A dynamic mode of mitotic bookmarking by transcription factors
Figures
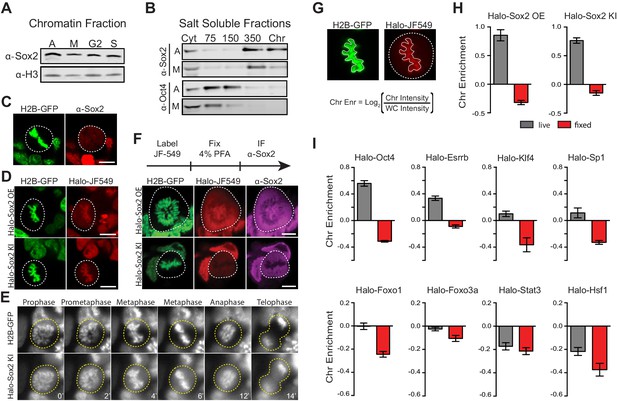
Transcription factors are not excluded from mitotic chromosomes.
(A) Biochemical fractionation of asynchronous (A) mouse ES cells and synchronized populations at mitosis (M), G2, and S phases was performed to isolate the chromatin-associated fraction. Sox2 and H3 were detected by Western blot analysis. (B) Salt fractionation of asynchronous (A) and mitotic (M) mouse ES cells. Sox2 and Oct4 were detected by Western blot analysis. Cyt, Cytoplasmic fraction. Chr, Chromatin fraction. (C) Immunofluorescence with α-Sox2 of mouse ES cells stably expressing H2B-GFP using confocal microscopy showing exclusion of Sox2 from mitotic chromosomes (D) Live-cell imaging using confocal microscopy of mouse ES cells stably expressing H2B-GFP with overexpressed Halo-Sox2 (Halo-Sox2 OE, top) and endogenously tagged Halo-Sox2 (Halo-Sox2 KI, bottom) (E) Epi-fluorescence time-lapse imaging of mouse ES cells stably expressing H2B-GFP and endogenously-tagged Halo-Sox2 KI (F) Live cells with overexpressed Halo-Sox2 (top) or endogenously-tagged Halo-Sox2 (bottom) were labeled with JF549 dye and subjected to standard immunofluorescence by fixation with 4% PFA and detection with α-Sox2. (G) Strategy for quantifying TF chromosome enrichment. (H) Chromosome enrichment levels for overexpressed and endogenously-tagged Halo-Sox2. n = 40 cells (I). Chromosome enrichment levels for indicated Halo-tagged transcription factors. n = 40 cells. Data are represented as mean ± SEM. Scale bars, 5 µm.
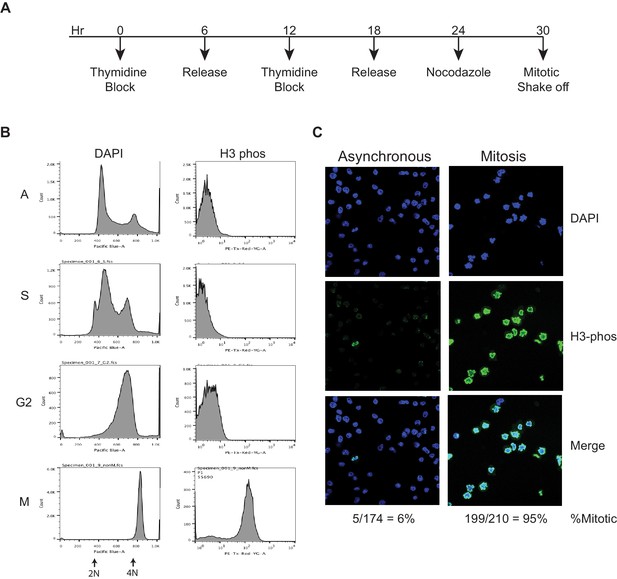
Synchronization of mouse ES cells.
