A microRNA negative feedback loop downregulates vesicle transport and inhibits fear memory
Figures
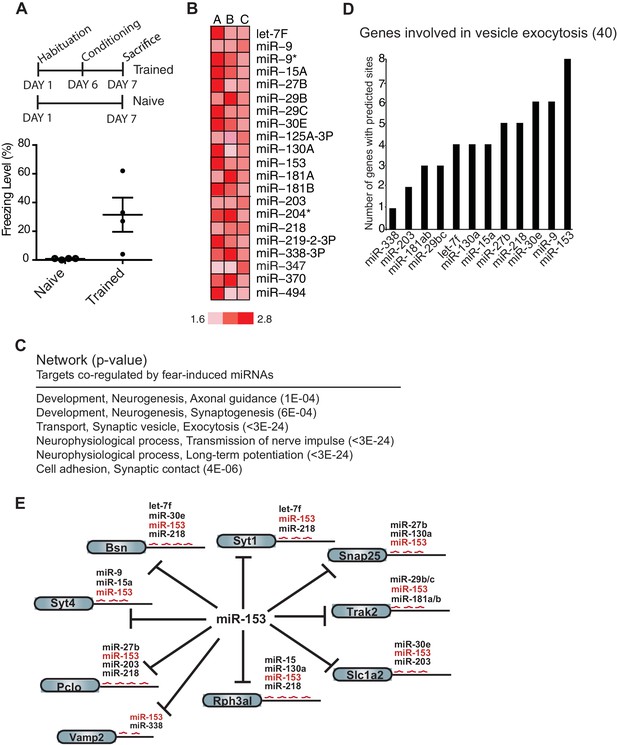
Expression profiling of miRNAs reveals 21 miRNAs that are induced in the hippocampus of adult rats 24 hr post-contextual fear conditioning.
(A) Schematic representation of contextual fear conditioning paradigm. Rats were trained to associate an aversive unconditioned stimulus (foot shock) with the environment (context). Freezing behavior was examined 24 hr after contextual fear conditioning training for a control group of rats (n = 4 for each group, naïve and trained), a subset of rats for which tissue was not harvested. Error bars indicate SEM. P value from pairwise unpaired t-test is indicated with asterisks, *p<0.05. (B) MiRNAs that displayed at least a 1.5-fold increase in expression between trained and naïve rats in three different experiments. (C) Network analysis using MetaCore (Thompson Reuters) identifies pathways involved in neuronal development, vesicle exocytosis and synaptic plasticity that are co-regulated by three or more of the fear induced miRNAs identified in panel B. P values were calculated for each canonical signaling pathway as compared to the number of occurrences from random sets of brain-expressed genes (see Materials and methods for a detailed description of brain-expressed gene lists). All 6 of the pathways are statistically significant compared to random sets of brain-expressed genes, ***p<0.0001. (D) MiR-153 and miR-9 are the top two miRNAs co-regulating targets involved in the vesicle exocytosis pathway. (E) Eight predicted targets from the vesicle exocytosis pathway that may be co-regulated by miR-153 and at least two other fear-induced miRNAs. The potential targeting fear-miRNAs are indicated above each target.
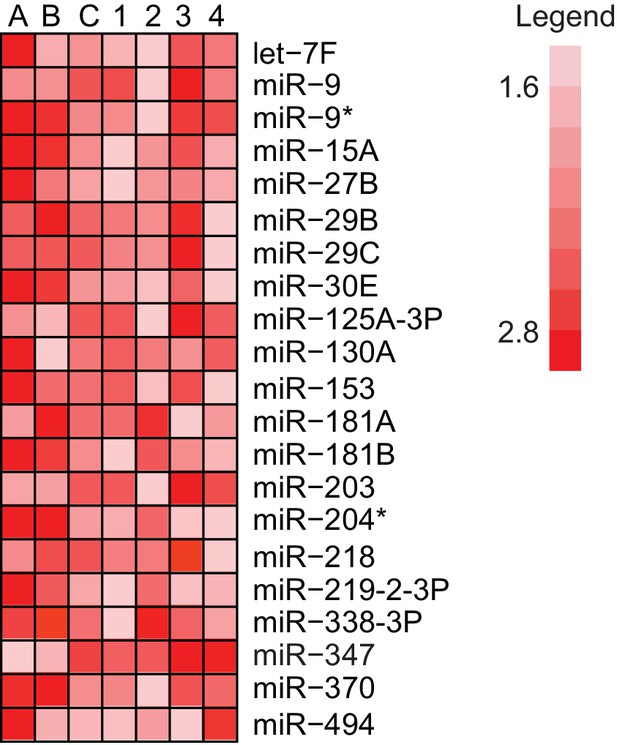
Identification of hippocampal fear-induced miRNAs.
MiRNAs that displayed at least a 1.5-fold increase in expression between trained and naïve rats in three different experiments with pooled hippocampal RNA from three individual trained rats relative to three individual naïve rats are labeled A–C. Expression ratios comparing a single trained and naïve rat are labeled 1–4.
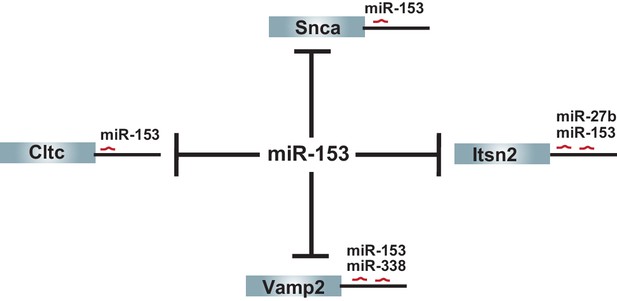
Additional targets shared between miR-153 and other fear-induced miRNAs.
The remaining 4 (Vamp2, Snca, Cltc, Itsn2) predicted targets of miR-153 from the vesicle exocytosis pathway that are regulated by miR-153 alone or at least one other fear-induced miRNA are shown.
