Multiple alleles at a single locus control seed dormancy in Swedish Arabidopsis
Figures
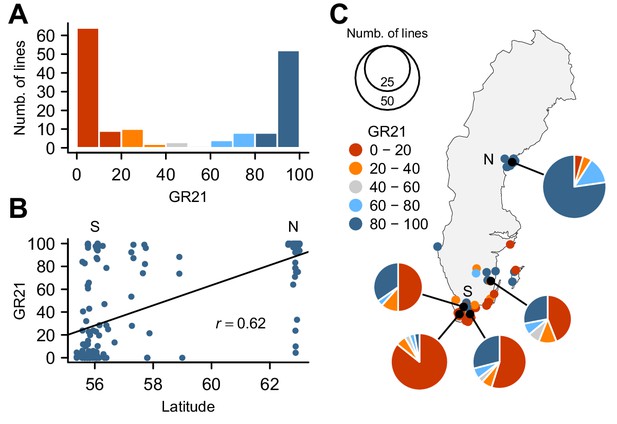
Natural variation for seed dormancy in Swedish A. thaliana.
(A) The distribution of the germination rate 21 days after seed harvest (GR21). (B) Correlation between GR21 and latitude. We define southern Sweden (S) as the region below 58°N and northern Sweden (N) as above 62°N. (C) Geographic distribution of GR21. An interactive version of this map is available online (https://goo.gl/3HeqX5).
-
Figure 1—source data 1
The 161 Swedish accessions used in this study.
Geographic information and GR21 values used to generate Figure 1 and perform GWAS.
- https://doi.org/10.7554/eLife.22502.003
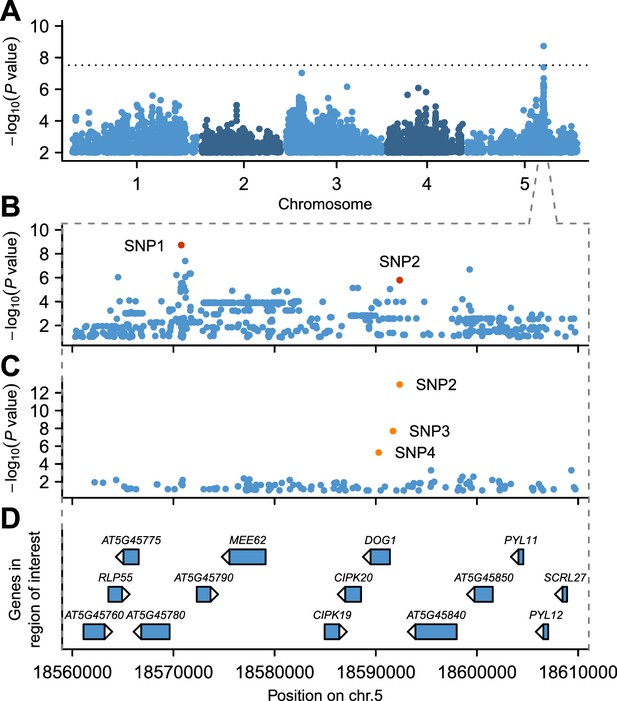
GWAS for primary seed dormancy on a set of 161 A. thaliana Swedish accessions.
(A) Manhattan plot of genome-wide association results for GR21. The dotted horizontal line indicates a significance level of 0.05 after Bonferroni correction for multiple testing. (B) Magnification of the peak region on chromosome 5. The most significant SNP (SNP1) was located at position 18,570,773. SNP2 (18,592,365) is in moderate LD with SNP1, despite a physical separation of almost 21 kb. Both SNPs are highlighted. (C) Output of a local multi-locus association scan using SNP2 as starting point. The resulting optimal model consisted of three highlighted SNPs at the DOG1 locus: SNP2, SNP3 (18,591,702) and SNP4 (18,590,289). (D) Annotated genes in the region under the peak. The GWAS results can be viewed interactively online (https://goo.gl/30EPt3).