(A) Time-course strategy for synchronizing mouse ES cells. Thymidine at 2 mM is added to cells for 6 hr followed by release with fresh media for 6 hr. This cycle of thymidine block and release is repeated, and followed by Nocodazole treatment (100 ng/µL) for 6 hr. Mitotic cells are collected by shake off. (B) Flow cytometry analysis of asynchronous (A) cells and synchronized populations as S, G2, and mitosis (M) phases. DAPI is used to mark DNA content, and an antibody against phosphorylated H3 is used to mark mitotic cells. This analysis shows fairly pure populations of synchronized cells. (C) Asynchronous and Mitotic cells are stained with DAPI and immunostained with antibody against phosphorylated H3, and visualized on a confocal microscope. By counting the H3-phosphorylated positive cells over total cells as marked by DAPI, we can measure the efficiency of mitotic enrichment by the synchronization strategy to be about 95%, providing an orthogonal measurement for the purity of mitotic cells.
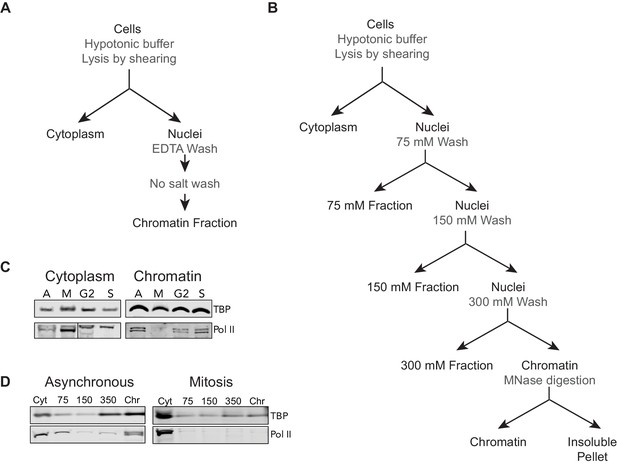
Biochemical and Salt fractionation strategies.
(A) For biochemical fractionation, asynchronous mouse ES cells or synchronized populations are swelled with hypotonic buffer and lysed by gentle shearing to separate nuclei from cytoplasmic fraction. The nuclei are washed repeatedly with EDTA-containing no-salt buffer to lyse the nuclei and remove the soluble nuclear fraction. The remaining chromatin-bound fraction is collected. (B) For salt fractionation, asynchronous mouse ES cells or synchronized mitotic cells are swollen with hypotonic buffer and lysed by gentle shearing to separate nuclei from cytoplasmic fraction as before. The nuclei are then incubated in low salt buffer containing 75 mM NaCl for 1 hr at 4°C. The nuclei are pelleted and the supernatant containing 75 mM soluble fraction is collected. The nuclei are then incubated in 150 mM salt buffer for 1 hr at 4°C. The nuclei are similarly pelleted and the supernatant containing the 150 mM soluble fraction is collected. This regime is repeated for 300 mM NaCl salt buffer. The final insoluble chromatin is resuspended in 300 mM NaCl buffer and incubated overnight at 37°C with 1 U of Micrococcal nuclease (MNase) to digest and solubilize chromatin. After overnight incubation, the insoluble material is pelleted by high speed centrifugation, and the soluble chromatin fraction is collected. (C) Biochemical fractionation of asynchronous (A) mouse ES cells and synchronized populations at mitosis (M), G2, and S phases was performed to isolate the chromatin-associated fraction. TBP (Abcam #ab51841) and Pol II (8WG16) were detected by Western blot analysis. (D) Salt fractionation of asynchronous (A) and mitotic (M) mouse ES cells. TBP and Pol II were detected by Western blot analysis. Cyt, Cytoplasmic fraction. Chr, Chromatin fraction.
