p27Kip1 promotes invadopodia turnover and invasion through the regulation of the PAK1/Cortactin pathway
Figures
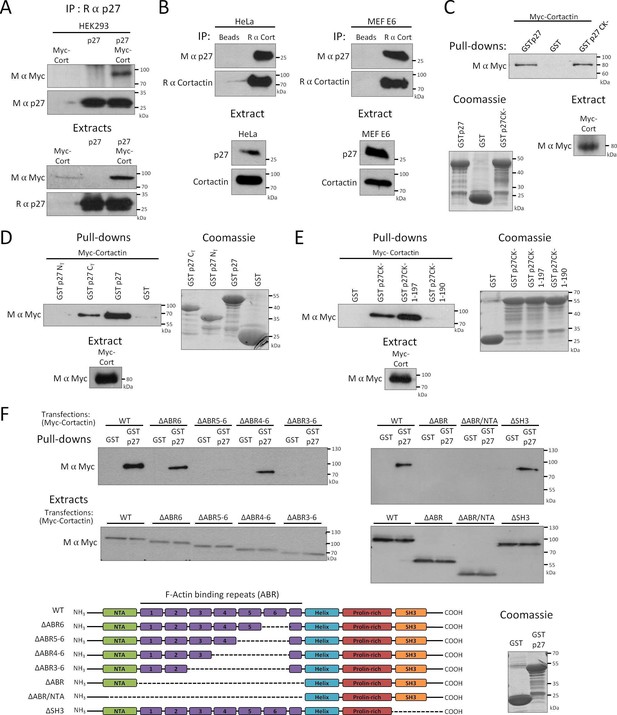
p27 binds to Cortactin.
(A–B) Co-immunoprecipitation of p27 and Cortactin: (A) p27 was immunoprecipitated using rabbit anti-p27 (C19) antibodies from HEK293 lysates transfected with plasmids encoding p27, Myc-Cortactin (Myc-Cort) or both. (B) Immunoprecipitation of endogenous Cortactin using rabbit anti-Cortactin (H191) antibodies from HeLa or E6 MEF lysates, beads alone were used as control. (A–B) Co-immunoprecipitated proteins were detected by immunoblot with mouse anti-c-Myc (9E10) (A) or mouse anti-p27 (F-8) antibodies (B). Immunoprecipitated proteins were visualized by reprobing the membrane with mouse anti-p27 (F-8) (A) or with rabbit anti-Cortactin (H-191) antibodies and anti-rabbit Ig light-chain secondary antibodies (B). Immunoblots of extracts show the level of proteins in each condition. (C–F) Pull-down assays: HEK293 cells were transfected with Myc-Cortactin (C–E) or various deletion mutants of Myc-Cortactin (F) (ΔABR6, ΔABR5-6, ΔABR4-6, ΔABR3-6, ΔABR, ΔABR/NTA or ΔSH3, described in the schematic representation of Cortactin, bottom panel). NTA: N-Terminal acidic domain; ABR: actin binding repeat; Helix: helical domain; SH3: Src-homology three domain. Lysates were subjected to pull-down assays using GST, GST-p27 or GST-p27CK- (C), or GST, GST-p27, GST-p27 NT (1-87) and GST-p27 CT (88-198) (D), or GST, GST-p27CK-, GST-p27CK- (1-197) and GST-p27CK- (1-190) (E) or GST and GST-p27 (F). The amounts of Myc-Cortactin bound to the beads and of transfected protein present in the extracts were detected by immunoblot using mouse anti c-Myc (9E10) antibodies. The amounts of GST or GST p27 and mutants used in the assays were visualized by Coomassie staining of the gels. (A–F) All panels show representative results of at least three independent experiments.
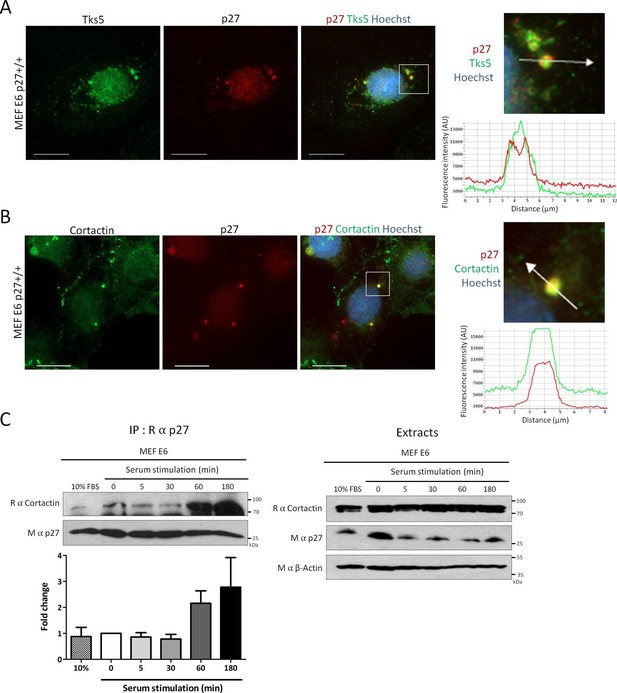
p27 colocalizes with Cortactin in invadopodia.
(A–B) p27+/+ MEFs were seeded on gelatin-coated coverslips for 24 hr. Cells were permeabilized with digitonin prior to fixation. Cells were labeled with mouse anti p27 (SX53G8.5) in red (A–B) and rabbit anti-Tks5 (M-300) (A) or rabbit anti-Cortactin (H-191) (B) in green. Images were acquired using a 60x objective and images displayed are cropped areas. Graphs displaying the fluorescence intensity (arbitrary unit) under the arrows in the enlarged panels were generated with the NIS Element software. Scale bars: 20 μm. (C) p27+/+ E6 MEFs were starved overnight in DMEM-0.1% FCS and then stimulated with growth medium for the indicated times. Cell lysates were subjected to immunoprecipitation using rabbit anti-p27 (C–19). Immunoprecipitated proteins and corresponding cell extracts were immunoblotted with rabbit anti-Cortactin (H–191) and mouse anti-p27 (F–8) antibodies. β-actin was used as loading control. The graph represents the mean fold change in amounts of Cortactin co-precipitated with p27 in each condition compared to time zero from five independent experiments. These differences were not statistically significant.