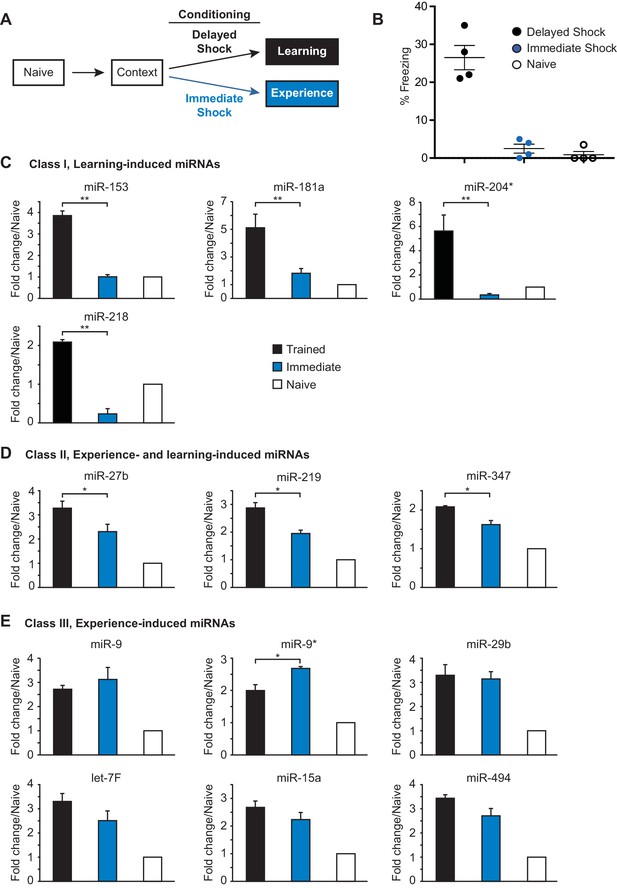
Identification of three classes of miRNA that are induced in the hippocampus with contextual fear conditioning.
(A) Summary of experimental strategy. (B) Behavioral test of the naïve, immediate shock, and delayed shock animals. (C–D) RT-qPCR experiments showing changes in miRNA levels in the hippocampi of animals that were subjected to contextual fear conditioning with either the application of immediate shock after introduction to a novel context (immediate, blue bars) or delayed shock (trained, black bars) relative to animals that were only handled (naïve, white bars). (C) Class I miRNAs were specifically induced in the trained group. (D) Class II miRNAs were induced in both the immediate and trained groups with stronger induction in the trained group. The cases where the differences in miRNA levels between immediate and trained groups were statistically significant are indicated (*p value<0.05). (E) Class III miRNAs were induced in both the immediate and trained hippocampi with no apparent increase in trained versus the immediate group. Each group contained nine animals, hippocampi from groups of three animals within each group were pooled for RNA isolation and RT-qPCR analysis.
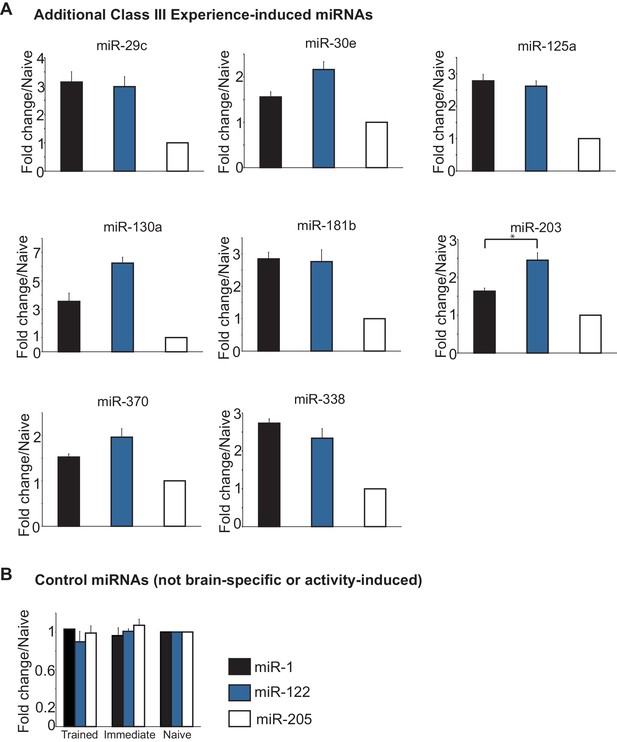
Class III miRNAs and control miRNAs.
(A) Additional Class III miRNAs that were induced in both the immediate and trained hippocampi with no apparent increase in trained versus the immediate group. (B) Control miRNAs represent miRNAs that are not specific for neuronal tissue and are not activity induced.
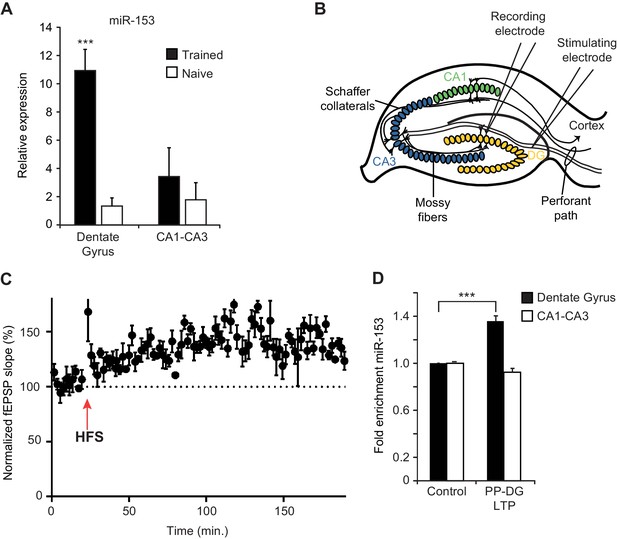
Expression of miR-153 is induced in the dentate gyrus by fear conditioning and LTP.
(A) RT-qPCR analysis of RNA levels showing average expression profiles of miR-153 from the dentate gyrus and CA1-CA3 regions of hippocampus in naïve and trained rats. Expression levels were normalized to snoRNA-202 RNA levels. Error bars indicate standard deviation. ***p<0.001 (B) Schematic representation of a hippocampal slice showing stimulating and recording electrode sites. (C) Three-hour time course of perforant path-dentate gyrus (PP-DG) LTP in slices from wild-type mice (n = 5). A 20 min baseline was recorded, after which LTP was induced with four epochs of high frequency stimulation (labeled as HFS with a red arrow) applied 15 s apart. fEPSP slope was plotted demonstrating robust LTP even 3 hr after the induction (data points are averaged every 1.5 min). Each point represents mean ± SEM. (D) RT-qPCR analysis showing average expression relative to control for RNA isolated from the dentate gyrus and CA1-CA3 regions of hippocampal slices following 3 hr of PP-DG LTP. Expression levels were normalized to control RNA from the same region of the hippocampus. Error bars indicate standard error.