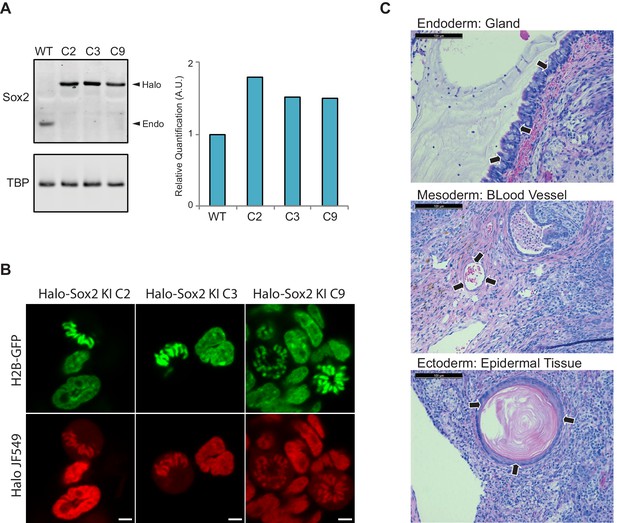
Endogenous knock-in of HaloTag to Sox2 locus using CRISPR/Cas9.
(A) We obtained three independent clones with homozygous knock-in of HaloTag to the Sox2 locus. We performed Western blot analysis of cell lysates from wild type ES cells (WT) and each individual knock-in clone for α-Sox2 and α-TBP for loading control. All of Sox2-detected molecules for each clone are migrating higher than the wild-type Sox2 suggesting that all molecules contain the HaloTag. Quantification of Western blot analysis is shown on the right. (B) Live-cell imaging of the three homozygous knock-in clones with cells labeled with Halo-dye JF549. (C) Halo-Sox2 KI cells were injected into SCID-Beige mice. Thirty days post inoculation kidney and testis tumors were harvested. Histological analyses of tumors show small areas of differentiated cells, representing endoderm (gland), mesoderm (blood vessel), and ectoderm (epidermal tissue).
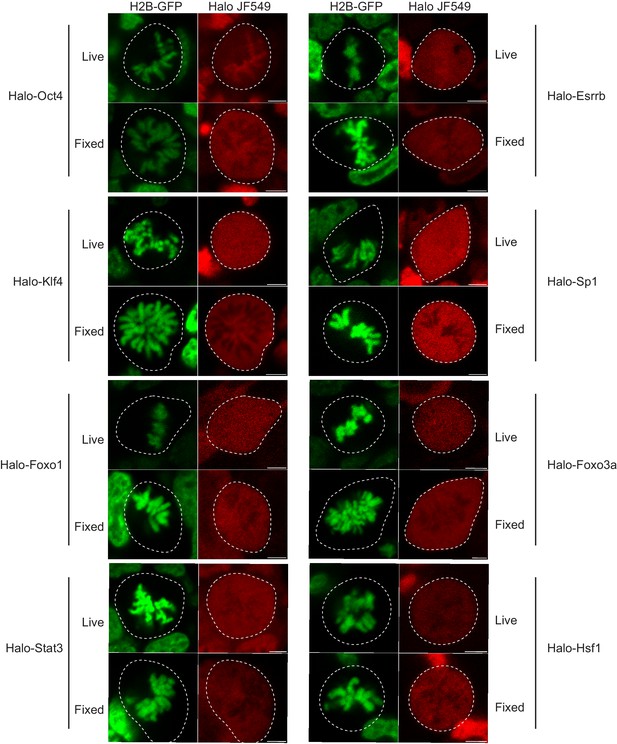
Live versus fixed images of Halo-tagged TFs in mouse ES cells.
Mouse ES cells stably expressing H2B-GFP are transfected with the indicated Halo-tagged TF. Cells were labeled with Halo dye JF549 and imaged using a confocal microscope under live conditions or after 15 min of fixation with 4% PFA. Quantification of chromosome enrichment is shown on Figure 1I.