-
Figure 2—source data 1
Quantification of co-immunoprecipitation between p27 and Cortactin in MEF E6 (Figure 2C) and HeLa cells (Figure 2—figure supplement 2).
- https://doi.org/10.7554/eLife.22207.005
-
Figure 2—source data 2
Statistical analyses for Figure 2C and Figure 2—figure supplement 2.
- https://doi.org/10.7554/eLife.22207.006
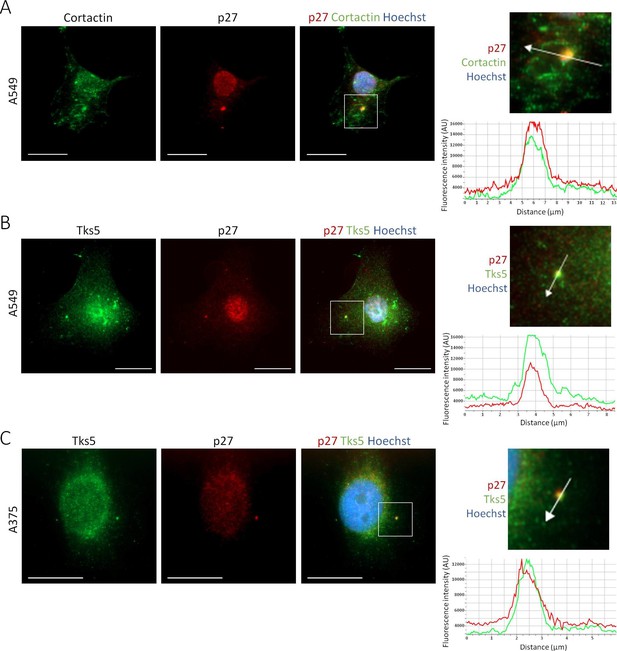
p27 colocalizes with Cortactin and Tks5 in different tumor cell lines.
(A–C) Twenty-four hr after seeding, A549 [A-B] or A375 cells [C] were permeabilized with digitonin prior to paraformaldehyde fixation. Cells were immunostained with mouse anti p27 (SX53G8.5) (red) [A-C] and with rabbit anti-Cortactin (H-191) [A] or rabbit anti-Tks5 (M-300) [B-C] (green) antibodies. Graphs display the fluorescence intensity (arbitrary unit) in the green and red channels over the distance depicted by the arrow in each enlarged area. Images were acquired using a 60x objective and images displayed are cropped areas. Measurements were made with the NIS Element software (Nikon). Scale bars: 20 μm.
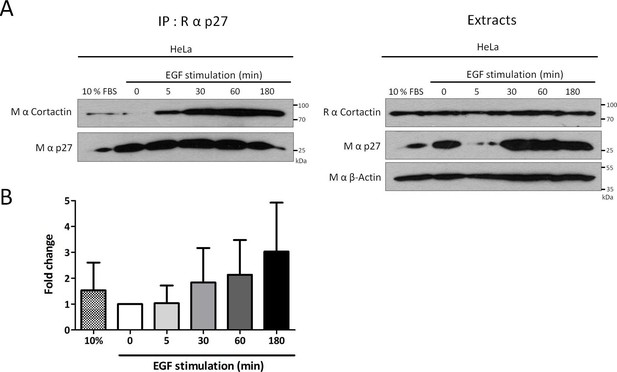
p27/Cortactin interaction in HeLa cells after EGF stimulation.
(A) HeLa cells were starved with DMEM 0.1% FCS overnight, stimulated with 100 ng/ml EGF (Peprotech) for the indicated times and cell lysates were submitted to co-immunoprecipitation using rabbit anti-p27 (C-19) antibodies. Co-immunoprecipitated proteins and total proteins in lysates were detected by immunoblot with anti-Cortactin and mouse anti-p27 (F-8) antibodies. β-actin was used as loading control. (B) The graph represents the mean fold change of Cortactin co-precipitated with p27 compared to time zero in three independent experiments. These differences were not statistically significant.
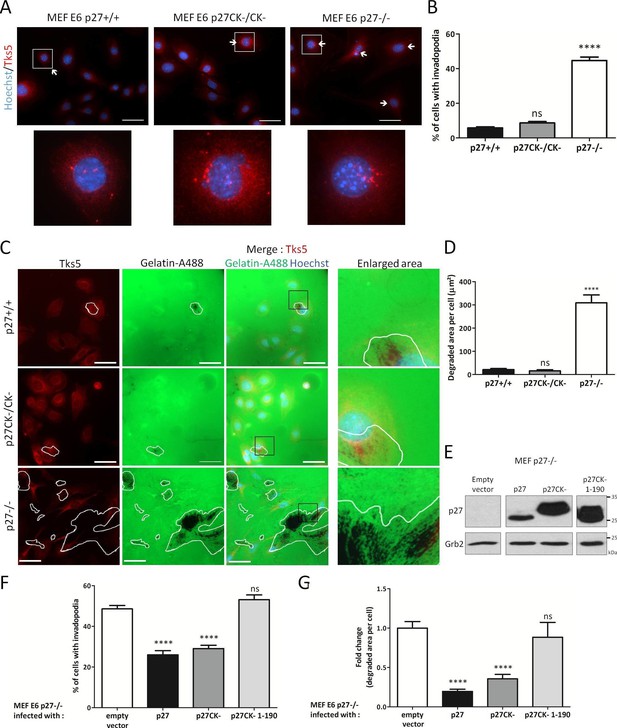
p27 regulates invadopodia formation and matrix degradation.