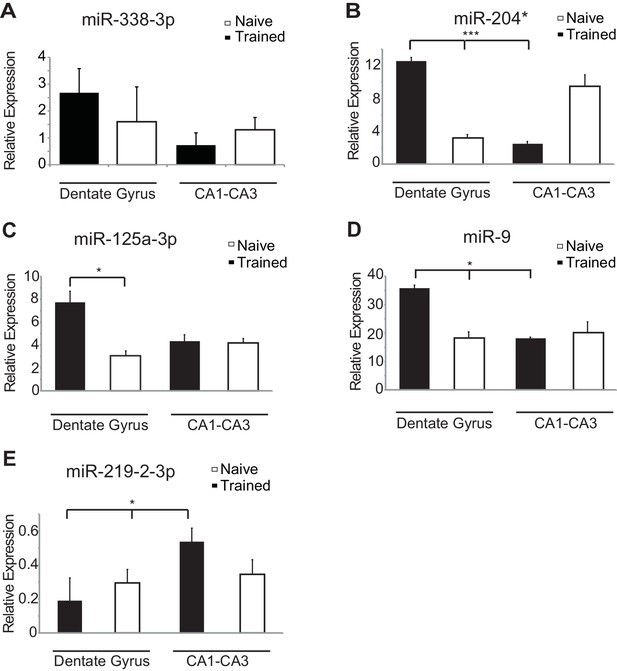
Region-specific expression of hippocampal fear-induced miRNAs.
RT-qPCR analysis of RNA levels for (A) miR-338–3p (B) miR-204* (C) miR-125a-3p (D) miR-9 (E) miR-219-2-3p miRNAs from the dentate gyrus and CA1-CA3 regions of hippocampus in naïve and trained rats. Expression levels were normalized to snoRNA-202 RNA levels, and values are reported relative to snoRNA-202 expression. Error bars indicate standard deviation. *p<0.05, ***p<0.001.
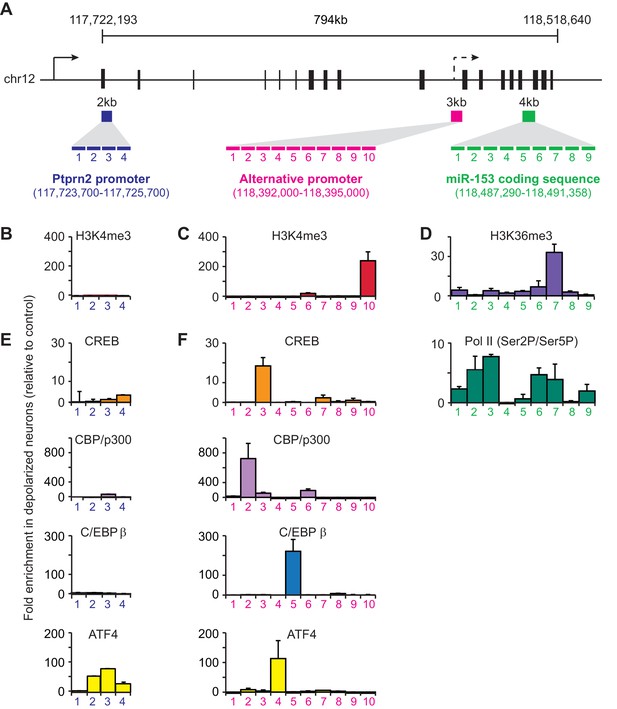
Transcriptional regulation of miR-153 expression may proceed through a cryptic promoter.
(A) Schematic representation of Ptprn2 gene with regions identified for each tiling primer set used to map H3K4me3 for Ptprn2 (purple box, B), H3K4me3 for the cryptic promoter (magenta box, C), and H3K36me3 for the miR-153 coding sequence (green box, D). (B–C) ChIP-qPCR experiments showing changes in the association of histone H3K4me3 (red) with the (B) Ptprn2 promoter and (C) alternative cryptic promoter area. (D) ChIP-qPCR experiments showing changes in association of histone H3K36me3 (purple) and Pol II (green) across the miR-153 coding sequence. Tiling primer sets spanning a 2 kilobase range (1primer set/500 base pairs) were used to map the Ptprn2 promoter (B); a 3 kilobase range (1 primer set/300 base pairs) were used to map the putative cryptic promoter (C); and a 4 kilobase range (1 primer set/440 base pairs) were used to map the miR-153 coding sequence (D). (E–F) ChIP-qPCR experiments showing changes in association of CBP/p300 (purple), CREB (phosphorylated at Ser133, orange), C/EBPß (blue) and ATF4 (yellow) with the (E) Ptprn2 promoter and (F) alternative cryptic promoter area. All experiments were performed with chromatin isolated from mature mouse hippocampal neurons (14 DIV) depolarized continuously for 3 hr with 55 mM KCl relative to untreated hippocampal neurons. The experiments were performed in triplicate and the data are presented as mean ± standard deviation.
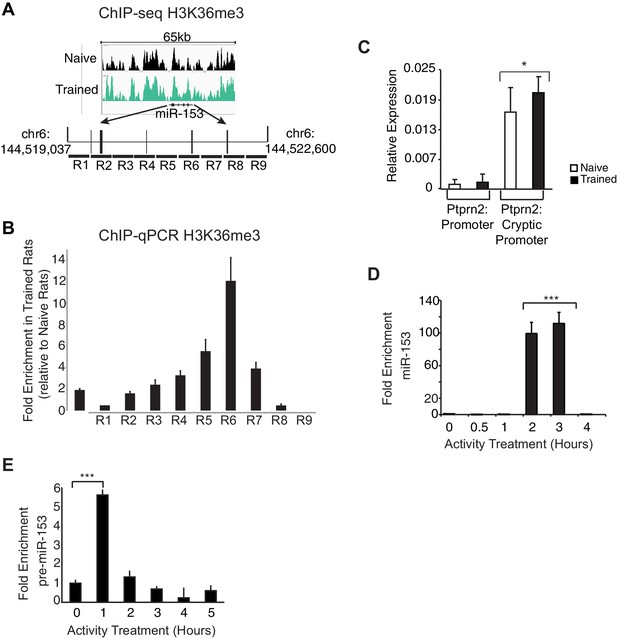
H3K36 trimethylation occupancy is increased across the miR-153 coding sequence after contextual fear-conditioning and miR-153 is transcriptionally induced from an alternative promoter within Ptprn2.