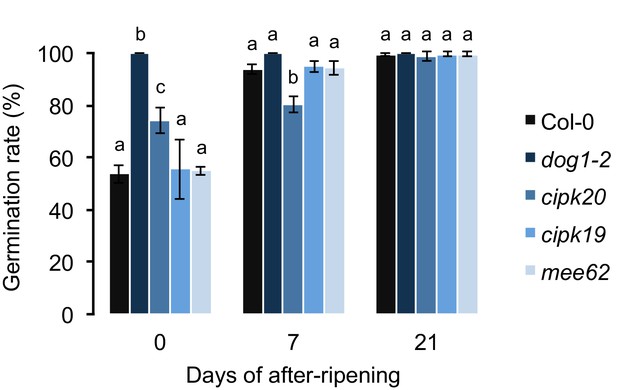
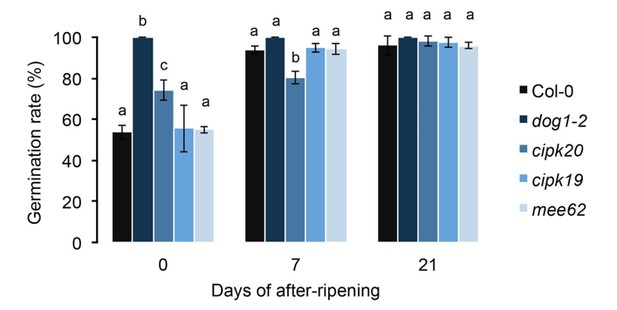
The effect of mee62, cipk19 and cipk20 knock-outs on dormancy.
Effects were assessed by measuring the germination rate of insertion mutants in response to various length of after-ripening (0, 7 and 21 days). Col-0 and the dog1-2 mutant were included as controls. At each time point, we analysed three biological replicates per genotype (error bars represent standard deviation). Bars with different letters indicate significant difference in multiple range comparison (one-way ANOVA followed by Tukey’s HSD test, p<0.05, n = 3).
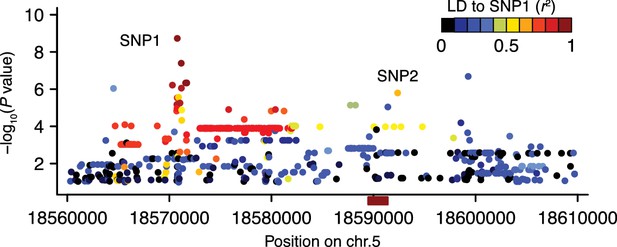
The linkage disequilibrium pattern in the region surrounding DOG1.
Manhattan plot of a local (same region as in Figure 2B–D) association scan for GR21 where the color scale indicate the extent of LD starting from the most strongly associated SNP (SNP1) at position 18,570,773. The r2 value between SNP1 and SNP2 is 0.61. The red box on the horizontal axis indicates the position of the DOG1 locus.
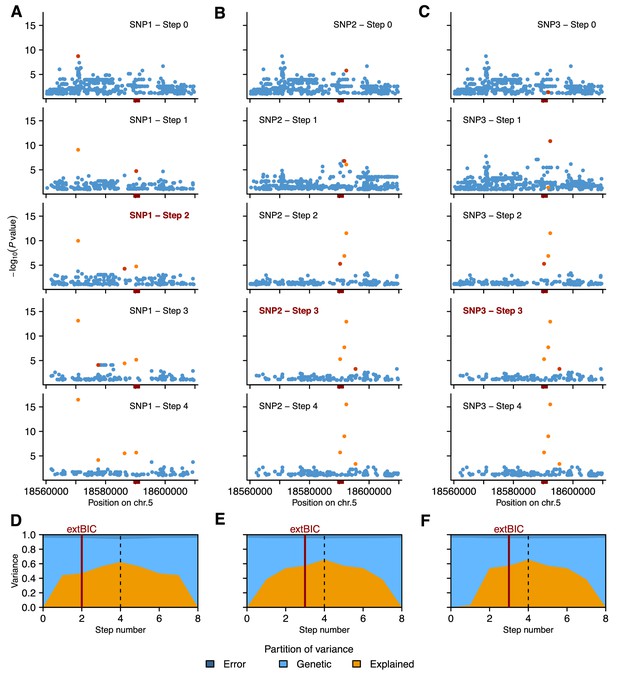
Details of local MLMM association scans for GR21.
We used three different SNPs as starting point: SNP1 (A and D), SNP2 (B and E) or SNP3 (C and F). (A and C) Manhattan plots of local MLMM association scans. The previous and the next SNPs added as cofactors to the model are highlighted in light and dark orange, respectively, for each of the four forward regression steps. The DOG1 locus is marked by the red box on the horizontal axis. (D and F) Partition of variance at each step of MLMM (four forward and four backward) into variance explained by the SNPs included in the model (Explained), kinship (Genetic) and noise (Error). The vertical red bar shows the optimal number of steps according to the extended Bayesian information criteria (extBIC).