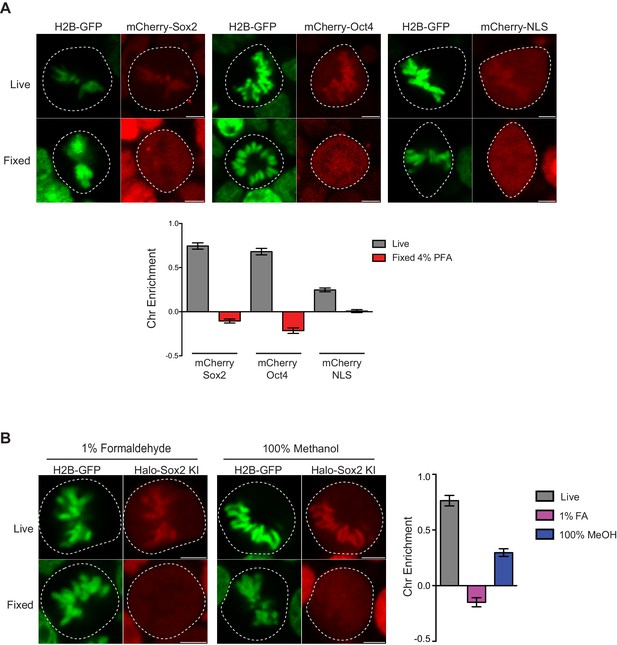
Controls for live versus fixed imaging.
(A) To ensure that the results were not biased by the HaloTag, we expressed mCherry-tagged Sox2, Oct4, or NLS in mouse ES cells stably expressing H2B-GFP and performed live and fixed imaging as before. mCherry-tagged factors are enriched on mitotic chromosomes in a similar manner as Halo-tagged factors. Quantification for chromosome enrichment as described in Figure 1G is shown. (B) We also performed fixation using 1% formaldehyde and 100% methanol on endogenously-tagged Halo-Sox2 KI cells. Quantification of chromosome enrichment is shown. Data is represented as mean ± SEM.
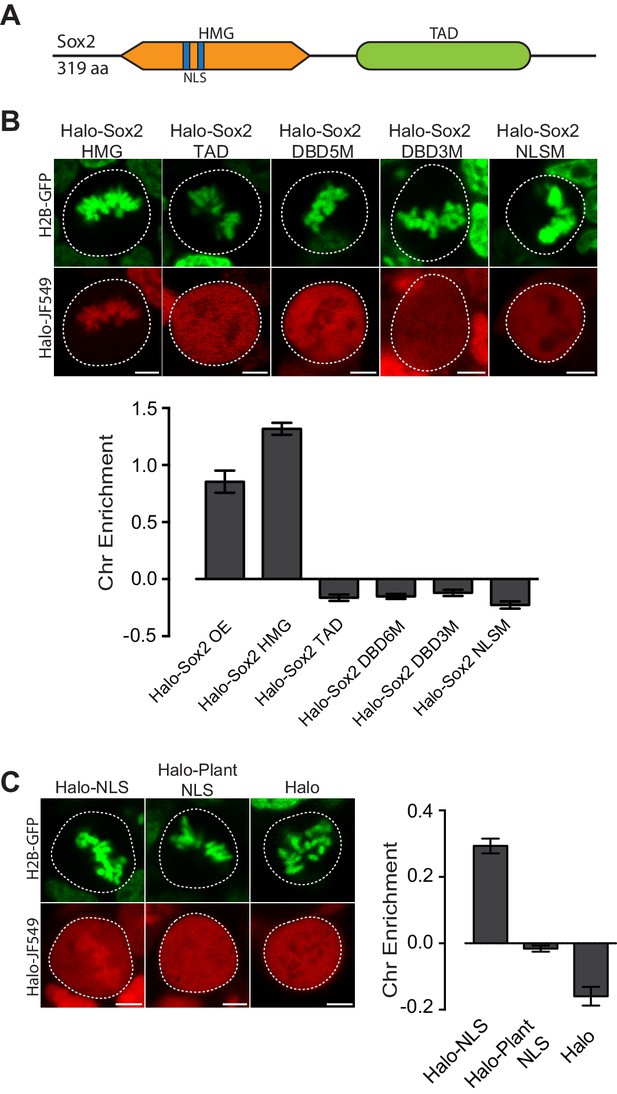
Mitotic enrichment of Sox2 requires DNA binding and nuclear import.