(A) p27+/+, p27CK−/CK− and p27−/− immortalized MEFs were seeded on Oregon green-gelatin (gelatin-A488) for 16 hr. Cells were stained with rabbit anti-Tks5 (M-300) to visualize invadopodia. (B) The percentage of cells forming invadopodia was determined in a minimum of 15 fields, representing a minimum of 330 cells per genotype, for each experiment. The graph shows the mean of 3 independent experiments. (C) Cells were seeded as in (A). Tks5 staining shows invadopodia (red) and areas of degraded fluorescent gelatin indicate invadopodia activity (green). (D) The areas of degraded gelatin were measured in at least 15 fields per genotype in each experiment. The graph shows the mean of 3 independent experiments. (E–G) p27−/− E6 MEFs were infected with either empty vector or p27, p27CK− or p27CK− 1–190 vectors and then seeded on Gelatin-A488 for 48 hr. (E) p27 levels after retroviral infection were determined by immunoblot with rabbit anti-p27 (N-20) antibodies; Grb2 was used as loading control. (F–G) After Tks5 staining, invadopodia forming cells (F) were quantified as in B and area of degraded gelatin (G) as in D. Scale bars: 50 μm; ‘ns’ not significant; ****p<0.0001. In A and C, images were acquired using a 40x objective and images displayed are cropped areas.
-
Figure 3—source data 1
Quantification of cells with invadopodia (Figure 3B); quantification of degraded gelatin area per cell (Figure 3C); quantification of cells with invadopodia after p27 re-expression (Figure 3F) and quantification of degraded gelatin area per cell after p27 re-expression (Figure 3G).
- https://doi.org/10.7554/eLife.22207.010
-
Figure 3—source data 2
Statistical analyses for Figure 3B,C,F and G.
- https://doi.org/10.7554/eLife.22207.011
-
Figure 3—source data 3
Immunoblot scans of Figure 3E.
- https://doi.org/10.7554/eLife.22207.012
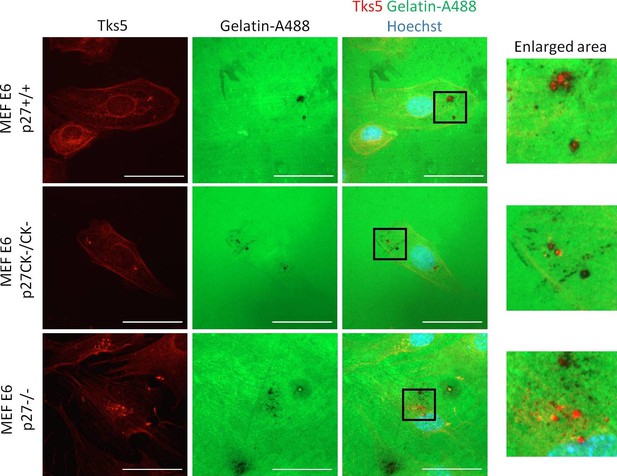
p27+/+, p27CK−/CK− and p27−/− MEFs form functional invadopodia.
HPV E6 immortalized p27+/+, p27CK−/CK− and p27−/− MEFs were seeded on Gelatin-A488 for 16 hr. Cells were fixed in paraformaldehyde and stained with rabbit anti-Tks5 (M-300; red) antibodies to visualize invadopodia. Images were acquired using a 60x objective and images displayed are cropped areas. Scale bars: 50 μm.
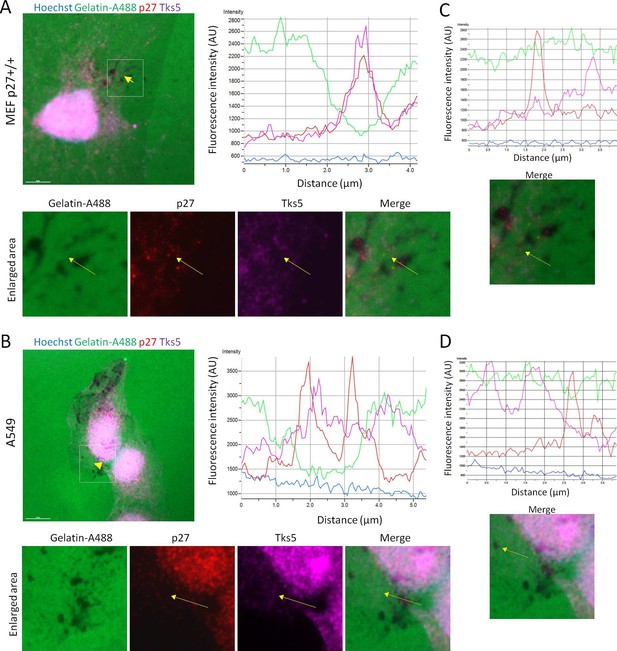
p27 colocalizes with Tks5 at sites of gelatin degradation.
HPV E6 immortalized p27+/+ MEFs (A) or A549 lung adenocarcionma cells (B) were seeded on Gelatin-A488 and treated with 1 μM FRAX597 to stabilize invadopodia after 2 hr. After 72 hr, cells were permeabilized with digitonin, fixed in paraformaldehyde and stained with mouse anti-p27 (SX53G8.5, red) and rabbit anti-Tks5 antibodies (M-300; purple) to visualize p27/Tks5 colocalization and sites of gelatin degradation. Images were acquired using a 60x objective and images displayed are cropped areas. Scale bars: 10 μm. Graphs display the fluorescence intensity (arbitrary unit) in each channel over the distance depicted by the arrows (NIS Element software, Nikon). (C–D) Profiles of Cortactin and p27 localization outside of gelatin degradation area in p27+/+ MEFs (C) and A549 cells (D). The graphs were generated by moving the arrows from panels A and B to regions without gelatin degradation.