(A) Genome browser tracks of H3K36me3 ChIP-seq data across the miR-153 coding sequence in. 18 the hippocampus of trained (green) and naïve (black) rats. Figure was generated using integrative genomics viewer. (B) ChIP-qPCR experiments showing changes in association of H3K36me3 (black) across the miR-153 coding sequence, for chromatin isolated from hippocampus of trained rats relative to naïve rats. Tiling primer sets spanning a 3.6 kilobase range (1 primer set/400 base pairs) were used to map the coding sequence. The experiments were performed in triplicate and the data are presented as mean ± standard deviation. (C) RT-qPCR analysis showing average expression profiles of Ptprn2 for RNA isolated from hippocampus of naïve and trained rats 24 hr after contextual fear conditioning. Exon downstream of the host gene promoter (left) and exon downstream of a cryptic promoter (right) are shown. Error bars indicate standard deviation. *p<0.05 (D) RT-qPCR analysis of miR-153 expression for RNA isolated from mature mouse hippocampal neurons (14 DIV) depolarized continuously for 0.5–4 hr with KCl (as described in Supplemental Materials and methods). Error bars indicate standard deviation. ***p<0.001 (E) RT-qPCR analysis showing average expression profiles of the precursor form of miR-153 (pre-miR-153) for RNA isolated from mature mouse hippocampal neurons (14 DIV) depolarized continuously for 1–5 hr with 55 mM KCl relative to untreated hippocampal neurons. Error bars indicate standard deviation. ***p<0.001.
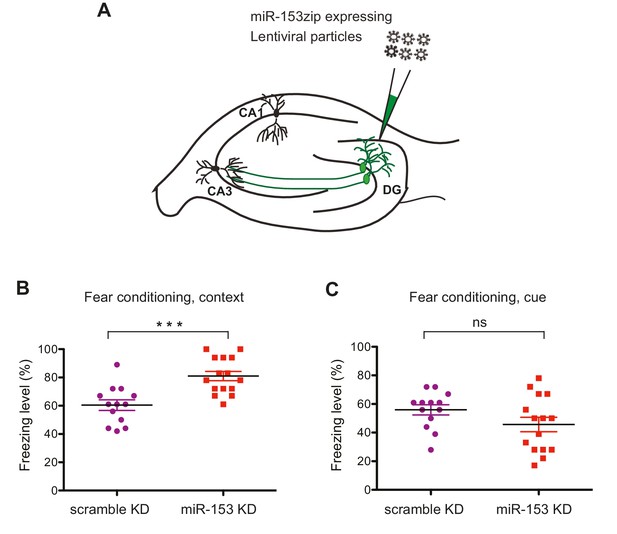
Knockdown of miR-153 enhances contextual fear-based memory.
(A) Schematic representation of the DG region of the hippocampus that is injected by lentiviruses expressing miRZip-153 or miRZip-scramble. (B) miRZip-153 (KD) and miRZip-scramble (KD) injected mice were tested with a contextual fear conditioning task. Freezing behavior was examined 24 hr after contextual fear conditioning training. Contextual fear conditioning training was performed after injection of miRZip-153 (KD) or miRZip-scramble (KD) into the dentate gyrus region of the hippocampus. p=0.001 (C) miRZip-153 (KD) and miRZip-scramble (KD)-injected mice were tested with a cued fear conditioning task. Freezing behavior was examined 24 hr after cued fear conditioning training. Cued fear conditioning training was performed after injection of miRZip-153 (KD) or miRZip-scramble (KD) into the dentate gyrus region of the hippocampus.
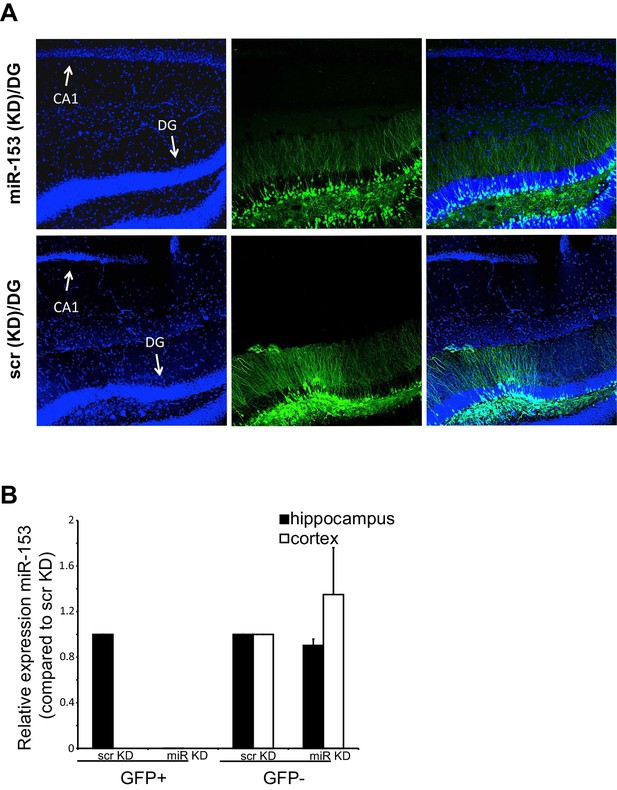
miR-153 (KD)-GFP and scrambled-GFP in the dentate gyrus of C57BL/6 mice.
(A) Confocal microscope images of hippocampi injected with either the miRZip-153 (KD) (top) or miRZip-scr (KD) scrambled control (bottom) lentivirus construct in the dentate gyrus area. The brain slices and labeled with antibodies against GFP (green) and Hoechst (blue). DG: dentate gyrus region, CA1: Cornu Ammonis region 1. Scale bar: 100 μm. (B) RT-qPCR analysis of RNA levels for miR-153 from hippocampus and cortex tissue isolated from miRZip-153 (KD) and miRZip-scr (KD) injected mice. Transcript levels are reported for FACS sorted GFP+ or GFP- neurons from miRZip-153 (KD) tissues relative to FACS sorted GFP+ or GFP- neurons from miRZip-scr (KD), a control scrambled miR lentivirus. Each experiment was performed in triplicate. Error bars indicate standard deviation.