(A) Schematic of Sox2 domains. HMG, High Mobility Group domain. TAD, Transactivation Domain. NLS, Nuclear Localization Signal. (B) Live-cell imaging of mouse ES cells stably expressing H2B-GFB and various Halo-tagged Sox2 truncations or mutations. Bottom, chromosome enrichment quantification for the various Halo-tagged Sox2 over-expressing (OE) constructs. Halo-Sox2 HMG construct is a truncation of Halo-Sox2 with the TAD region deleted. Halo-Sox2 TAD is a truncation of Halo-Sox2 with the HMG domain deleted. Halo-Sox2-DBD5M and Halo-Sox2 DBD3M are the full length Halo-Sox2 with 5 and 3 point mutations to abrogate DNA binding, respectively. Halo-Sox2 NLSM is the full length Halo-Sox2 with point mutations to abolish NLS function. n = 40 cells. (C) Live-cell imaging of mouse ES cells stably expressing H2B-GFP and HaloTag fused to SV40 NLS, plant-specific NLS, or by itself. Right, chromosome enrichment levels for the indicated HaloTag constructs. n = 40 cells. Data are represented as mean ± SEM. Scale bars, 5 µm.
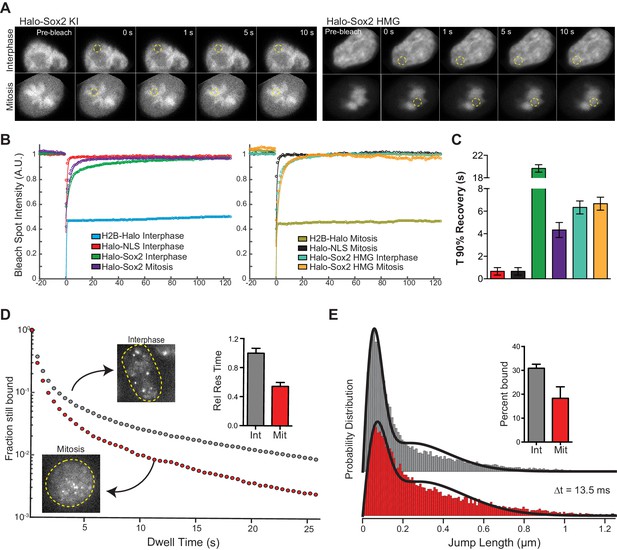
Sox2 interaction with mitotic chromosomes is highly dynamic.
(A) FRAP analysis of HaloSox2 KI and HaloSox2 HMG cells for interphase and mitosis. (B) Quantification of fluorescence recovery at the bleach spot for the indicated Halo-tagged construct in interphase and mitosis. n = 30 cells. (C) From (B), the average time to reach 90% recovery for the indicated (color-coded) Halo-tagged construct. (D) Dwell time histogram of the fraction of endogenously-tagged Halo-Sox2 molecules remaining bound for interphase (gray) and mitotic (red) cells. Representative images are shown. Inset, quantification of the relative Sox2 residence time as percentage of interphase cells. n = 30 cells. (E) Jump length histogram for three consecutive images (Δt = 13.5 ms) of the endogenously-tagged Halo-Sox2 molecules for interphase (gray) and mitotic (red) cells. A 2-state model is used to fit the histogram (solid line), and the fraction bound is calculated (inset). n = 24 cells. Data are represented as mean ± SEM.
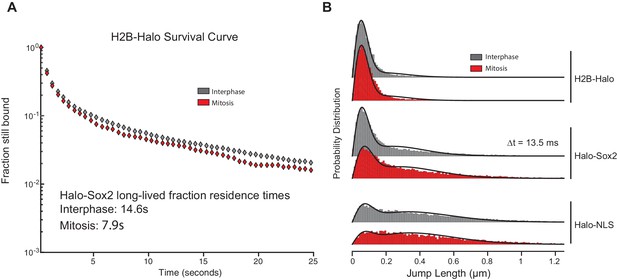
Controls for single particle tracking experiments.