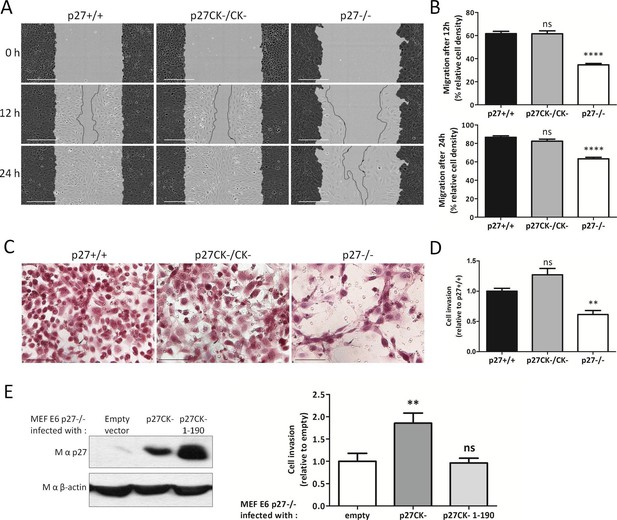
p27 promotes cell migration and invasion.
(A) Representative images of scratch wound migration assays with p27+/+, p27CK−/CK− and p27−/− immortalized MEFs. Dark grey areas show the initial wound masks and dotted lines the migration fronts. Scale bars: 300 μm. (B) Mean cell migration at 12 hr and 24 hr post wounding for each genotype of five independent experiments. Percent of area in which the cells migrated, or wound closing (relative wound density) was calculated with the Incucyte software. ‘ns’: not significant; ****p<0.0001. (C) Representative images of p27+/+, p27CK−/CK− and p27−/− immortalized MEFs that invaded through a layer of Collagen I in transwell invasion assays and migrated to the bottom side of the transwell membrane after 48 hr. Scale bars: 100 μm. (D) The graph shows the mean number of cells that invaded through Collagen I quantified by XTT staining, expressed relative to p27+/+ cells, of three independent experiments. **p<0.01. (E) p27−/− E6 MEFs were infected with either empty vector, p27CK- or p27CK- 1–190 vectors and used in transwell invasion assays as in (C–D). p27 levels after retroviral infection were determined by immunoblot with mouse anti-p27 (SX53G8.5); β-actin was used as loading control. The graph shows the mean number of cells that invaded through Collagen I quantified by XTT staining, expressed relative to p27−/− cells, of four independent experiments.
-
Figure 4—source data 1
quantification of relative wound density (Figure 4B); quantification of invasion (Figure 4D); and quantification of invasion rescue by p27 re-expression (Figure 4E).
- https://doi.org/10.7554/eLife.22207.016
-
Figure 4—source data 2
Statistical analyses for Figure 4B,D and E.
- https://doi.org/10.7554/eLife.22207.017
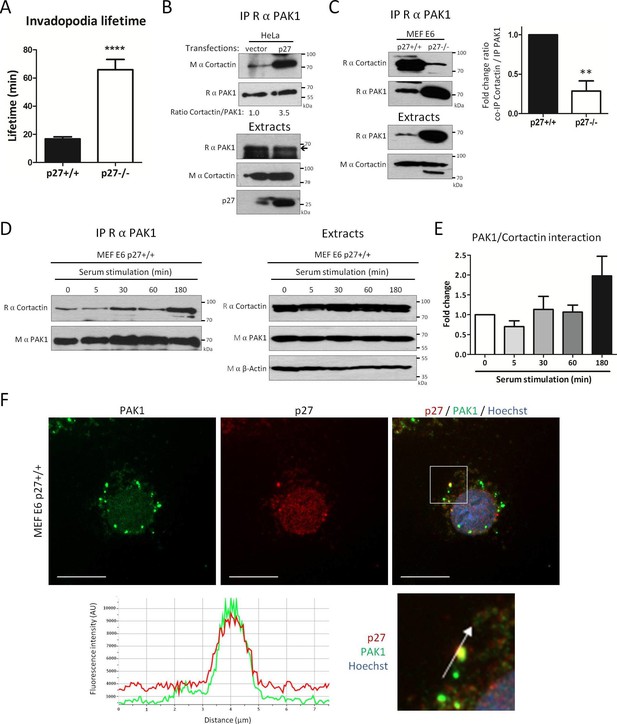
p27 promotes binding of Cortactin to PAK1.
(A) Live p27+/+ and p27−/− immortalized MEFs expressing eGFP-Tks5 were imaged by videomicroscopy to measure invadopodia lifetime, using Tks5 as an invadopodia marker. The graph represents the average invadopodia lifetime of 20 invadopodia per genotype per experiment from three independent experiments. (B–C) Co-immunoprecipitations using rabbit anti-PAK1 (N-20) of HeLa cell lysates transfected with empty vector or p27 encoding vector (B) or p27+/+ and p27−/− E6 MEF lysates (C). Co-immunoprecipitated Cortactin was detected with mouse (B) or rabbit (C) anti-Cortactin antibodies. Immunoprecipitated PAK1 was visualized by reprobing the membranes with rabbit anti-PAK1 (N-20) and anti-Rabbit Ig light-chain secondary antibodies. Immunoblots of extracts show the level of proteins in each condition. In (C), the graph shows the mean fold change in the ratio of Cortactin to PAK1 co-immunoprecipitated, expressed relative to p27+/+ cells, in three independent experiments. **p<0.01. (D) p27+/+E6 MEFs were starved overnight in DMEM 0.1% FCS and then stimulated with growth medium for the indicated times. PAK1 was immunoprecipitated from cell lysates using rabbit anti-PAK1 (N-20). Immunoblots of immunoprecipitates (left panels) and extracts (right panels) were probed successively with rabbit anti-Cortactin (H-191) and anti-rabbit Ig light chain secondary antibodies and then with mouse anti-PAK1 (A6) antibodies. β-actin was used as loading control. (E) The graph shows the mean amount of Cortactin bound to PAK1 at each time-point from four independent experiments, normalized to time zero. These differences were not statistically significant. (F) p27+/+ MEFs were seeded on coverslips and permeabilized with digitonin prior to fixation. Cells were labeled with rabbit anti-PAK1 (N-20, green) and mouse anti p27 (SX53G8.5, red) antibodies. Images were acquired using a 60x objective and images displayed are cropped areas. The graph displaying the fluorescence intensity (arbitrary unit) under the arrow in the enlarged panel was generated with NIS Element software. Scale bars: 20 μm.