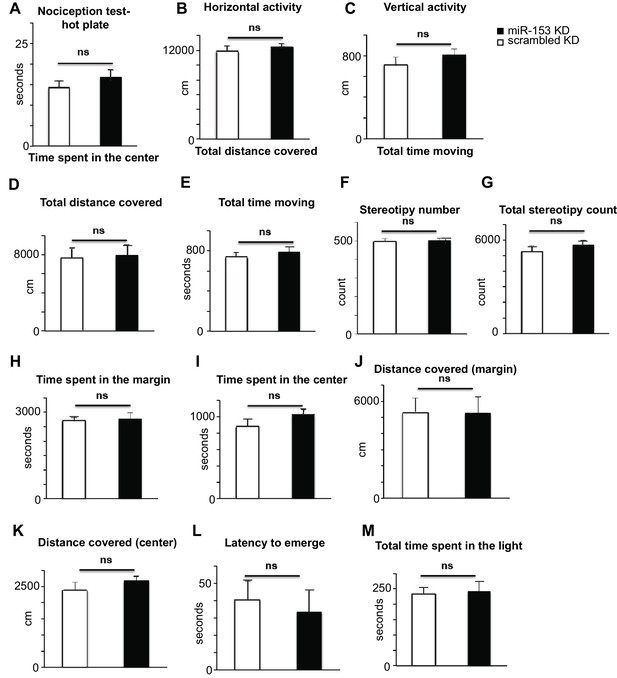
Behavioral characterization of miR-153 (KD)-GFP and scrambled-GFP injected mice.
(A) miRZip-153 (KD) mice exhibit normal pain sensitivity behavior as measured by length of time required for observation of nociceptive response (rear paw licking). miRZip-153 (KD) mice demonstrate normal (B) horizontal activity, (C) vertical activity, (D) distance traveled, as well as (E) total time moving, (F) total movement number and (G) frequency of stereotypic behavior in the open field test (60 min observation). (White – miRZip-scr (KD) control mice; black – miRZip-153 (KD) mice; n = 15 + 13 animals. Error bars indicate SEM, ns: not significant, *p<0.05, **p<0.01, ***p<0.001). miRZip-153 (KD) mice exhibit normal activity and anxiety-related behavior in the open field as measured by the time spent in (H) the margin and (I) the center (60 min observation). miR-153 (KD) mice exhibit normal activity and anxiety-related behavior in the open field as measured by total distance covered in (J) the margin and (K) the center (60 min observation). miR-153 (KD) mice exhibit normal anxiety-related behavior in the Light-Dark exploration test as measured by (L) the time of the first exit into the brightly lit area and (M) the total time spent in the brightly lit area.
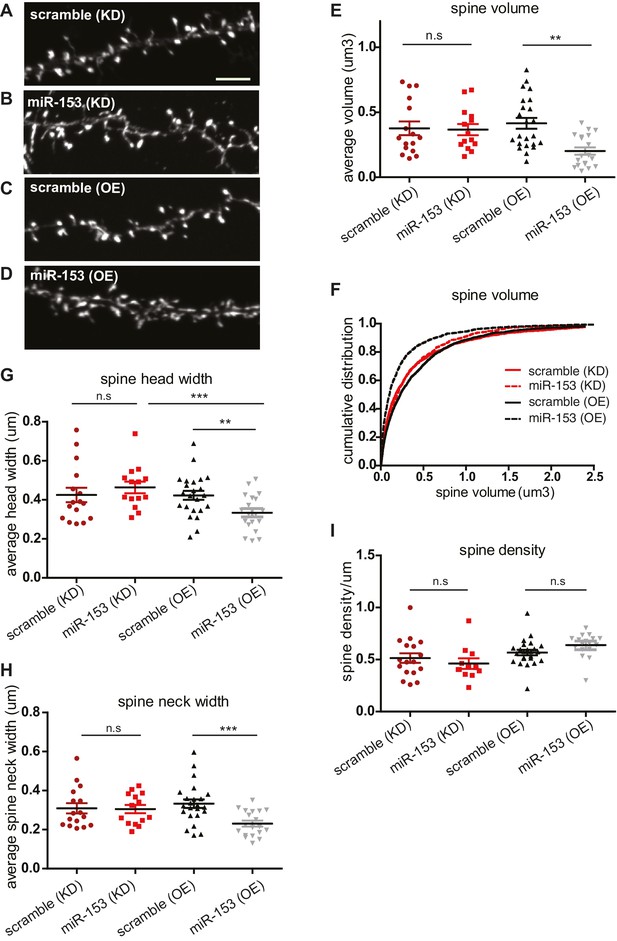
Overexpression of miR-153 decreases dendritic spine volume in hippocampal neurons.
(A–D) Lifeact-mRuby images, used to visualize spines, of dendritic regions from representative neurons (DIV18) transfected with scramble (KD) control vector (A), miR-153 (KD) inhibiting vector (B), scramble (OE) control vector (C), miR-153 (OE) overexpressing vector (D). Scale bar, 5 μm. KD represents knock-down and OE represents overexpression. (E) Average volume of spines (n > 800 spines) from neurons (n = 14–23) transfected as in (A–D). The means for these parameters were calculated for each neuron separately and then the averages from all neurons together were plotted as average numbers for each condition. Error bars indicate SEM. P values of pairwise unpaired t-tests are indicated with asterisks scramble (KD) vs miR-153 (KD) (p=0.84) and scramble (OE) vs miR-153 (OE) (p=0.003). *p<0.05, **p<0.01 (F) Cumulative distributions of spine volume were plotted for each group of hippocampal neurons (DIV18) transfected as above. At least 800 spines were measured from two independent experiments and three coverslips each experiment per condition. Spine volume was decreased in miR-153 overexpressing neurons (p<0.0001, D = 0.21), but not changed in neurons with miR-153 inhibited (p=0.37, D = 0.041). (G) Average spine head width calculated and represented as in (E). Error bars indicate SEM. P values of pairwise unpaired t-tests are indicated with asterisks; scramble (KD) vs miR-153 (KD) (p=0.43), scramble (OE) vs miR-153 (OE) (p=0.008) and miR-153 (KD) vs miR-153 (OE) (p=0.001). *p<0.05, **p<0.01, ***p<0.001 (H) Average spine neck width calculated and represented as in (E). Error bars indicate SEM. P values of pairwise unpaired t-tests are indicated with asterisks scramble (KD) vs miR-153 (KD) (p=0.9) and scramble (OE) vs miR-153 (OE) (p=0.0006). ***p<0.001 (I) Average spine density of hippocampal neurons calculated as in (E). Error bars indicate SEM. P values of pairwise unpaired t-tests are indicated with asterisks. n.s., not significant.