(A) For photobleaching corrections, we performed slow-tracking SPT for H2B-Halo in interphase (gray) and mitosis (red) and plotted the semi-log histogram of H2B-Halo dwell times. These results show little difference in H2B-Halo residence time between interphase and mitotic cells. (B) Probability distribution of jump lengths for H2B-Halo, Halo-Sox2 KI, and Halo-NLS in interphase (gray) and mitosis (red) at △t = 13.5 ms.
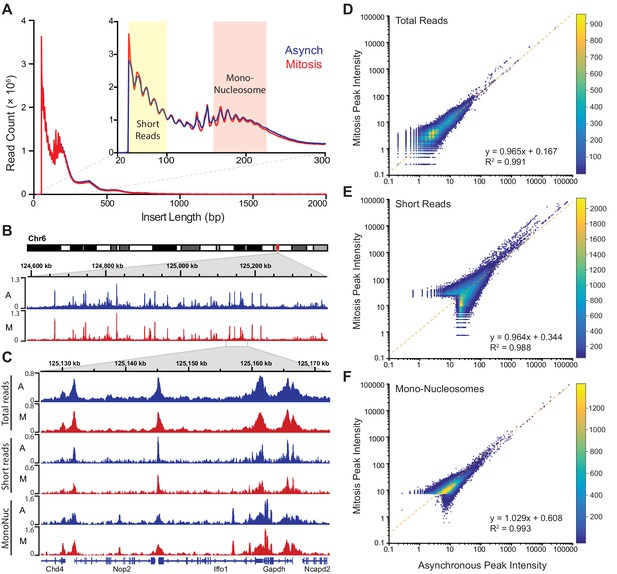
Global accessibility is maintained in mitotic chromosomes.
(A) Fragment length distribution of ATAC-seq reads for asynchronous (blue) and mitotic (red) cells. Inset, magnification of fragment length distribution under 300 bp showing size cut-offs for short reads (under 100 bp) and mono-nucleosome sized fragments (180–247 bp). (B) Asynchronous and mitotic ATAC-seq profiles for an 800 kb region in chromosome 6. (C) Comparison of total, short, and mono-nucleosome sized reads for asynchronous and mitotic samples in a 40 kb region in chromosome 6. (D–F) Heatmap scatter plots of peak intensities for asynchronous vs mitotic samples in total reads (D), short reads (E), and mono-nucleosome sized reads (F). Linear regression fit and R2 values are shown.
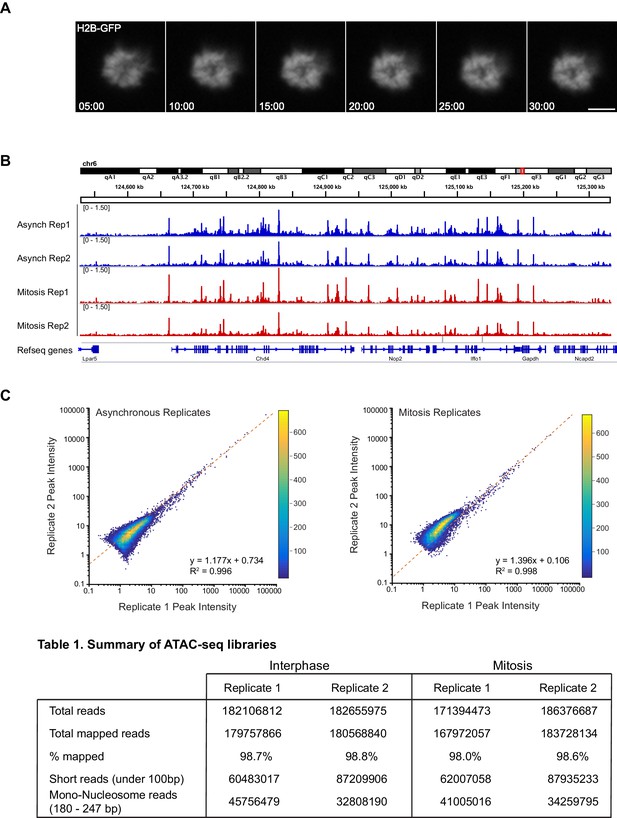
ATAC-seq replicates for asynchronous and mitotic samples.