-
Figure 5—source data 1
Quantification of invadopodia lifetime (Figure 5A); quantification of co-immunoprecipitation between Cortactin and PAK1 in MEFs (Figure 5C); and quantification of co-immunoprecipitation between Cortactin and PAK1 after serum stimulation (Figure 5E).
- https://doi.org/10.7554/eLife.22207.019
-
Figure 5—source data 2
Statistical analyses for Figure 5A.
- https://doi.org/10.7554/eLife.22207.020
-
Figure 5—source data 3
Statistical analyses for Figure 5C.
- https://doi.org/10.7554/eLife.22207.021
-
Figure 5—source data 4
Statistical analyses for Figure 5E.
- https://doi.org/10.7554/eLife.22207.022
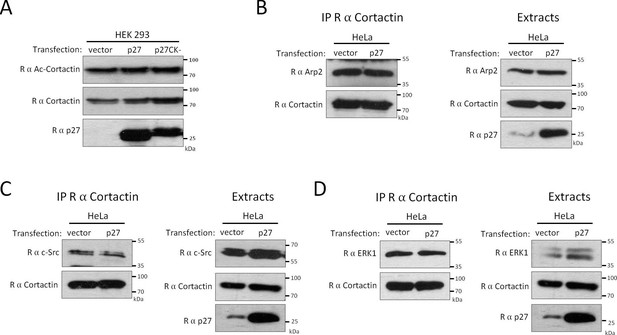
p27 does not affect Cortactin acetylation or the recruitment of c-Src, Arp2 and ERK1 to Cortactin.
(A) HEK 293 cells were transfected with either empty vector, p27 or p27CK- coding vectors. Lysates were used for immunoblot with rabbit anti-Acetyl-Cortactin (BD-Transduction Laboratories), anti-Cortactin (H-191) and anti-p27 (C-19) antibodies. (B–D) Co-immunoprecipitations were performed using rabbit anti-Cortactin (H-191) in HeLa cell lysates transfected with either empty vector or p27 coding vector. Co-immunoprecipitated proteins were detected with rabbit anti-Arp2 (H-84) (B), anti-c-Src (SRC2) (C) or anti-ERK1 (K-23) (D). The immunoprecipitated proteins were visualized by reprobing the membrane with rabbit anti-Cortactin (H-191) and anti Rabbit Ig light-chain secondary antibodies. Immunoblots of extracts show the levels of proteins of interest in each condition. Experiments were performed at least three times, except only twice for [D].
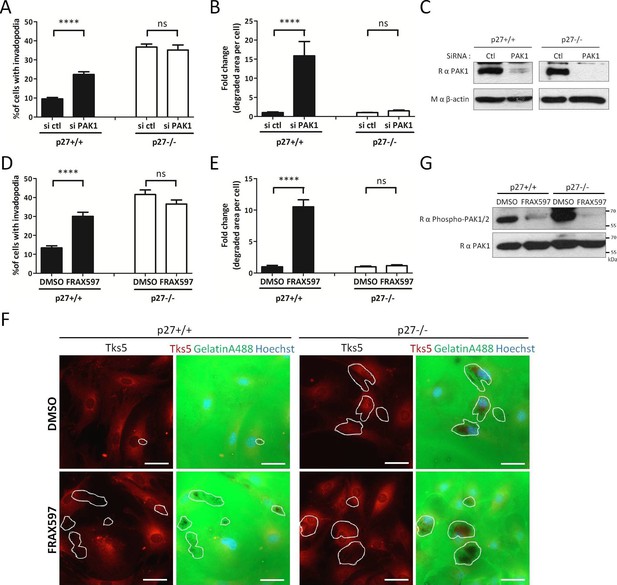
p27 regulates invadopodia formation downstream of PAK1.
(A–B) p27+/+ and p27−/− E6 MEFs were seeded on gelatin-A488 and transfected with either control siRNAs or PAK1 siRNAs for 48 hr. Cells were stained with rabbit anti-Tks5 (M-300) to visualize invadopodia. At least ten fields per conditions were used to count cells forming invadopodia (A), representing a minimum of 212 cells per genotype for each experiment, or to measure the area of degraded gelatin, expressed in fold-change compared to vehicle treated conditions (B). (C) PAK1 siRNA efficacy at 48 hr was evaluated by immunoblot with rabbit anti-PAK1 (N-20) antibodies. β-actin was used as loading control. (D–F) p27+/+ and p27−/− E6 MEFs were seeded for 48 hr on gelatin-A488 and after 1 hr were treated with DMSO or 1 μM FRAX597, a PAK1-3 inhibitor. Quantification of cells forming invadopodia (D) and gelatin degradation (E) was performed as in (A–B), with a minimum of 215 cells counted per genotype per experiment. (F) Images were acquired using a 40x objective and images displayed are cropped areas. Scale bars: 50 μm. (G) FRAX597 inhibitor efficacy was evaluated by immunoblot with rabbit anti-phospho-Ser144-PAK1/phospho-Ser141-PAK2 and rabbit anti-PAK1 (N-20) antibodies. (A–B; D–E) The graphs show the mean of at least three independent experiments. ****p<0.0001.
