. See Figure 1—source data 1, Figure 2—source data 1 and Figure 3—source data 1 for details.
Additional files
-
Supplementary file 1
Sequences of siRNAs used in this study.
- https://doi.org/10.7554/eLife.21350.017





