Lysosomal membrane glycoproteins bind cholesterol and contribute to lysosomal cholesterol export
Figures
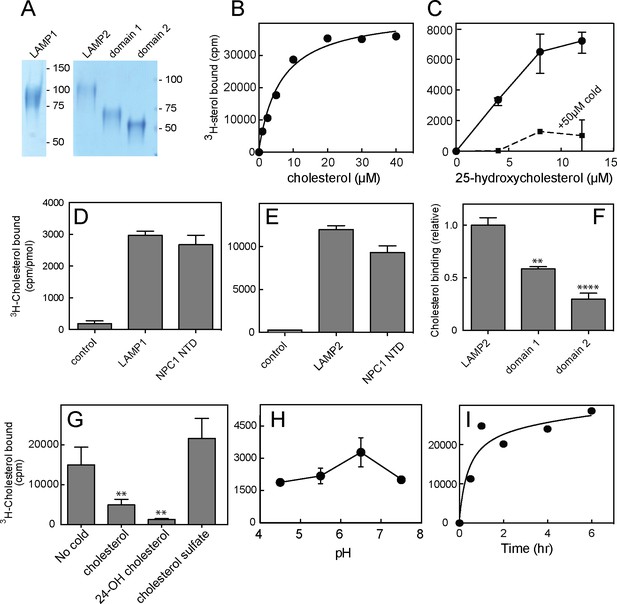
Cholesterol binding to LAMP proteins.
(A) Coomassie-stained SDS-PAGE of purified, secreted human LAMP1, LAMP2 or LAMP domains from LAMP2. B,C. 3H-cholesterol or 3H-25 hydroxycholesterol binding to soluble LAMP2 (full length protein). Also shown in C is binding in the presence of 50 µM cold hydroxycholesterol. D,E, 3H-cholesterol binding to indicated, soluble proteins compared with the soluble NPC1 N-terminal domain. (F), Cholesterol binding to LAMP2 domains 1 and 2 compared with full length, soluble LAMP2 protein. P values were determined relative to the full length soluble LAMP2 protein. (G) Sterol competition (30 µM) for 3H-cholesterol binding to soluble LAMP2. P values were determined relative to no cold addition. H, I. pH dependence and kinetics of 3H-cholesterol binding to LAMP2. In B,C,E,H and I, a representative experiment is shown; C and E show the average of duplicates. In C, the background counts in reactions containing the control protein, GFP-binding protein, were subtracted; in D and E, the control was TIP47 protein. D,F, and G show the combined results of two experiments in duplicate. Numbers at right (A) indicate mass in kD for this and all subsequent figures. Reactions contained D,E,G,H,I, 500 nM total cholesterol; F, 5 µM total cholesterol; B and C were carried out using increasing concentrations of the indicated sterol.
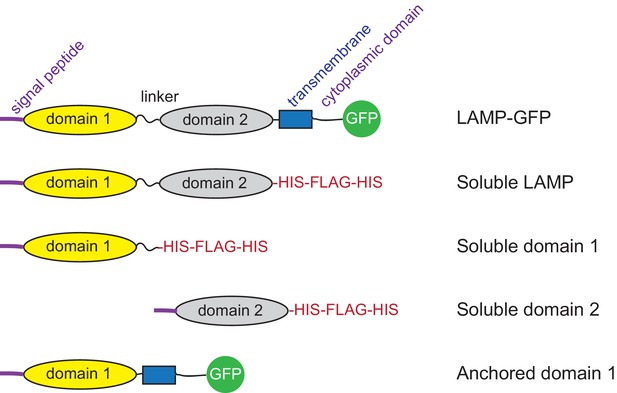
Diagram of constructs used.