-
Figure 6—source data 1
Quantification of invadopodia forming cells (Figure 6A) and degraded gelatin area (Figure 6B) after PAK1 silencing; quantification of invadopodia forming cells (Figure 6D) and degraded gelatin area (Figure 6E) after FRAX597 treatment; quantification of invadopodia forming cells (Figure 6—figure supplement 1A) and degraded gelatin area (Figure 6—figure supplement 1B) after FRAX1036 and G-5555 treatment.
- https://doi.org/10.7554/eLife.22207.025
-
Figure 6—source data 2
statistical analyses for Figure 6A,B,D and E and Figure 6—figure supplement 1A and B.
- https://doi.org/10.7554/eLife.22207.026
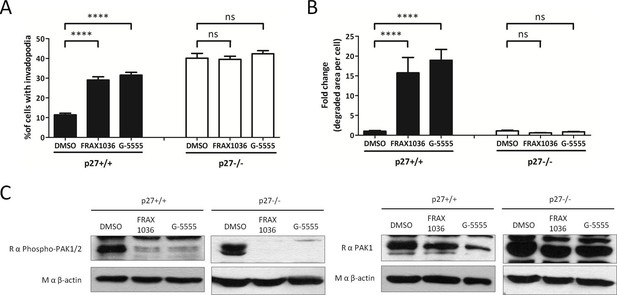
p27 regulates invadopodia formation in a PAK1 dependent manner.
(A–B) p27+/+ or p27−/− E6 MEFs were seeded for 48 hr on Gelatin-A488 and treated 1 hr after seeding with either DMSO,1 μM FRAX1036 or 1 μM G-5555, two PAK1-3 inhibitors. Cells were stained with rabbit anti-Tks5 (M-300) to visualize invadopodia. The graphs show the mean of 3 independent experiments. For each experiment, at least ten fields per conditions were used to count cells forming invadopodia, representing a minimum of 168 cells per genotype (A), or to measure the area of degraded gelatin, expressed in fold-change compared to vehicle treated conditions (B). (C) The efficacy of PAK inhibitors was evaluated by immunoblot with rabbit anti-phospho-Ser144-PAK1/phospho-Ser141-PAK2 and Rabbit anti-PAK1 antibodies. Reprobing of membranes with a mouse anti-β actin antibody was used to control protein loading.
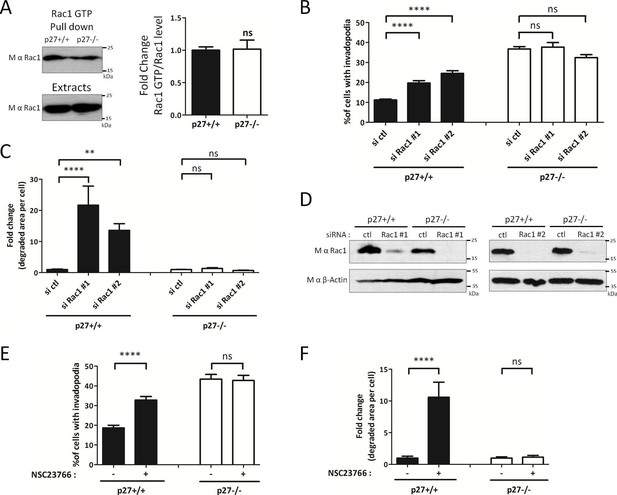
p27 regulates invadopodia formation downstream of Rac1.
(A) Cells were seeded for 24 hr and Rac1-GTP levels measured by GTP pull-downs assays using GST-PAK1-CD beads. The amounts of Rac1-GTP bound to the beads and of total Rac1 in the extracts were detected by immunoblot with mouse anti-Rac1. The graph shows the mean ratio of GTP-Rac1/total Rac1 from six independent experiments. (B–D) Cells were transfected with control (ctl) or Rac1 #1 or #2 siRNAs for 3 days. Cells were then seeded on Gelatin-A488 for 48 hr and for monitoring siRNA efficiency. Cells were stained with rabbit anti-Tks5 (M-300) to visualize invadopodia. At least ten fields per condition were used to count cells forming invadopodia, representing a minimum of 197 cells per genotype for each experiment (B) or to measure the area of degraded gelatin, expressed in fold-change compared to control siRNA treated conditions (C). (D) Rac1 silencing was evaluated by immunoblot with mouse anti-Rac1. β-actin was used as loading control. (E–F) Cells were processed as described in Figure 6D and E except that the Rac1 inhibitor NSC23766 at 100 μM was used instead of FRAX597. A minimum of 153 cells were counted per genotype for each experiment. (B–C; E–F) The graphs show the mean of at least three independent experiments. ****p<0.0001; **p<0.01.
-
Figure 7—source data 1
Quantification of Rac1 GTP/Rac1 levels (Figure 7A); quantification of invadopodia forming cells (Figure 7B) and degraded gelatin area (Figure 7C) after silencing of Rac1; quantification of invadopodia forming cells (Figure 7E) and degraded gelatin area (Figure 7F) after NSC23766 treatment; quantification of invadopodia forming cells (Figure 7—figure supplement 1A) and degraded gelatin area (Figure 7—figure supplement 1B) after RhoA silencing; and quantification of invasion after Y27632 treatment (Figure 7—figure supplement 1D).