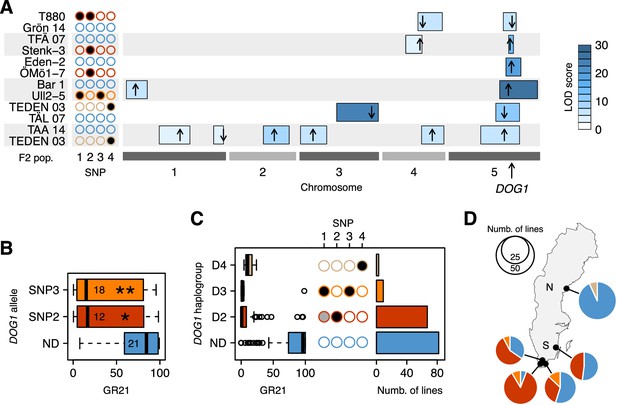
Functional validation and geographic distribution of DOG1 alleles.
(A) Summary of QTL mapping for GR21 in six different F2 populations. Open and closed circles correspond to the reference (non-dormant) and non-reference (all dormant) alleles at SNP1-SNP4, respectively. Circles are colored as in Figure 3C, to represent the haplotypes carried by parental lines. Horizontal boxes filled with shades of blue represent 95% confidence intervals of QTLs along the five A. thaliana chromosomes (Figure 3—source data 1). The position of the highest LOD score within a given interval is marked with a black arrow, whose direction indicates the allelic effect of the first parent (up increases GR21; down decreases GR21). Only QTLs with a significant LOD score are depicted. (B) Transgenic complementation of the non-dormant dog1-2 mutant with two dormant DOG1 alleles carrying either SNP2 or SNP3, or a non-dormant allele (ND) as control (Figure 3—source data 2). A t-test was performed to compare the effect of the dormant alleles to that of the non-dormant allele (p=0.0147 and p=0.0046 for the alleles tagged by SNP2 and SNP3, respectively). (C) GR21 variation within the four DOG1 haplogroups defined by SNP2-SNP4. The frequency of SNP1-SNP4 within each haplogroup is indicated with open (SNP is absent), grey (SNP segregates) and closed (SNP is fixed) circles. The barplot indicates the haplogroup frequency in the 161 Swedish lines. (D) Geographic distribution of DOG1 haplogroups. As in Figure 1, N and S stand for northern and southern Sweden, respectively. An interactive version of this map is available online (https://goo.gl/nfNQjh).
-
Figure 3—source data 1
QTL mapping results.
LOD scores and positions of 95% confidence interval of the major QTLs presented in Figure 3A.
- https://doi.org/10.7554/eLife.22502.009
-
Figure 3—source data 2
Phenotypes of the DOG1 transgenic lines.
GR21 values of the transgenic lines used to generate Figure 3B.
- https://doi.org/10.7554/eLife.22502.010
-
Figure 3—source data 3
Information about Bar 1, an extra Swedish accession used as parent in one of the F2 mapping population.
- https://doi.org/10.7554/eLife.22502.011
-
Figure 3—source data 4
SNPs and haplotypes at the DOG1 locus for the 161 Swedish accessions used in this study.
We provide the genotype of the accessions at SNP1-SNP4 as well as their DOG1 haplotype.
- https://doi.org/10.7554/eLife.22502.012
-
Figure 3—source data 5
Phenotypes and genotypes of the F2 individuals used in the QTL mapping approach.
This file also contains the NCBI SRA BioProject and BioSamples accession numbers.
- https://doi.org/10.7554/eLife.22502.013
-
Figure 3—source data 6
DOG1 PCR primers used in the complementation experiment.
The underlined sequences correspond to the flanking sequences of the EcoRI restriction site in the pGreenII 0029 that was used for transformation.
- https://doi.org/10.7554/eLife.22502.014
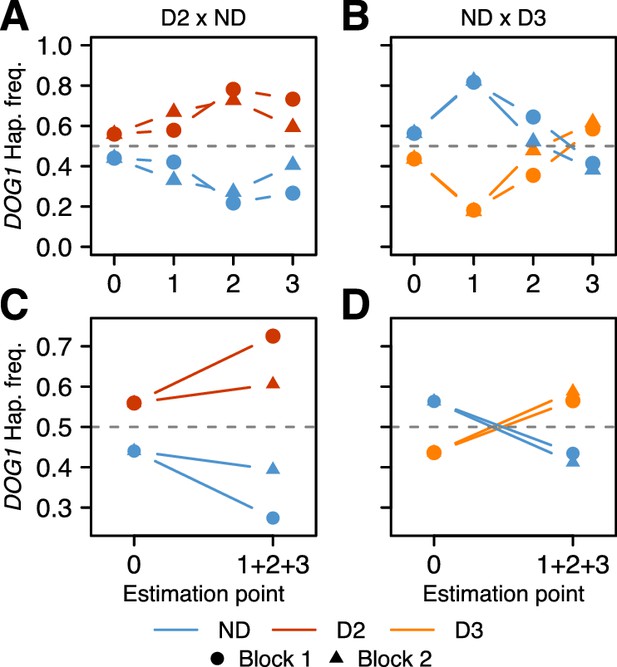
Changes in DOG1 haplotype frequency in southern Sweden field experiments.