https://doi.org/10.7554/eLife.21635.004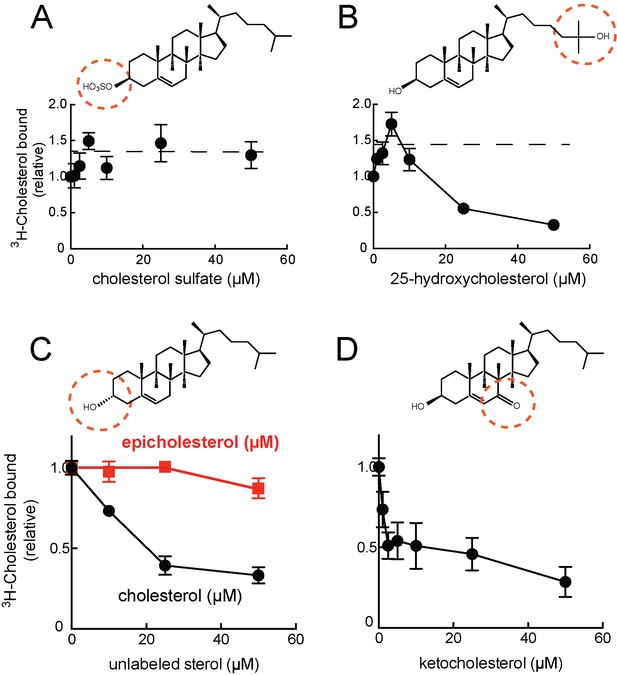
Cholesterol binding to LAMP2 is competed by 25-hydroxycholesterol (B), cholesterol (C), 7-ketocholesterol (D) but not cholesterol sulfate (A) or epicholesterol (C).
The structures above indicate the regions of the sterol that differ from cholesterol. In C, the background obtained in reactions containing GFP was subtracted. All panels used 50nM 3H-cholesterol and the indicated amounts of competitors.
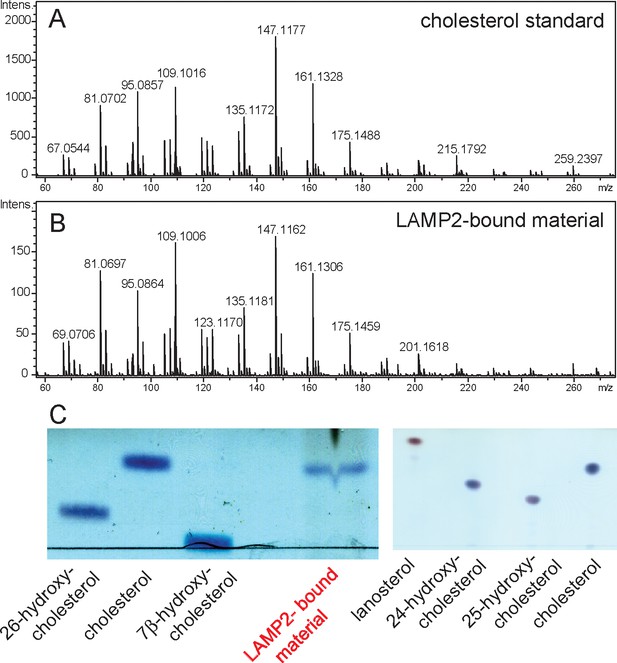
Mass spectrometry identification of small molecules released from LAMP2 after chloroform:methanol (2:1) extraction.
(A) masses from cholesterol standard; (B) masses of LAMP2-bound material; (C) Copper sulfate/phosphoric acid detection of indicated markers after thin layer chromatography compared with material eluted from soluble LAMP2 (50 µg). Shown are the results of a representative experiment carried out twice.