- https://doi.org/10.7554/eLife.22207.029
-
Figure 7—source data 2
Statistical analyses for Figure 7A,B,C,E,F, and Figure 7—figure supplement 1A,B and D.
- https://doi.org/10.7554/eLife.22207.030
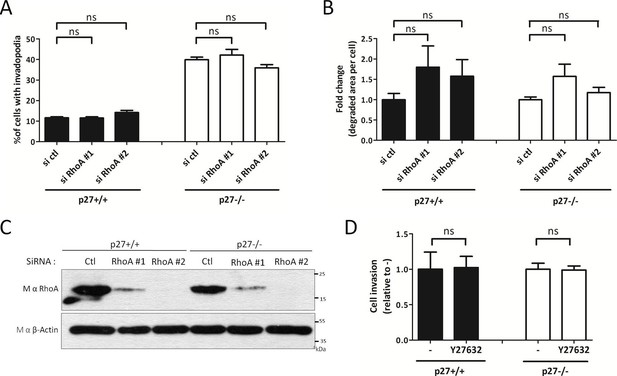
RhoA regulation by p27 is not involved in invadopodia formation.
(A–C) p27+/+ and p27−/− immortalized MEFs were transfected with siRNA control (ctl) or siRNA RhoA #1 or #2. After 3 days, cells were seeded on Gelatin-A488 for matrix degradation and for controlling siRNA efficiencies, for 48 hr. Cells were stained with rabbit anti-Tks5 (M-300) to visualize invadopodia. For each experiment, ten fields per condition, representing a minimum of 256 cells per genotype, were used to count invadopodia forming cells (A) or the area of degraded gelatin per cell (B). The graphs show the means of at least three independent experiments. (C) siRNA efficiency was evaluated by immunoblot with mouse anti-RhoA (26C4) antibodies. β-actin was used as loading control. (D) p27+/+ and p27−/− immortalized MEFs were subjected to invasion assays as in Figure 4C–D in presence or absence of the ROCK inhibitor Y27632 (10 μM). The graph shows the fold change in invasion relative to vehicle treated cells (-) from three independent experiments.
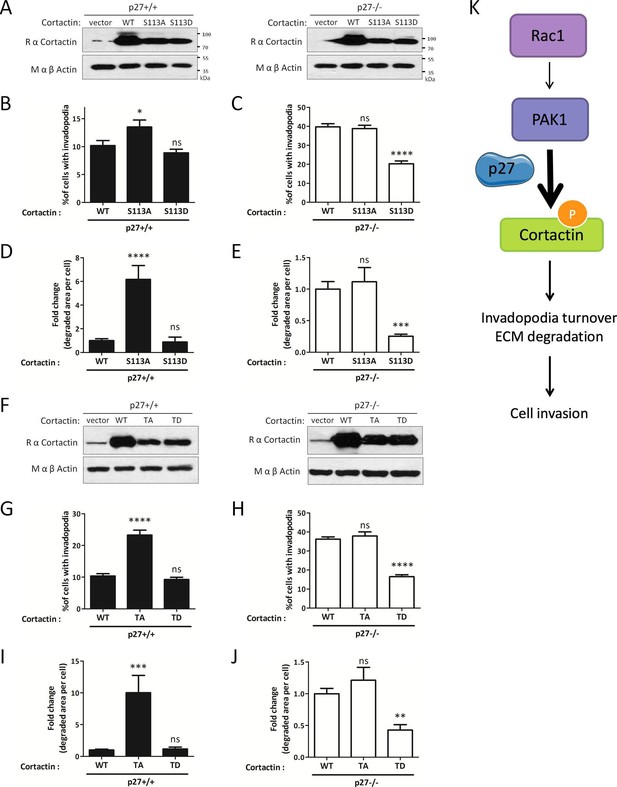
Mimicking phosphorylation of Cortactin restores invadopodia dynamics in p27−/− cells.
(A–E) p27−/− E6 MEFs were infected with empty vector or with vectors encoding wild type Cortactin (WT), S113A-Cortactin (S113A) or S113D-Cortactin (S113D). (A) Cortactin levels after retroviral infection were determined by immunoblot with rabbit anti-Cortactin (H-191) antibodies. β-actin was used as loading control. (B–E) Cells were seeded on gelatin-A488 for 48 hr. After Tks5 staining, cells forming invadopodia (B–C), or the area of degraded gelatin, expressed in fold-change compared to WT Cortactin transfected cells (D–E), were quantified in at least ten fields per condition in each experiment, representing a minimum of 179 cells per genotype. The graphs show the means of at least three independent experiments. (F–J) p27−/− E6 MEFs were infected with empty vector or with vectors encoding WT Cortactin, Cortactin TA (S113A/S150A/S282A) or Cortactin TD (S113D/S150D/S282D). (J) Cortactin levels after retroviral infection were determined as in (A). (G–J) Cells were processed as in (B–E) to quantify cells forming invadopodia (G–H), or the area of degraded gelatin (I–J), with a minimum of 222 cells counted per genotype per experiment. The graphs show the means of 3 independent experiments. (K) Schematic representation of the Rac1/PAK1/phospho-Cortactin pathway involved in invadopodia turnover and matrix degradation and its proposed regulation by p27. ****p<0.0001; ***p<0.001; **p<0.01; *p<0.05.
-
Figure 8—source data 1
Quantification of cells forming invadopodia (Figure 8B–C) and degraded gelatin area (Figure 8D–E) after infection with S113 phospho-mutants of Cortactin; quantification of cells forming invadopodia (Figure 8G–H) and degraded gelatin area (Figure 8I–J) after infection with triple phospho-mutants of Cortactin; quantification of P-Ser Cortactin/Cortactin ratio (Figure 8—figure supplement 1B).
- https://doi.org/10.7554/eLife.22207.033
-
Figure 8—source data 2
Statistical analyses for Figure 8.