(A) Time lapse imaging of Nocodazole-arrested mouse ES cells expressing H2B-GFP during the 30 min incubation with Tn5 transposase of ATAC-seq experiment. The cells remain mitotic throughout the tagmentation procedure. (B) ATAC-seq profiles at a locus in chromosome six for replicates 1 and 2 of asynchronous and mitotic samples show close concordance of each replicates. (C) Heatmap scatter plot of peak intensities for replicates of asynchronous (left) and mitotic (right) samples.
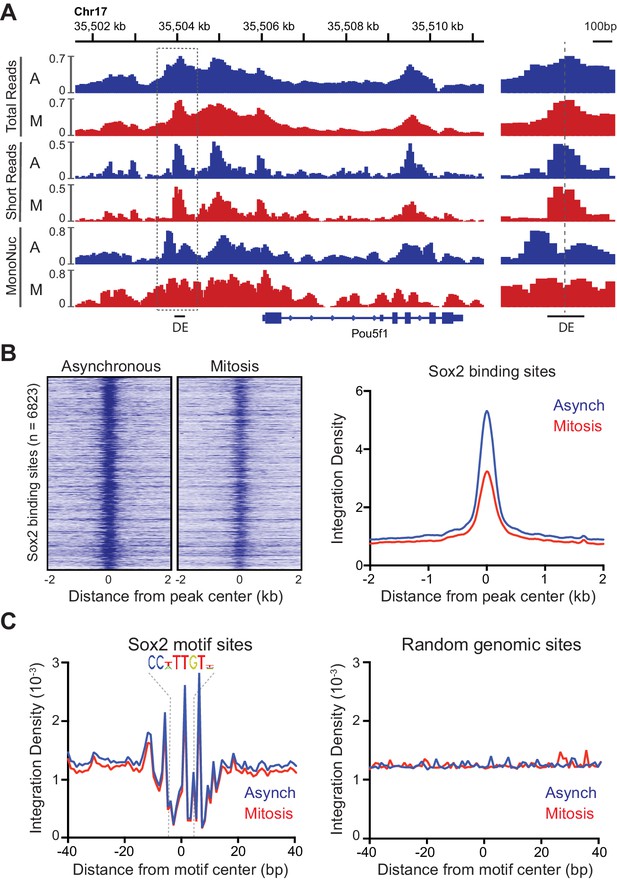
Accessibility of Sox2 binding sites in mitosis.
(A) Comparison of total, short, and mononucleosome sized reads for asynchronous and mitotic samples at the Pou5f1 gene. The boxed region centered at the distal enhancer (DE) is shown in greater detail on the right. (B) Heatmaps using the short reads were for asynchronous and mitotic ATAC-seq samples for all Sox2 bound sites (Chen et al., 2008) GEO Accession number GSE11431. Right, the average integration density for all Sox2 binding sites is plotted for asynchronous (blue) and mitotic (red) samples. (C) Aggregate ATAC-seq footprint for Sox2 (left) and for matched random genomic regions (right).
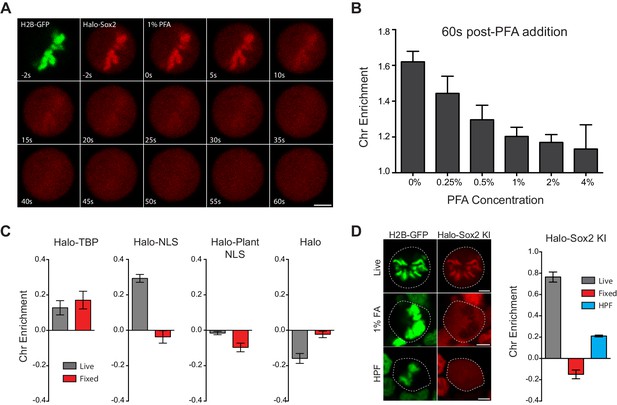
Mechanism of formaldehyde-based mis-localization of TFs.