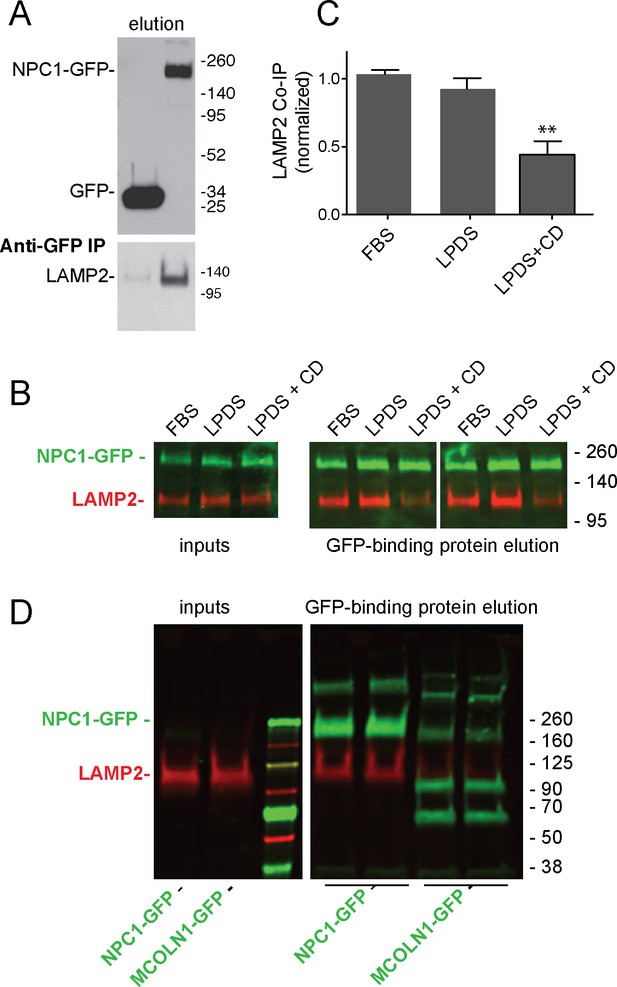
LAMP2 interacts with NPC1 protein.
(A) Anti-GFP immunoprecipitation from HEK293T cells grown in FBS containing medium expressing GFP or mouse NPC1-GFP. The blot was developed with ECL. Upper panel, anti-GFP immunoblot (100% elution); lower panel, anti-LAMP2 immunoblot to detect endogenous, full length protein (100% elution). B,C, co-immunoprecipitation of LAMP2 and NPC1-GFP in cells grown in FBS, LPDS (5%) or LPDS + cyclodextrin (1 mM) for 24 hr. Left panels, 1% inputs; right panels, 50% elutions. Error bars represent SEM for two combined experiments carried out in duplicate; P value is from comparison with LPDS by two-tailed Student’s t-test. B shows duplicate reactions to document reproducibility. D, Anti-GFP immunoprecipitation of HEK293T cells grown in FBS expressing GFP-MCOLN1 or mouse NPC1-GFP. GFP-MCOLN1 occurs as a ~90 kD form, a proteolytically processed form, and as higher oligomers (Vergarajauregui et al., 2011). Left panels, inputs (2%); right panels, total elution carried out in duplicate. GFP proteins are green; LAMP2 is presented in red. Shown is a representative experiment carried out twice in duplicate.
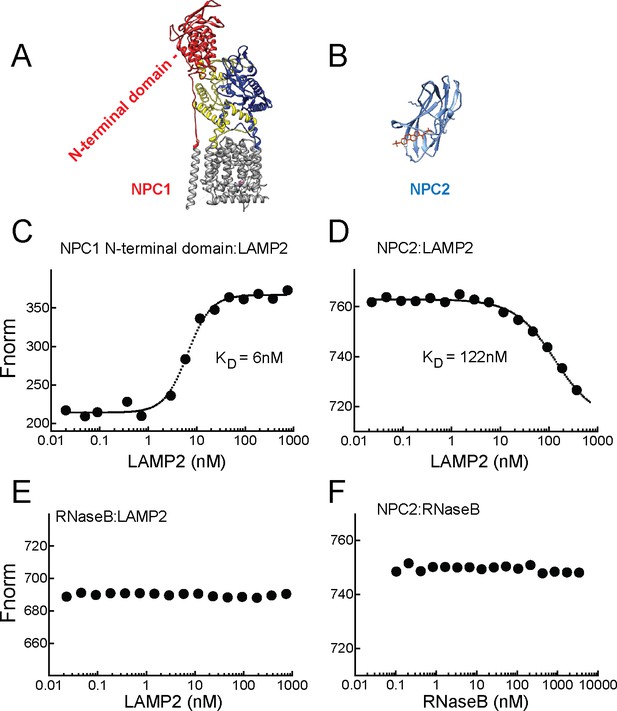
LAMP2 binds NPC1 and NPC2 proteins.