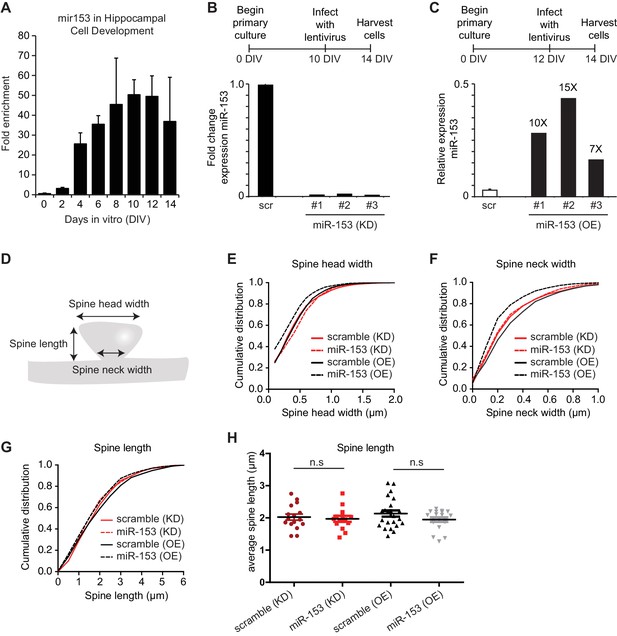
miR-153 negatively regulates dendritic spine size.
(A) Expression profile of miR-153 during primary hippocampal differentiation in vitro. (B–C) Schematic representation of treatment of miR-153 lentiviral-infected mature hippocampal cultures. RT-qPCR analysis of RNA levels showing expression profile of miR-153 in mature hippocampal neurons (14 DIV). Expression profiles are shown for neurons infected with either (B) miR-Zip-153 (KD) or miR-Zip-scr (KD), a control scrambled miR lentivirus (black bars). (C) Expression profiles for neurons infected with either Lv-miR-153 (OE) or Lv-scr (OE), a control scrambled miR lentivirus (white bar). Each sample was measured in triplicate and error bars indicate standard deviation. The scr (OE) replicates are represented as a single bar (white). (D) Schematic representation of a spine and the features that were measured in this analysis together with the total spine volume and the density of spines. (E–G) Cumulative distributions of (E) spine head width, (F) spine neck width, (G) spine length were plotted for each group of hippocampal neurons (DIV18) transfected as in Figure 3A–D. At least 800 spines were measured from two independent experiments and three coverslips each experiment per condition. (E) Spine head width was decreased when miR-153 was overexpressed (p<0.0001, D = 0.14) and increased when miR-153 was blocked (p<0.001, D = 0.09). (F) Spine neck width was decreased, when miR-153 was overexpressed (p<0.0001, D = 0.2) and not changed, when miR-153 was blocked (p=0.55, 0.04). Statistical significance was assessed by Kolmogorov-Smirnov test. (G) Spine length showed a decrease when miR-153 was overexpressed (p<0.0001, D = 0.14), but with no change when miR-153 was blocked (p=0.176, D = 0.05). Statistical significance was assessed by the Kolmogorov-Smirnov test. (H) Average spine length of hippocampal neurons (n = 14–23) transfected as above. It was calculated for each neuron separately and then the averages from all neurons together were plotted as average numbers for each condition. Error bars indicate SEM. P values of pairwise unpaired t-tests are indicated with asterisks scramble (KD) vs miR-153 (KD) (p=0.69) and scramble(OE) vs miR-153(OE) (p=0.14). n.s. not significant.
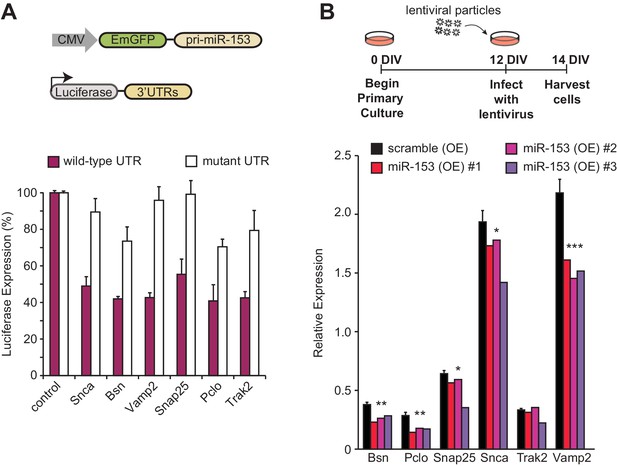
miR-153 regulates the expression of targets involved in the vesicle exocytosis pathway.
(A) Cells with luciferase reporter constructs containing wild-type (purple bars) or mutant (white bars) Bsn, Pclo, Snap25, Snca, Trak2, or Vamp2 mouse 3'-UTR region were co-transfected with the miR-153 expression plasmid. A miR-153 scrambled expression plasmid served as a control (leftmost bars). 3’-UTR mutations were in the seed sequence for the miR-153 binding site for each gene. HEK293T cells were co-transfected with both the reporter gene and miRNA expression vectors, and luciferase activity was measured 48 hr later. The experiments were performed in triplicate and error bars indicate standard deviation. (B) RT-qPCR analysis of RNA levels for genes (Bsn, Pclo, Snap25, Snca, Trak2, and Vamp2) from miRZip-153 (OE) infected 14 DIV hippocampal neurons (red, magenta and purple bars), relative to neurons infected with miRZip-scramble (OE), a control scrambled miR lentivirus (black bar). Each experiment was performed in triplicate. Error bars indicate standard deviation. *p<0.05, **p<0.01, ***p<0.001.
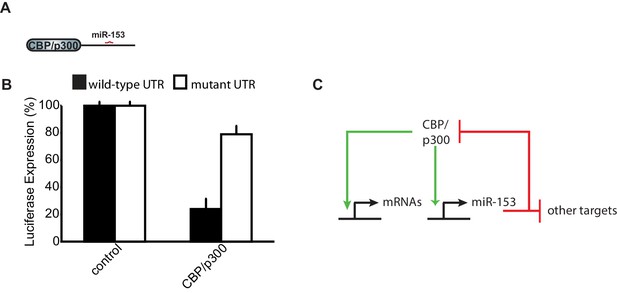
CBP/p300 is a target of miR-153.
(A) Schematic representation of CBP/p300 gene with miR-153 binding site indicated. (B) Cells containing the luciferase reporter constructs containing wild-type (black bars) or mutated CBP/p300 mouse 3'-UTR region (white bars) were co-transfected with miR-153. A miR-153 scrambled expression plasmid served as a control (left two bars). HEK293T cells were co-transfected with both the reporter gene and miRNA expression vectors, and luciferase activity was measured 48 hr later. The experiments were performed in triplicate and error bars indicate standard deviation. (C) Schematic representation of the feedback loop formed between miR-153 and CBP/p300. Green arrows indicate activation or positive feedback; red arrows indicate repression or negative feedback.