(A) Time-lapse two color imaging of endogenously tagged Halo-Sox2 mouse ES cells stably expressing H2B-GFP after adding 1% PFA. (B) Quantification of chromosome enrichment at 60 s after PFA addition with the indicated concentrations of PFA. n = 10 cells. (C) Quantification of chromosome enrichment of indicated HaloTag-fused constructs and HaloTag only in live and fixed conditions. n = 30 cells. (D) High Pressure Freezing and Freeze Substitution was performed on Halo-Sox2 KI cells stably expressing H2B-GFP. Comparison of chromosome enrichment quantification for HPF samples with live and fixed cells are shown. n = 10 cells. Data are represented as mean ± SEM. Scale bars, 5 µm.
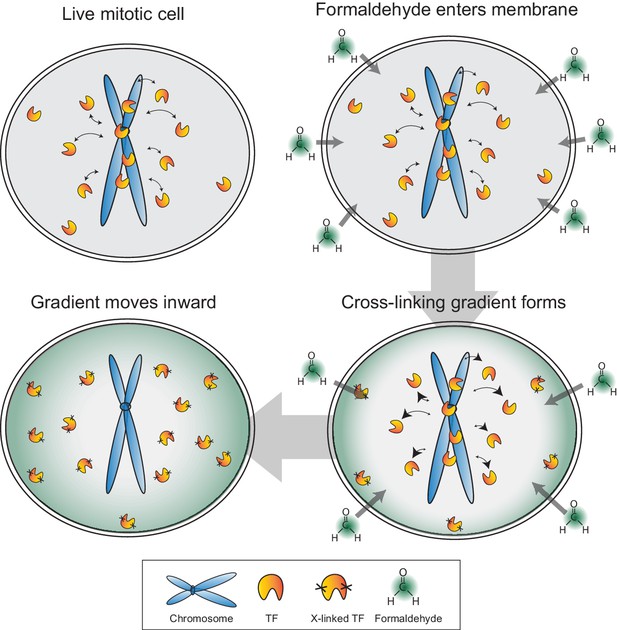
Model for formaldehyde-based mis-localization of transcription factors.
In live mitotic cells, TFs interact dynamically with mitotic chromosomes with intrinsic kon and koff rates, but can also sample the entire cellular space. During fixation, formaldehyde molecules enter the cell membrane and immediately cross-link with the nearest protein, resulting in a wave of cross-linking gradient that moves from the cell membrane inward. Such a gradient would cross-link cytoplasmic TFs first and result in an effective decrease in kon rates. As the gradient moves to the center, the result is an apparent exclusion of TFs from mitotic chromosomes.
Videos
SPT for residence time analysis of Halo-Sox2 KI cells in interphase.
Related to Figure 3. Imaging immobile Halo-Sox2 molecules in interphase ES cells at 2 Hz. Movie fps = 20. one pixel = 160 nm.
SPT for residence time analysis of Halo-Sox2 KI cells in mitosis.
Related to Figure 3. Imaging immobile Halo-Sox2 molecules in mitotic ES cells at 2 Hz. Movie fps = 20. one pixel = 160 nm.
SPT for fraction bound measurements of Halo-Sox2 KI cells in interphase.
Related to Figure 3. Imaging fast Halo-Sox2 molecules in interphase ES cells at 223 Hz. Movie fps = 20. one pixel = 160 nm.
SPT for fraction bound measurements of Halo-Sox2 KI cells in mitosis.
Related to Figure 3. Imaging fast Halo-Sox2 molecules in mitotic ES cells at 223 Hz. Movie fps = 20. one pixel = 160 nm.
Time-lapse imaging with PFA addition.
Related to Figure 6. Mitotic Halo-Sox2 KI cells were imaged at 2 Hz. 1% PFA was added after 10 s of imaging. Movie fps = 20. one pixel = 160 nm.
Additional files
-
Source code 1
Slimfast.
- https://doi.org/10.7554/eLife.22280.022
-
Source code 2
Mitotic chromosome enrichment.
- https://doi.org/10.7554/eLife.22280.023