A,B, Structures of NPC1 (Gong et al., 2016; pdb 3jD8) and NPC2 (Xu et al., 2007; pdb 2 hka) proteins; C,E, Microscale thermophoresis (E) or fluorescence (C) obtained with mixtures of soluble, AF647 labeled-NPC1 N-terminal domain or AF647-RNase B with increasing concentrations of soluble LAMP2 in 1 µM cholesterol. D,F, Microscale thermophoresis of AF647 labeled-NPC2 with increasing concentrations of soluble LAMP2 or RNase B in 1 µM cholesterol sulfate. For C–F, a representative experiment carried out at least twice is shown.
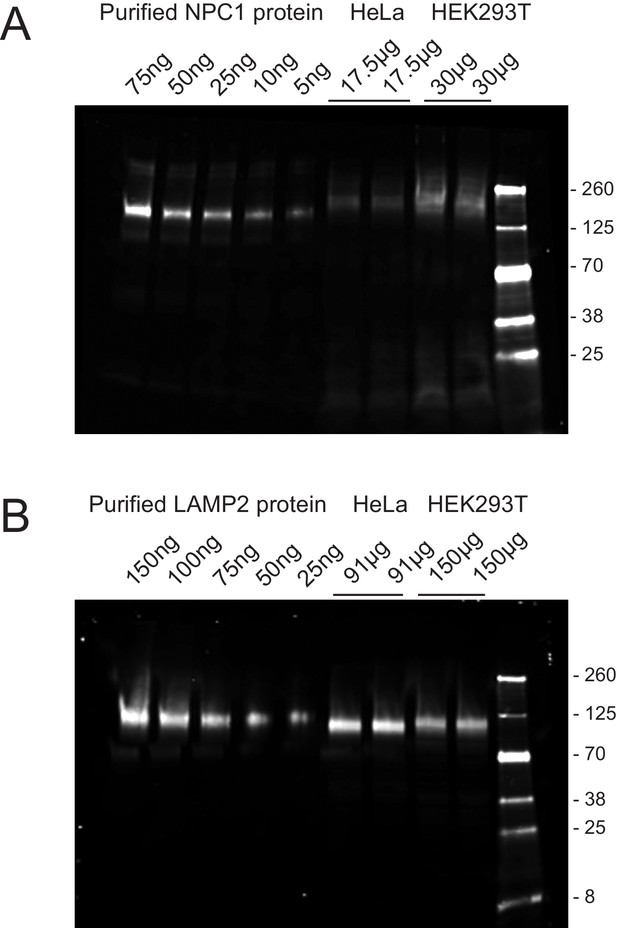
Quantitation of NPC1 (A) and LAMP2 (B) proteins in HeLa and HEK293T cells.
The values at the top of each gel represent the amount of purified protein or total cell extract analyzed, determined by BCA protein assay. The immunoblot was developed using rabbit anti-NPC1 or mouse anti-LAMP2 antibodies followed by detection with IRDye800CW goat anti-mouse or donkey anti-rabbit antibodies. Shown is an example of an experiment carried out in duplicate; subconfluent cultures were analyzed. Purified NPC1 is slightly smaller than that in the extract because it was deglycosylated (see Materials and methods).