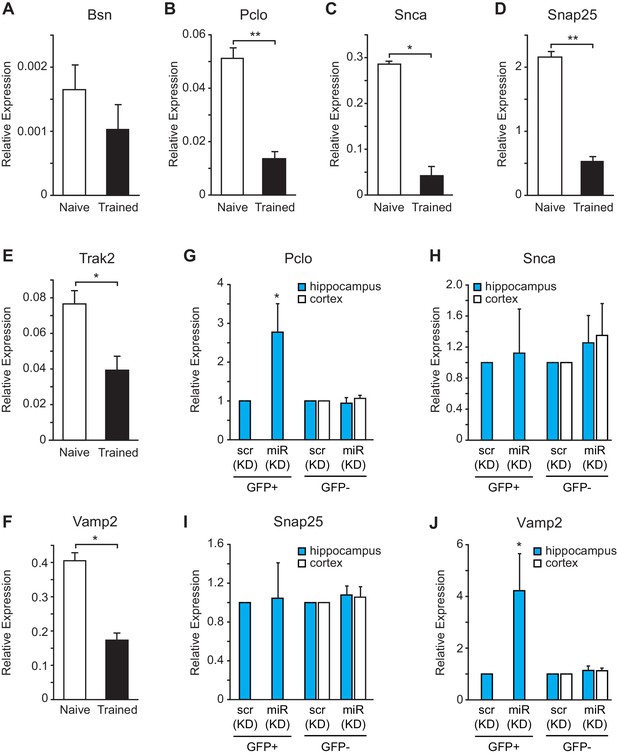
miR-153 regulates targets involved in the vesicle exocytosis pathway that are suppressed after fear conditioning.
(A–F) RT-qPCR analysis of RNA levels for vesicle exocytosis genes Bsn (A), Pclo (B), Snca (C), Snap25 (D), Trak2 (E), and Vamp2 (F) from hippocampus of naïve and trained rats. (G–J) RT-qPCR analysis of RNA levels for vesicle exocytosis genes Pclo (G), Snca (H), Snap25 (I), and Vamp2 (J) from hippocampus and cortex tissue isolated from miRZip-153 (KD) and miRZip-scr (KD) injected mice. Transcript levels are reported for FACS sorted GFP+ or GFP− neurons from miRZip-153 (KD) tissues relative to FACS sorted GFP+ or GFP− neurons from miRZip-scr (KD), a control scrambled miR lentivirus. Each experiment was performed in triplicate. Error bars indicate standard deviation. *p<0.05.
Fluorescence-activated cell sorting (FACS) plots of hippocampal neurons isolated from wild-type mice transduced in the dentate gyrus with miRZip lentiviruses.
(A) Schematic representation of the methods used to prepare samples for FACS sorting and RT-qPCR analysis from wild type mice transduced in the dentate gyrus with lentiviruses. (B–C) Representative examples of FACS sorting of GFP positive but DAPI negative neurons from wild type mice transduced in the dentate gyrus with miRZip-scr-GFP (B) or miRZip-153-GFP (C) lentivirus.
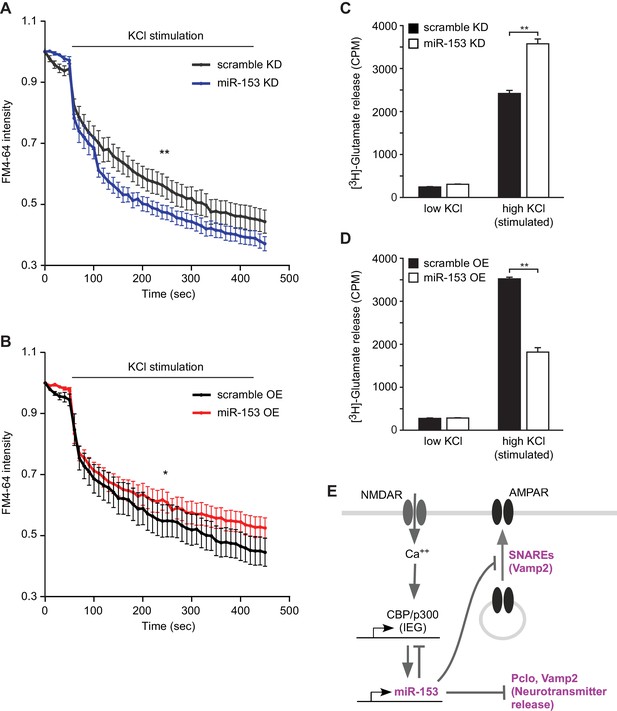
miR-153 regulates vesicle exocytosis and glutamate neurotransmitter release.
(A–B) Analysis of vesicle exocytosis by FM4–64 imaging in primary neurons. (A) Fold fluorescence depletion of FM4-–64 dye of scramble (KD) and miR-153 (KD) groups of (n = 10–12) neurons per group, **p=0.008, two-tailed Mann Whitney U test. (B) Fold fluorescence depletion of FM4-–64 dye of scramble (OE) and miR-153 (OE) groups of (n = 10–12) neurons per group, *p=0.014, two-tailed Mann Whitney U test. (C–D) [H3]-glutamate release in primary neurons. (C) [H3]-glutamate release as determined by measuring the radioactivity content in neurons transduced with miRZip-153 knockdown (KD) lentivirus (white bars) as compared to primary neurons transduced with miRZip-scr (KD) lentivirus (black bars) after depolarization with 55 mM KCl (high KCl). (D) [H3]-glutamate release as determined by measuring radioactivity content in neurons transduced with miR-153 overexpression (OE) lentivirus (white bars) as compared to primary neurons transduced with scramble overexpression (OE) lentivirus (black bars) after depolarization with 55 mM KCl (high KCl). Each experiment was performed in triplicate. Error bars indicate standard deviation. *p<0.05. (E) Schematic summary illustrating the role of miR-153 as a negative feedback regulator of the pathways that mediate changes in synaptic strength and neurotransmitter release.