(A and C) Frequencies in T880 (D2) x Grön 14 (ND), in which SNP2 segregates. (B and D) Frequencies in Bar 1 (ND) x Ull2-5 (D3), in which SNP3 segregates. (A–B) Frequency changes across time. Starting frequencies were estimated prior to seed dispersal and adjusted to take into account seed viability (0). Frequencies were then estimated in September (1) and November (2) by genotyping seedlings. Finally, seedlings generated from soil samples were used to estimate frequencies in the seed bank (3). (C–D) Difference between starting (0) and ending haplotype frequencies (weighted sum of 1 + 2 + 3).
-
Figure 4—source data 1
Genome-wide genotypes of the field experiments samples.
This file also contains sampling information (block, frame, sampling point etc.) and the NCBI SRA BioProject and BioSamples accession numbers.
- https://doi.org/10.7554/eLife.22502.016
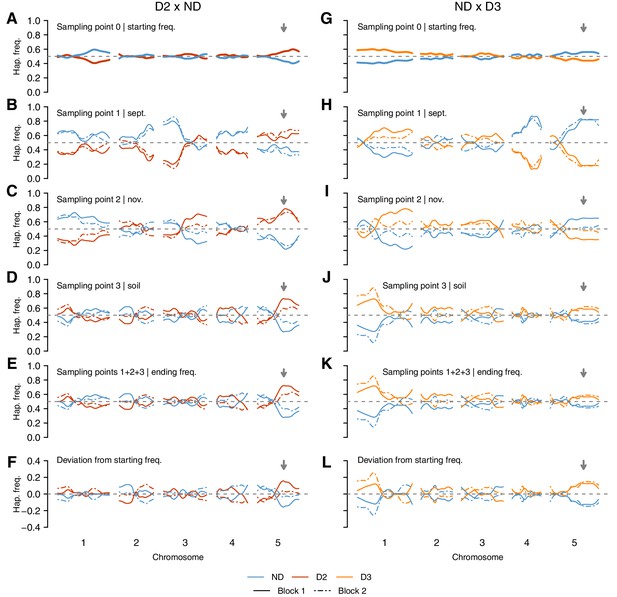
Genome-wide changes in DOG1 haplotype frequencies in southern Sweden field experiments.
(A–F) Haplotype frequencies in T880 (D2) x Grön 14 (ND), in which SNP2 segregates. (G–L) Haplotype frequencies in Bar 1 (ND) x Ull2-5 (D3), in which SNP3 segregates. (A and G) Starting frequencies (sampling point 0) were estimated prior to seed dispersal and adjusted to take into account seed viability. (B–D and H–J) Intermediate frequencies estimated in September (sampling point 1) and November (sampling point 2) by genotyping seedlings. Seedlings generated from soil samples were used to estimate frequencies in the seed bank (sampling point 3). (E and K) Ending haplotype frequencies (weighted sum of 1 + 2 + 3). (F and L) Deviation from starting frequencies at the end of the experiment ((1 + 2 + 3) − 0). The vertical grey arrow indicates the position of the DOG1 locus. .
Additional files
-
Supplementary file 1
Correlation between GR21 and climate variables.
Partial Mantel correlations between climate variables and GR21, where the kinship between accessions is included as a covariate in the model to correct confounding caused by population structure. The Mantel r values are the Spearman partial correlation coefficients.
- https://doi.org/10.7554/eLife.22502.018
-
Supplementary file 2
Local association scan using pairwise combination of SNPs.
Only the top 15 combinations are displayed (see Materials and methods, GWAS with pairwise combinations of SNPs).
- https://doi.org/10.7554/eLife.22502.019
-
Supplementary file 3
(A) Field experiments summary and sample inventory, per block and population.
The number of soil samples was adjusted (Soil adj.) to take into account the size of the sampled area and seed transportation over the course of the experiment (see Materials and methods). The mean fitnesses were calculated by dividing the total number of live samples by the number of dispersed viable seeds (Total / Dispersed * Viability). (B) Summary of allele frequency changes at the DOG1 locus, per population and block. (C) Summary of genotype frequency changes at the DOG1 locus, per population and block.(D) Relative fitnesses of DOG1 alleles in field experiments, per population and block.
- https://doi.org/10.7554/eLife.22502.020