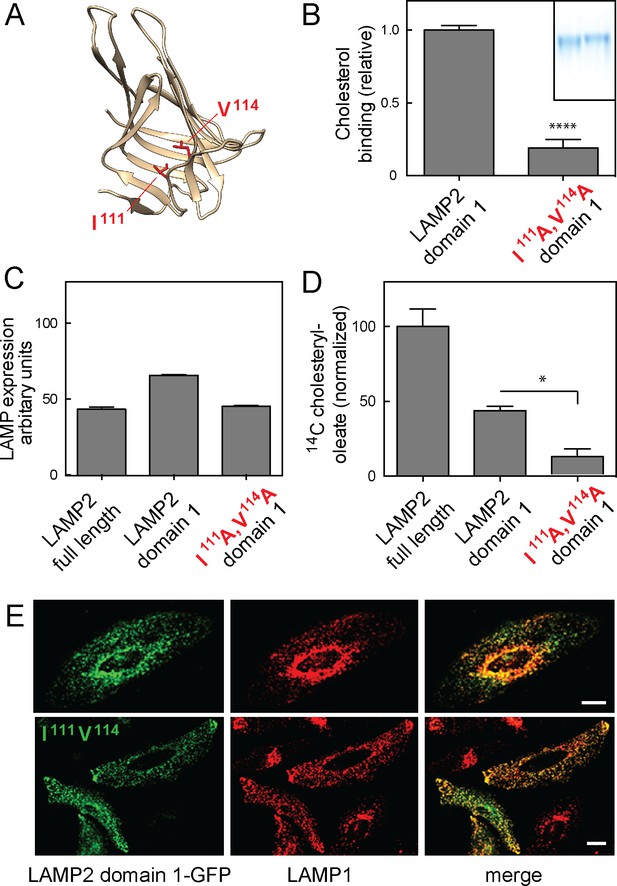
Cholesterol binding to LAMP2 domain 1 is required for its ability to rescue cholesterol export from LAMP-deficient lysosomes.
(A), predicted structure model of LAMP2 domain 1; residues I111 and V114 are highlighted in red. (B) Relative 3H-cholesterol binding to soluble LAMP 2 domain 1 or LAMP2 domain 1-I111A/V114A. Shown is combined data from 5 independent experiments carried out in duplicate in the presence of 50 nM 3H-cholesterol. Inset, SDS-PAGE analysis of wild type (left) and domain 1-I111A/V114A (right) proteins analyzed. P value was determined by two-tailed Student’s t-test. (C) flow cytometry analysis of mean fluorescence of GFP rescue constructs in lentivirus-tranduced cells (>20,000 cells analyzed). (D) Cholesteryl oleate synthesis in MEF cells lacking LAMP1 and LAMP2 after rescue with either full length, membrane anchored LAMP2, membrane anchored LAMP2 domain 1, or membrane anchored LAMP2 domain 1-I111A/V114A. C-terminally GFP-tagged, rescue proteins were stably expressed using lentivirus transduction; shown is the combined result of 2 independent experiments, normalized for the amount of mature protein in each sample (Figure 6—figure supplement 1) relative to the amount of rescue seen with full length LAMP2 protein. P-values are in relation to full length for domain 1, or to domain 1 for the mutant protein, and were determined by one way ANOVA. E, confocal light microscopic analysis of GFP rescue construct localization (green) and endogenous LAMP1 protein (red) in transiently transfected HeLa cells; bars represent 20 µm.
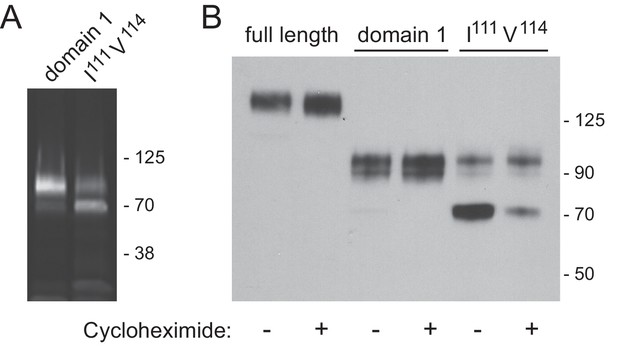
Immunoblot analysis of LAMP2 constructs from lentivirus transduced LAMP1/LAMP2-knock out MEF cells.
(A) total cell extract from cells expressing membrane anchored, GFP-LAMP2 domain 1 or GFP-LAMP2 domain 1-I111A/V114A; (B) migration of the constructs indicated, before or after 4 hr cycloheximide treatment (50 µg/ml). Panel A was developed as in Figure 4B; panel B was developed as in Figure 4A.