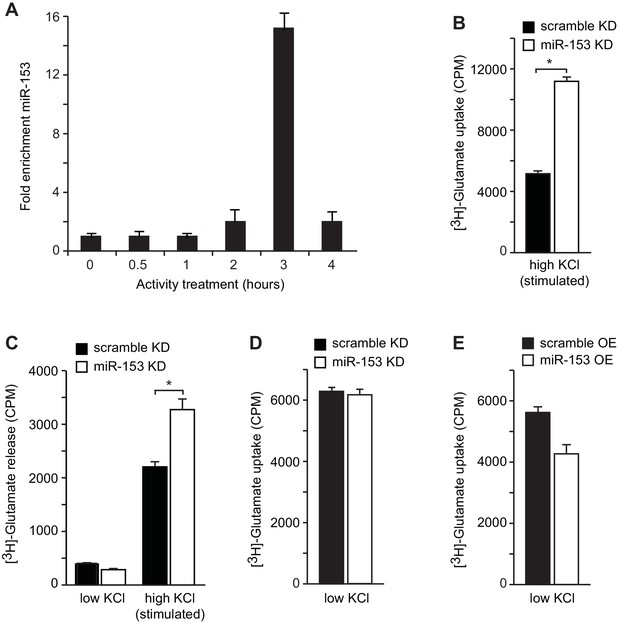
Neurotransmitter uptake and release measurements in H19-7 hippocampal neuronal cells.
(A) RT-qPCR analysis of miR-153 expression for RNA isolated from differentiated H19-7 cells depolarized continuously for 0.5–4 hr with KCl (as described in Supplemental Materials and methods). Experiments were performed in triplicate and error bars indicate standard deviation. (B) [H3]-glutamate uptake as determined by measuring the radioactivity content in H19-7 miRZip-153 knockdown (KD) cells as compared to H19-7 miRZip-scr (KD) cells after depolarization with 55 mM KCl (high KCl). (C) [H3]-glutamate secretion by 5 mM KCl (low KCl) or 55 mM KCl (high KCl) as determined by measuring the radioactivity in the supernatant of H19-7 miRZip-153 (KD) cells as compared to H19-7 miRZip-scr (KD) cells. (D) [H3]-glutamate uptake as determined by measuring the radioactivity content in primary neurons transduced with miRZip-153 knockdown (KD) lentivirus as compared to primary neurons transduced with miRZip-scr (KD) lentivirus. (E) [H3]-glutamate uptake as determined by measuring radioactivity content in primary neurons transduced with miR-153 overexpression (OE) lentivirus as compared to primary neurons transduced with scramble overexpression (OE) lentivirus. Each experiment has been performed in triplicate. Error bars indicate standard deviation. *p<0.05.
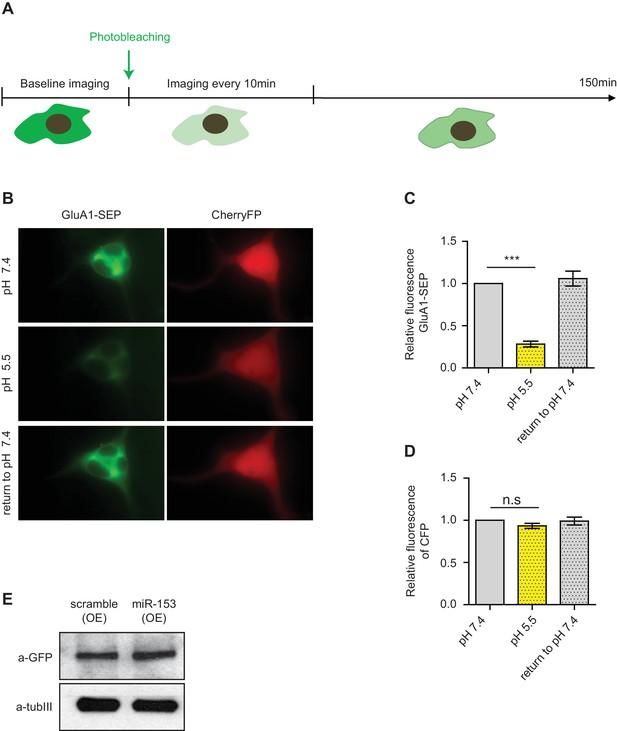
Fluorescence recovery after photobleaching (FRAP) of SEP-GluA1 AMPA receptors in neuroblastoma N2A cells.
(A) Schematic representation of the design of the FRAP experiment. (B) Images of N2A cells that were perfused with p.H 7.4 ACSF followed by a brief exposure to p.H 5.5 and then returned to p.H 7.4. (C) Relative levels of SEP-GluA1 at neutral and acidic conditions for N2A cells treated as in (B). (D) Relative levels of mCherryFP at neutral and acidic conditions. (E) Immunoblot for GluA1 total levels in scramble (OE) and miR-153 (OE) cells.
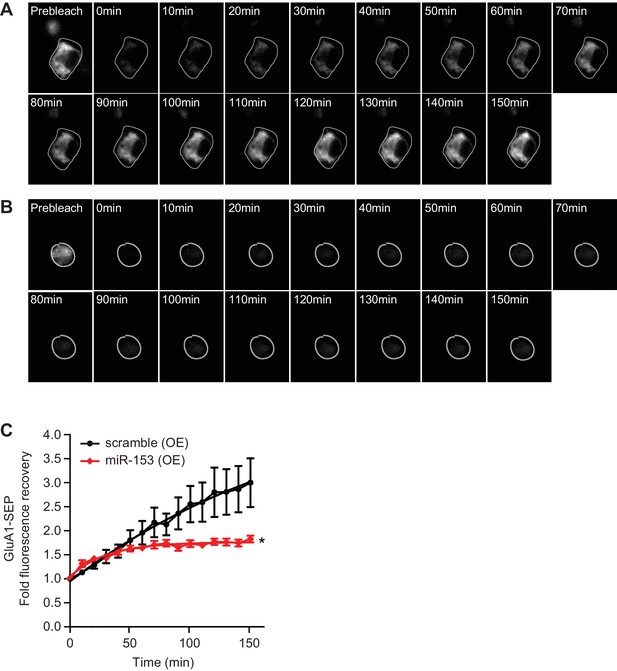
miR-153 regulates AMPAR transport in neuroblastoma N2A cells.
(A) Time lapse images following photobleaching of SEP-GluA1 in scramble (OE) cells. (B) Time lapse images following photobleaching of SEP-GluA1 in miR-153 (OE) cells. (C) Fold fluorescence recovery of SEP-GluA1 of scramble (OE) and miR-153 (OE) groups of cells (n = 5–8 cells per group, p=0.028, two-tailed Mann Whitney U test).
Additional files
-
Supplementary file 1
(A) List of 353 targets with predicted binding sites for 3 or more of the fear-induced miRNAs. (B) List of targets from the vesicle exocytosis pathway with predicted binding sites for 3 or more fear-induced miRNAs. (C) Predicted Targets of miR-153 from top five Metacore networks.
- https://doi.org/10.7554/eLife.22467.024