- https://doi.org/10.7554/eLife.22207.034
-
Figure 8—source data 3
Statistical analyses for Figure 8—figure supplement 1B.
- https://doi.org/10.7554/eLife.22207.035
-
Figure 8—source data 4
Mascot search results for Cortactin for Figure 8—figure supplement 2.
- https://doi.org/10.7554/eLife.22207.036
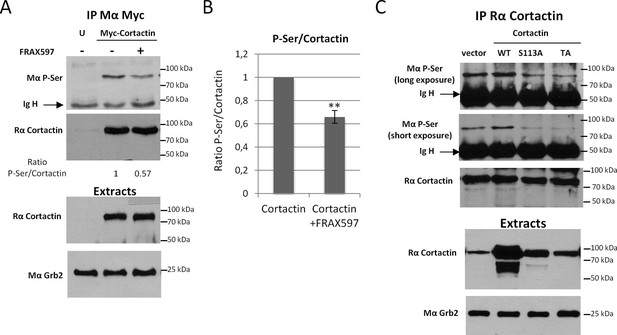
Cortactin is phosphorylated on S113/S150 and/or S282 in vivo.
(A) HEK 293 cells were tranfected with Myc-tagged Cortactin for 24 hr or left untransfected (U) and 3 μM FRAX597 was added for 12 hr where indicated. Cortactin was immunoprecipitated with anti-Myc antibodies (9E10) and resolved on SDS-PAGE and transferred on PVDF membranes. The amount of Ser-phosphorylated Cortactin was detected by immunoblot with a monoclonal antibody against phospho-serine (BD) and the total amount of immunoprecipitated Cortactin by reprobing with anti-Cortactin (H191) antibody. P-Ser and Cortactin band intensities were quantified with Image J to calculate ratios, normalized to one for the control condition. Extracts were probed for Cortactin and Grb2 was used as loading control. (B) Average ratio of P-Ser signal/Total Cortactin signals from three independent experiments. Error bars are s.e.m. **p<0.01. (C) HEK 293 cells were tranfected with either empty vector, wild-type Cortactin (WT), S113A Cortactin or S113A/S150A/S282A (TA) Cortactin for 24 hr. Cortactin was immunoprecipitated with anti-Cortactin (H191) antibodies and processed as in (A). Serine phosphorylation of Cortactin was evaluated with anti-Phospho-Ser antibodies (BD). This data is representative of three independent experiments.
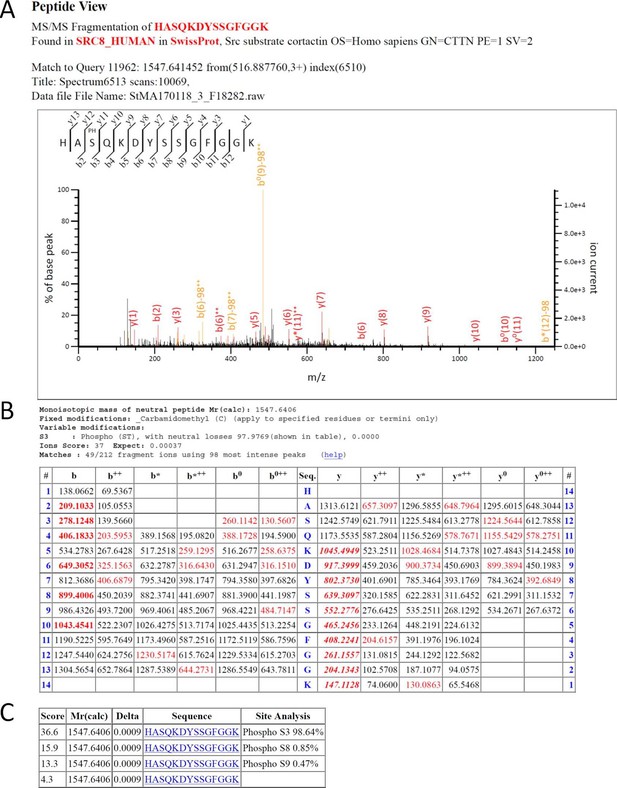
Cortactin is phosphorylated on S150 in vivo upon PAK activation.
MS/MS analysis of Cortactin immunoprecipitated from HEK 293 cells transfected either with Myc-Cortactin or Myc-Cortactin and Myr-SH3-2, the second SH3 domain of Nck1 fused to the myristoylation signal of Src, which activate PAK kinase. S150 phosphorylation was only detected from lysates in which PAK activtiy was stimulated by the presence of the Src-SH3-2 domain. (A) MS/MS spectrum annotation of the residues 148–161 from Uniprot database reference SRC8_HUMAN or Q14247 (Src substrate Cortactin) protein. MS-Fragmented peptide correspond to the m/z selection of 516.888, matching [HASQKDYSSGFGGK +80 Da (HPO3)]3+ among 691 spectrum matching 98 distinct sequences of the same protein. Measured fragment matching masses bearing the modified serine residue showing a loss of neutral mass (H3PO4) are indicated in yellow. Not all the annotations could fit in the figure. The table below gathers all the matching fragments. (B) Table of theoretical fragment masses for the residues 148–161 of Uniprot database reference SRC8_HUMAN or Q14247 (Src substrate Cortactin) protein. In red are experimental masses of m/z detection of 516.888 matching [HApSQKDYSSGFGGK]3+ from the above spectrum. (C) Table of possible matches of the same set of experimental data with the four possible and closest theoretical sequences. The best score matches the phosphorylation of the serine 150 among the residues 148–161 of Uniprot database reference SRC8_HUMAN or Q14247 (Src substrate Cortactin) protein. Site analysis percentage corresponds to the probability output from the PhosphoRS algorythm.





